Abstract
The derivation and long-term maintenance of human embryonic stem cells (hESCs) has been established in culture formats that are both dependent and independent of support (feeder) cells. However, the factors responsible for preserving the viability of hESCs in a nascent state remain unknown. We describe a mass spectrometry-based method for probing the secretome of the hESC culture microenvironment to identify potential regulating protein factors that are in low abundance. Individual samples were analyzed several times, using successive mass (m/z) and retention time-directed exclusion, without sampling the same peptide ion twice. This iterative exclusion -mass spectrometry (IE-MS) approach more than doubled protein and peptide metrics in comparison to a simple repeat analysis method on the same instrument, even after extensive sample pre-fractionation. Furthermore, implementation of the IE-MS approach was shown to enhance the performance of an older quadrupole time of flight (Q-ToF) MS. The resulting number of identified peptides approached that of a parallel repeat analysis on a newer LTQ-Orbitrap MS. The combination of the results of both instruments proved to be superior to that achieved by a single instrument in the identification of additional proteins. Using the IE-MS strategy, combined with complementary gel- and solution-based fractionation methods, the hESC culture microenvironment was extensively probed. Over 10 to 12 times more extracellular proteins were observed compared with previously published surveys. The detection of previously undetectable growth factors, present at concentrations ranging from 10−9 to 10−11 g/ml, highlights the depth of our profiling. The IE-MS approach provides a simple and reliable technique that greatly enhances instrument performance by increasing the effective depth of MS-based proteomic profiling. This approach should be widely applicable to any LC-MS/MS instrument platform or biological system.
Human embryonic stem cells (hESCs)1 are non-transformed cell lines that can proliferate indefinitely in culture, although maintaining the potential to form all primary human cell types (pluripotency) (1,2). These cells, which originate from the inner cell mass of pre-implantation blastocysts, represent a unique source of human cells for cell replacement therapies and for creating model human systems for understanding disease and development (3). Like other mammalian ESCs, hESCs were originally derived and propagated on replication-deficient mouse embryonic fibroblast (MEF) feeder cells in serum (2,4), with varying efficiencies (5). At the heart of this variability is a lack of understanding of the regulatory pathways and growth factors that govern hESC self-renewal and pluripotency (6). This ambiguity restricts the application of hESCs in both research and therapeutic applications.
We hypothesize that under optimal hESC culture conditions, there exist autocrine and paracrine growth factors, produced both by the feeder cells and the hESCs themselves, that establish the complex microenvironment required to retain hESC potential in culture. Previous genomic-based studies suggested the presence of such networks of hESC transcriptional regulation (7); however, these networks were not correlated to the extracellular microenvironment that ultimately controls hESC fate. Moreover, prior attempts to identify proteins within the hESC microenvironment using MS-based approaches produced few potential candidate regulators and provided little new insight or tangible improvements upon hESC line derivation and culture (6, 8–11).
Several studies of extracellular proteomes (secretomes) (12–17) identified a small number of extracellular proteins but failed to identify growth factors that were known to be present. One of the main problems inherent in these and other large scale MS-based proteomic studies was that a “single-pass” analysis strategy was employed. Each peptide-containing sample/fraction was analyzed once, generally using liquid chromatography coupled to a mass spectrometer (LC-MS/MS). Because there may be hundreds of thousands peptides present in such complex biological mixtures; many, if not the majority, of the peptides in these samples are not selected for MS/MS analysis. Consequently, numerous proteins go unidentified.
In a typical LC-MS/MS experiment in data-dependent acquisition (DDA) mode, the most abundant peptides ions are selected preferentially for MS/MS fragmentation, resulting in the identification of the most abundant proteins in a given mixture. To overcome this limitation, a number of strategies have been developed. These include organellar separation (18), as well as several pre-fractionation and enrichment strategies (19). Despite these enhancements, many proteins remain unidentified, simply because the dynamic range of the experiment is reduced by the vast excess of peptides from high-abundance proteins present. This is further compounded by the intrinsic limitation of duty cycle and dynamic range of mass spectrometry instrumentation.
In this study, we devised a novel MS-based proteomic method to profile the microenvironments of hESCs in vitro. Using this approach, we characterized proteins in MEF feeder cell conditioned medium (CM) and culture medium conditioned by either H1 or H9 (hESC-CM) that were grown in the absence of feeders. The creation of a functionally validated, serum-free, CM platform allowed for the detection of low-abundance protein growth factors at concentrations <10−9 g/ml from relatively large volumes of sample (50–100 ml). To maximize proteome coverage, we employed an optimized iterative exclusion (IE) analysis method (20), wherein each fraction was analyzed 3–5 times. Round 1 consisted of a simple LC-MS/MS experiment. Subsequent rounds were carried out with the same type of LC-MS/MS experiment, except that ions selected in all previous rounds were excluded based on an optimized accurate mass and retention time (RT) window. To further increase coverage and evaluate its utility with complex samples, our IE-MS approach was combined with both gel and solution-based pre-fractionation approaches. This targeted approach provides the most comprehensive insight into the hESC culture microenvironment to date, highlighted by the identification of a large number of previously undetected growth factors. Overall, this MS-based method circumvents common problems in protein identification, such as bias against low-abundance peptide ions and poor reproducibility between replicate samples.
EXPERIMENTAL PROCEDURES
hESC Culture—
hESC lines (H1 and H9) were maintained in feeder-free culture in MEF-CM (21) or in the absence of conditioning with 36 ng/ml basic fibroblast growth factor (bFGF) (22). hESCs were plated on Matrigel-coated plates (BD Biosciences). The medium was changed daily, and the cells were passaged every 5–7 days through dissociation with 200 units/ml collagenase IV (Invitrogen). Cell counting, flow cytometry, and teratoma formation and histological analysis were also performed as described previously (22).
MEF- and hESC-CM—
hESC medium consisted of knockout Dulbecco's modified Eagle medium, 1% nonessential amino acids, 1 mm l-glutamine (all from Invitrogen), 0.1 mm β-mercaptoethanol (Sigma Aldrich), and knockout serum replacer medium (Invitrogen) (either 0 or 20%, depending on whether medium was defined as “serum-free”). Recombinant human bFGF (4 ng/ml; Invitrogen) was added prior to conditioning of hESC medium by MEFs. MEFs were prepared according to previously established protocols (21) prior to conditioning, while both H1 and H9 hESCs were cultured normally to 80% confluence for 24 h before conditioning in the serum-free hESC media. Serum-free (no serum, serum supplement, replacement, or alternative) hESC medium was prepared and conditioned on two independent batches of irradiated MEFs, prepared using the methods of Xu et al. (21). Alternatively, the same serum-free medium was conditioned for 24 h on each H1 and H9 hESCs grown in feeder-free conditions on Matrigel (22). For hESCs, standard volumes of medium were conditioned for 24 h on cells at ∼80% confluence and ∼50% positive for pluripotent markers. To remove cell debris, all CM was filtered through a 0.22-μm sterile membrane and stored at −80 °C. In evaluating the serum effect, bovine serum albumin removal was performed using the Montage albumin removal kit according to the manufacturer's instructions (Millipore). For functional evaluation, serum-free MEF-CM was supplemented with 20% knockout serum replacement medium prior to culturing the hESCs; the “non-conditioned medium” was standard hESC medium that was not exposed to MEFs.
CM Protein Extraction, Fractionation, and Digestion—
Approximately 50 μg of protein (determined by Bradford assay) was extracted from CM batch for each pre-fractionation and digestion strategy employed (Fig. 1). Based on a CM protein concentration of 1–2 μg/ml, 50–100 ml of MEF- or hESC-CM was concentrated to 250 μl using an Amicon 15-ml centrifugal filter device (Millipore), washed twice with filtered de-ionized water, and dried in a vacuum centrifuge. For gel-enhanced fractionation, samples were reconstituted in 1× Laemmli loading buffer and resolved on a 1.5-mm, 10 or 12% SDS-PAGE mini-gel. The gel was stained with Coomassie and the entire lane representing the concentrated sample divided into ∼20 sections (fractions). For the multi-dimensional protein identification technology (MuD-PIT) analysis, desalted tryptic peptides (solid phase extraction) in 10% formic acid (FA) were loaded on a strong cation exchange column (Bio SCX Series II, 0.8 × 50 mm; Agilent) in 5% acetonitrile (ACN), 0.1% FA, collecting the flow-through as the first fraction. Twelve more fractions were created by sequentially injecting 20 μl of the following KCl fractions in 0.1% FA and collecting the flow-through: 7.5, 15, 30, 45, 60, 75, 90, 120, 150, 300, 500 mm KCl in 5% ACN and 500 mm KCl in 30% ACN. All MuD-PIT fractions were dried in a vacuum centrifuge.
Fig. 1.
Schematic description of the sample preparation and analyses of hESC- and MEF-CM. The extraction, fractionation, and MS-based analytical strategy applied to the proteins in CM from feeder cells (MEF-CM) or hESCs cultured-free of feeder cells (hESC-CM). Biological duplicates of MEF-CM and hESC-CM were analyzed. 50 μg of protein from each replicate was analyzed using Gel-enhanced (GeLC-MS), MuD-PIT, and no-pre-fractionation analysis. Nano-LC-MS/MS was then performed multiple times on each fraction utilizing our m/z and RT-driven IE-MS strategy.
Proteolytic Digestion with Trypsin—
For gel-enhanced analyses, each gel section was digested manually (23). Briefly, gel bands were cubed into smaller pieces (∼2 mm2) and destained by washing in 1 m ammonium carbonate (NH4CO3) containing 20% ACN. For cysteine reduction, the gel pieces were dehydrated with 100% ACN and rehydrated with 10 mm dithiothreitol in 100 mm NH4CO3 for 30 min. The dithiothreitol solution was removed and the gel pieces alkylated by adding 100 mm iodoacetamide in 100 mm NH4CO3 for 30 min. The gel pieces were washed and dehydrated with 100% ACN, then rehydrated with 50 mm NH4CO3. For digestion, the gel pieces were first dehydrated with 100% ACN, then rehydrated with modified porcine trypsin (Promega, Madison, WI) (20 μg/ml in 50 mm NH4CO3) on ice for 15 min. Excess trypsin solution was removed, the gel pieces were covered with 50 mm NH4CO3, and the samples were maintained at 37 °C for 18 h. To extract the resulting peptides, the supernatant was collected and gel pieces were extracted three times with 10% FA and once with 100% ACN. Samples were then evaporated to dryness with a SpeedVac and re-suspended in 10% FA for LC-MS/MS analysis.
For the MuD-PIT and no pre-fractionation protocols, samples were reconstituted in 8 m urea with 50 mm NH4CO3, reduced with 10 mm dithiothreitol, alkylated with 30 mm iodoacetamide, diluted 1:4 with 50 mm NH4CO3, and digested with trypsin (1:25 enzyme/substrate ratio) at 37 °C overnight. To remove urea and other salts, as well as to concentrate samples for analytical SCX fractionation, tryptic peptides were extracted using a 1-ml C18 solid phase extraction cartridge (Waters, Milford, MA), eluted with 50% (v/v) ACN, 0.1% (v/v) FA, and re-concentrated in a vacuum centrifuge. Dried fractions were reconstituted in 10% FA for LC-MS/MS analysis or SCX fractionation.
IE-MS (LC-MS/MS) Analysis—
All dried fractions were reconstituted in 10% FA prior to injection. For gel-enhanced analysis, excised band samples were identified as having low, medium, or high complexity based on clear, light, or dark Coomasie staining, respectively. For MuD-PIT analysis, sample complexity was based on in-house standards where samples were divided into low complexity (0–15 and 500 mm), medium complexity (30, 150, and 300 mm), and high complexity (45–120 mm) fractions. Depending upon the anticipated complexity of the sample, ∼1/4, 1/5, or 1/8 of each fraction was analyzed using a 60-, 90-, or 150-min LC method, respectively. Separation using LC (5–40% ACN, 0.1% FA gradient) was performed on a NanoAcquity UPLC (Ultra Performance Liquid Chromatography - Waters) with a 15 cm x 75 μm C18 reverse phase column. Peptide ions were detected in DDA mode by tandem MS (Q-ToF Ultima; Waters).
We found that the IE strategy yielded the best results when high quality MS data were acquired and the LC separation was performed with precision. To generate the highest quality MS/MS spectra we used the following DDA parameters: survey scan (MS only) range m/z 400–1500, 1 s scan time, 1–4 precursor ions selected based on intensity (25 cps) and charge state (+2, +3, and +4). For each MS/MS scan, the m/z range was extended to m/z 50–2000, a scan time of 1 s used in early exclusions, with an increase to a scan time of 1–4 s in later exclusions (signal-dependent, total ion chromatogram 6000 cps). The MassLynx charge state-dependent collision energy profile was used. Selected precursors were then excluded for the next 45 s. This method was optimized to ensure the highest rate of successful MS/MS spectral assignment. In general, the majority of MS/MS spectra (∼75%) passed our quality threshold in one scan for the first 2–3 rounds of IE analysis. The result is an average of ∼1000 MS/MS spectra per 90-min acquisition where ∼50% of these spectra will lead to a successfully identified peptide when searched.
Creating Exclusion Lists—
For the IE analysis of each fraction, the m/z and RT values were manually extracted from the “.RAW” data folder (“auto.txt” file) for all of the ions selected in the previous MS/MS experiment. All previously selected ions were excluded; not just those identified as peptides. This ensures that ions with high spectral intensity were not analyzed more than once, even if they were not identified as peptides via conventional MS/MS analysis. To create an exclusion window centered on the major isotopes and avoid excluding masses below the mono-isotopic peak, an m/z shift of 0.7 was added to each m/z value selected for MS/MS. These ions were excluded (MassLynx DDA exclude functionality; Waters) from all analyses performed after that fraction using a m/z tolerance window of ±0.8 and RT window of ±45 s. This iterative process was repeated at least three to five times, depending on fraction abundance. This process resulted in the compilation of exclusion lists containing thousands of entries.
Comparing IE and Repeat Injection LC-MS/MS—
To compare the performance of IE analysis to a simple repeat injection experiment, we analyzed three large fractions of MEF-CM from an SDS-PAGE sample. Approximately 50 μg of protein from MEF-CM was separated by SDS-PAGE and ∼1/6 of this was used to create each MEF-CM fraction for the parallel analysis. Gel sections were digested with trypsin and peptides extracted. For each round of parallel analysis, ∼10% of each fraction was analyzed using identical LC conditions (the same 90-min gradient, 15 cm × 75 μm C18 reverse phase column, similar flow rate and identical amounts of digest injected on-column) and MS settings (1 s MS scan, followed by 4 × 1 s MS/MS scans; Q-ToF Ultima, Waters) over five rounds either simply repeating the same analysis (repeat injection) or employing the IE-MS strategy. These settings enabled the acquisition of as many MS/MS spectra as possible without sacrificing data quality.
To compare conventional Q-ToF IE-MS analysis with repeat injection analysis on a faster scanning mass spectrometer, a similar comparison was carried out using an LTQ-Orbitrap (ThermoFisher) for the repeat injection analysis. Fractionated MEF-CM digests were created, and 1/10 of each fraction was analyzed using the same 90-min LC gradient. The IE-MS Q-ToF analysis was carried out as described previously. For LTQ-Orbitrap repeat injection analysis, the data-dependent acquisition mode was enabled and each 1s survey scan (Resolution: 60,000) was followed by three MS/MS scan (0.35 s each) with dynamic exclusion for a duration of 30 s on the LTQ linear ion trap mass spectrometer. Multiply charged ions with intensity values above 10000 counts were selected for MS/MS sequencing. The normalized collision energy was set to 25%. Although not standard, IE-MS applications are also possible on the LTQ-Orbitrap with the implementation of a custom script into the control software2.
MS Data Interpretation and Gene Ontology Assignment—
The acquired MS/MS spectra were processed by using MassLynx 4.0 with the MaxEnt 3 PeptideAuto function using default parameters to generate peak list files (pkl). Files were extracted and searched against both forward and reverse human or mouse international protein index databases (version 3.22; Human 57846, Mouse 51477 entries) using Spectrum Mill (A03.03; Agilent Technologies, Santa Clara, CA). To account for contaminating proteins, both mouse and human international protein index databases were appended with a short list of common protein contaminants (i.e. albumin, keratin, trypsin; Agilent Spectrum Mill contaminants). For the data extractions, MS/MS spectra had to contain a minimum of one amino acid sequence tag. All database searches allowed for a fixed modification of Cys with iodoacetamide, variable oxidation of methionine, a digest with trypsin, and up to two missed cleavages, with a spectral peak intensity minimum of 60%. As suggested by the manufacturer, a minimum peptide score of 6 was selected to ensure that sufficient peptide fragment ions were matched in comparison to unmatched fragment ions for the proposed peptide. This threshold gives an acceptable (5.7%) false positive rate. For protein scoring a minimum protein score of 13 was chosen. A threshold level was set so that proteins with a minimum of two matching peptides required significant peptide scores for a positive identification.
The following settings for a database search for the Q-ToF data were employed: mass tolerance of 150 ppm for MS spectra and 100 ppm for MS/MS spectra. For the LTQ-Orbitrap data, the following settings were used: ESI-linear ion trap, a mass tolerance of 0.03 Da for MS spectra, and 0.5 Da for MS/MS spectra. For the Q-ToF data, using the scoring thresholds mentioned previously, the dataset exhibited a false positive peptide identification rate of 5.72% by using a reverse database search performed by a function of Spectrum Mill. False positive peptides (peptides that exhibited a forward minus reverse score of less than zero) were removed from the list of identified peptides. Therefore a 0% false positive rate is effectively used in the finalized dataset for both hESC and MEF-CM.
Dataset redundancy was handled in Spectrum Mill using grouping functions built into the software. Protein scores and the number of distinct peptides were calculated, such that only peptides with the highest MS/MS search score were counted. If peptides were listed for more than one protein these proteins were grouped together, thereby grouping proteins that may appear under different names or accession number. Only the highest scoring member of a protein group was displayed and used in calculations. However, in cases where distinct peptides uniquely identified a different isoform, the software included each protein isoform and its distinct peptides in its report.
Gene ontology was assigned to all identified proteins in all samples by using BioMart. Only those proteins with assigned cellular component information were included in the ontology analysis. Proteins were sorted by cellular component using the key words described in supplemental Fig. 3. Biological function was assigned to all extracellular proteins using the following parent keyword searches of gene ontology or protein description: Extracellular Structure (extracellular matrix, basement membrane, collagen, basal lamina), Growth Factor (growth factor, growth factor binding, cytokine, chemokine, hormone, receptor binding, NOT membrane), Proteolysis (proteolysis, protease, peptidase), and Growth & Development (NOT “Growth Factor”, growth, differentiation, development, morphogenesis, organogenesis).
Validation of IE-MS Findings: Quantitation of Insulin-like Growth Factor II (IGF-II) and TGFβ-1 in CM—
Mouse IGF-II in MEF-CM was assessed using the Duo Set ELISA kit (R&D Systems). IGF-II in hESC-CM was determined with the Non-Extraction IGF-II ELISA kit (Diagnostic Systems Laboratories) or by Western blot using antibody clone S1F2 and IGF-II standard (both Upstate). IGF-II Western blots were performed on ∼3 ml of concentrated serum-free CM. TGFβ-1 content was assessed by ELISA using OptEIA TGFβ-1 Set (BD BioSciences). MEF and hESC-CM samples were not diluted prior to the IGF-II ELISA. The rest of the assays were performed according to manufacturers’ recommendations.
RESULTS
IE Increases the Identification of Low-abundance Peptides—
Conventional LC-MS/MS experiments are limited by dynamic range restrictions that prevent a more complete analysis of a given proteome. For example, based on the international protein index, the human proteome consists of ∼6 × 105 unique tryptic peptides with masses between 700 and 6000 Da. However, multiple charge states, missed cleavages, and mass modifications could increase this complexity another 10-fold, resulting in ∼6 × 106 potentially observable m/z species in a typical MS experiment. In the MS analysis of a complex biological sample, even if only a small portion (∼10%) of these peptides were actually observable/present and they were further reduced in complexity 10-fold through fractionation, ∼6 × 104 uniquely observable m/z species would still be in each MS analysis. Currently, the fastest MS instrumentation can perform up to a maximum of ∼10,000 in a single 90-min run; however, lower quality MS/MS are typically obtained. This raises the probability that a large proportion of peptides/proteins, particularly those of lower spectral abundance, will go undetected in any given MS-based proteomic analysis.
To overcome this limitation, we devised a strategy to improve upon a simple repeat analysis of the same proteomic sample wherein we directed the MS to ignore ions previously selected for MS/MS (Fig. 2a). This was accomplished using very large exclusion lists derived from m/z values and LC RTs from previous rounds. The software then directed the MS to ignore ions of higher spectral abundance that were already fragmented in previous rounds. This allowed previously uncharacterized ions of lower spectral abundance to be targeted in later rounds (Fig. 2a).
Fig. 2.
A schematic of the iterative exclusion-liquid chromatography-tandem mass spectrometry (IE-LC-MS/MS) approach. a, during repeat data-dependent MS analysis of the same sample, peptide ions of higher spectral abundance are preferentially selected for MS/MS. In later rounds of analysis, at a given RT, previously selected peptide ions are excluded based on m/z to force the analysis of ions of lower spectral abundance. b, to minimize the exclusion footprint and maximize IE analysis coverage a shift of 0.7 ± 0.8 is added to the previously acquired ion's m/z to create a narrow, but all-encompassing generic exclusion window for a peptide ion's isotopic envelope.
Our method also prevents MS/MS analyses of ions within the isotopic envelope of excluded ions, because these experiments provide no new protein sequence information. These ions are often of high enough intensity that they can potentially trigger an MS/MS event. Thus, we add a mass shift of 0.7 to all previously selected ions and then exclude additional MS/MS events within a window of ± 0.8 (Fig. 2b). This also ensures that potential ions, with m/z slightly below the mono-isotopic peaks, are not excluded in subsequent rounds of LC/MS-MS analysis.
To demonstrate the benefits of the IE-MS strategy, we performed a parallel evaluation of the IE-MS method with a repeat injection analysis (five injections with random selection of the most abundant ions for MS/MS) starting from the same single-pass (single injection) analysis. Three large protein fractions of MEF-CM prepared by gel electrophoresis (Fig. 3) were used as the analytes in this comparative study. Equal amounts of sample were analyzed using identical 90-min LC-MS/MS methods. Compared with a single-pass analysis, the repeat injection method yielded a ∼1.5-fold increase in the number of unique proteins and peptides identified. However, use of the IE-MS strategy further improved this to a ∼2-fold increase (Fig. 3, a and b). This improvement was a result of the IE-MS strategy preventing the repeat selection of the same peptide (and non-peptide) ions, while allowing the MS to select ions of lower abundance in later analysis rounds (Fig. 3, c–e).
Fig. 3.
Direct comparison of IE-MS analysis to basic repeat injection. Three large MEF-CM fractions from gel separation were each analyzed by five rounds of LC-MS/MS using identical instrumental and sampling parameters. One set was simply analyzed five times (repeat injection) where the second set, starting with the same initial run, was analyzed using the IE strategy. The mean and S.D. (n = 3) of the relative number of unique (a) proteins and (b) peptides cumulatively identified over each round of analysis for each method. For the first and fifth analysis rounds, the total numbers of unique peptides or proteins identified in combined MEF-CM or fractions is indicated for each method. The resulting data for the three different fractions were pooled for each method and filtered to only proteins identified with >2 unique peptides. c, frequency of how many rounds (out of 5 total) a unique peptide was identified using repeat injection or IE-MS analysis. Heat maps for (d) repeat injection and (e) iterative exclusion where each row corresponds to a protein identification sorted by total spectral intensity show which round of analysis two or more unique peptides were identified (red square), and whether a particular protein was unique to that analysis strategy (blue square).
In the repeat injection analysis, almost 50% of unique peptides identified were found in at least four out of five of the analytical rounds. In contrast, the IE-MS approach provided a more efficient MS workflow, as ∼75% of unique peptides were identified two times or less over five rounds (Fig. 3c). The majority of repeat identification in the IE-MS method (Fig. 3c) were the result of the acquisition of different charge states as well as some poorly resolved peptides by LC (data not shown). This enhanced efficiency not only increases the number of identified peptides and proteins (Fig. 3, a and b), but also increases overall protein sequence coverage as demonstrated by an increased unique peptide to protein ratio in later analysis rounds (Fig. 3, a and b and Fig. 5, a–d). Consequently, focusing on proteins identified with high confidence (i.e. with >2 unique peptides per protein), the IE strategy revealed 30% more unique proteins compared with repeat injection, specifically identifying species of lower spectral abundance in later analysis rounds (Fig. 3, d and e). The heat maps in Fig. 3, d and e illustrate that during IE-MS analyses, as the rounds of iterative exclusion increased, more unique peptides (proteins) were identified compared with the repeat analysis method. As peptide ion abundance decreases, the IE strategy (Fig. 3e) continues to add to the total number of unique peptides and proteins identified. In many cases, the increase in the number of identified proteins was the result of additional peptides that were found in later rounds of IE-MS analysis adding to single peptide hits found in earlier rounds. Conversely, the number of peptides identified by repeat injection (Fig. 3d) displayed a more random pattern with little correlation between spectral abundance and the analysis round in which they were identified. In addition, there were a few proteins uniquely identified using the repeat injection technique that were mainly a result of erroneous precursor mass assignment or low quality MS/MS (data not shown). Taken together, these data clearly demonstrate the utility of the IE-MS strategy for increasing proteome coverage particularly with respect to species of low spectral abundance.
Fig. 5.
The benefit of IE in LC-MS/MS analysis of complex mixtures with sample pre-fractionation. Following five rounds of IE-MS analysis, the mean and S.D. (n = 4) of the relative number of unique proteins and peptides identified in the MEF and hESC-CM replicates relative to the initial analysis using (a) no pre-fractionation, (b) MuD-PIT, (c) gel-enhanced, or (d) datasets from all methods combined. d, the average mass spectral intensity of newly identified unique peptides relative to the initial analysis for each pre-fractionation approach. For the first and fifth IE-MS rounds, the total numbers of unique peptides or proteins identified in combined MEF-CM or hESC-CM replicates is indicated for each method.
Importantly, in later rounds of IE-MS analysis, as the number of assigned MS/MS increases with corresponding lower spectral abundance the quality of the MS/MS spectra comes into question. Generally, MS/MS quality depends on the intensity of the precursor ion. As the number of iterative runs increases the overall precursor intensity decreases. Hence, the frequency of assigned spectra would be reduced as the runs are proceeding. To counterbalance this in the practical application of IE-MS analysis, the duration of the MS/MS acquisition in later rounds could be increased accordingly. To this end, we coupled MS/MS scan time with a signal-dependent quality threshold when utilizing IE-MS for routine applications (see under “Experimental Procedures”).
To assess the effectiveness of IE-MS analysis with a conventional analysis on the most modern MS instrumentation, a similar comparison was then carried out using the IE-MS strategy on a Q-ToF MS (Global, Waters) compared with repeat injection using an identical sample and LC conditions on a faster scanning LTQ-Orbitrap (Fig. 4). Not surprisingly, a single-pass analysis on the LTQ-Orbitrap yielded up to 50% more unique peptides and proteins (Fig. 4, a and b) compared with the same run on the Q-ToF. However, after one to two more rounds of IE-MS analysis, the Q-ToF was able to achieve a comparable level of unique protein and peptide coverage as the single LTQ-Orbitrap run. This pattern continued over five rounds of analysis, further increasing these numbers and producing complementary results, which paralleled LTQ-Orbitrap analysis. These results indicate that the effective increase in performance realized when applying IE-MS analysis on older instrumentation can provide a number of identified peptides/proteins approaching that of newer instruments, like the LTQ-Orbitrap. Moreover, a 1.5–2.0-fold increase in LTQ-Orbitrap peptide and protein statistics (Fig. 4, a and b) observed over five rounds of repeat injection analysis, combined with the complimentary results seen with the IE-MS Q-ToF analysis, indicates that a single-pass analysis on even the most modern instrumentation is still far from comprehensive. Additionally, it seems that in striving toward comprehensive MS-based proteomics, repeat analyses will be necessary with greater success achieved with a directed strategy such as the IE-MS technique.
Fig. 4.
Comparison of IE-MS Q-ToF to repeat LTQ-Orbitrap analysis. Three large MEF-CM fractions from gel separation were each analyzed by five rounds of LC-MS/MS using identical amounts and LC conditions. One set was simply analyzed five times on a fast scanning LTQ-Orbitrap (1 s MS, 3 × 0.33 s MS/MS, 2 s duty cycle - repeat injection), whereas the second set was analyzed using the IE strategy on a slower scanning Q-ToF (1s MS, 4 × 1s MS/MS, 5 s duty cycle). The mean and S.D. (n = 3) of the relative number of unique (a) peptides and (b) proteins cumulatively identified over each round of analysis for each method or based on the two datasets combined. For the first and fifth analysis rounds, the total numbers of unique peptides or proteins identified in combined MEF-CM or fractions is indicated for each method.
Using a Serum-free Culture Medium Enables Identification of Endogenous Growth Factors—
This MS–based study aimed at identifying protein growth factors in CM, such as bFGF, that are known to be present at low ng/ml concentrations in hESC cultures (25). Generally, analyses of pure protein (peptides) by MS have limits of detection in the femtomole (10−15 moles) range. We conservatively projected a limit of detection of 1 pmol (10−12 moles) for proteins (peptides) within our complex mixtures. Under these constraints, up to 100 ml of CM could be required to obtain detectable amounts of protein growth factors (supplemental Fig. 1).
Unfortunately, hESC CM samples typically contain high concentrations of serum supplement proteins (∼20 mg/ml) that are 106-fold greater in concentration than those of the putative growth factors. Consequently, the detection of the protein growth factors in the presence of the serum supplement proteins would not be feasible given the 103-104 dynamic range of current MS-based proteomic technologies (26). Therefore, the removal of the serum supplement prior to the conditioning process was essential. As demonstrated in supplemental Fig. 1b, the removal of the serum supplement provided the crucial enrichment of secreted proteins, thereby allowing for the detection of ng/ml concentrations of growth factors in the absence of interfering serum proteins.
Although it was apparent that the creation of a CM formula that was free of serum was necessary for the detection of putative growth factors, the serum-free CM required validation to ensure that this formulation would have no adverse effects on the hESCs (i.e. differentiation, cell death, etc.) (supplemental Fig. 1c). In contrast to non-conditioned medium, medium conditioned by feeder cells (MEFs) in the absence of serum displayed little difference compared with standard MEF-CM and maintained expansion of cells with the hESC phenotype (SSEA3/4 and Tra-1–81 expression) over multiple passages (supplemental Fig. 2, a and b). After 12 passages, these cells formed teratomas in vivo that contained human cell types from all three embryonic germ layers, indicating continued pluripotency (supplemental Fig. 2c). Simultaneously, hESCs that underwent short-term exposure (24 h) to serum-free medium in feeder-independent culture maintained expression of pluripotent hESC markers (supplemental Fig. 2d). Together, these data suggest that the production of unknown factors continues despite the absence of serum, creating a microenvironment that supports the self-renewal and pluripotency of hESCs.
IE-MS Analysis and Pre-fractionation Are Complementary—
The ultimate goal of this research was the characterization of the supportive protein microenvironment of hESCs created under feeder cell-dependent and independent conditions. For a more comprehensive analysis that would demonstrate the broad applicability of our IE strategy, we used both in-solution and polyacrylamide gel electrophoresis (PAGE)-based protein separations to help characterize the proteomes of each sample. To this end, de-salted protein concentrates were subjected to three different sample preparation methods (Fig. 1); (1) no pre-fractionation as an analytical baseline; (2) gel-enhanced (also known as GeLC-MS) (27); and (3) MuD-PIT analysis (28). Each of the resulting fractions was subjected to a minimum of three rounds of IE-LC-MS/MS analysis using successive exclusion lists excluding up to 104 ions over multiple rounds.
The combination of the IE-MS method with pre-fractionation is presented in Fig. 5. The number of unique peptides identified in the hESC-CM increased by ∼75% after four rounds of IE-MS analysis without pre-fractionation (Fig. 5a). This improvement increased to ∼120% in the MuD-PIT and 135% in the gel-enhanced approach (Fig. 5, b and c). When the results of all three pre-fractionation strategies were combined (Fig. 5d), IE-MS analysis provided a 2-fold improvement in both peptide and protein metrics compared with a single round of analysis by the gel-enhanced method.
As anticipated, in addition to increased protein/peptide numbers, the average spectral intensity (MS signal) of the newly identified peptides in final exclusion round was as small as 10% of those peptides in the initial analysis (Fig. 5e). Together, these data demonstrate that, even when employing both extensive and complimentary sample pre-fractionation (29), IE-MS methods are still highly valuable. This technique increases protein/peptide metrics by effectively maximizing instrumental dynamic range, directing the selection of lower spectral intensity ions generally missed in a single-pass analysis.
The Protein Microenvironment of hESCs Revealed—
Following multiple LC-MS/MS analyses using stringent MS/MS identification criteria (see under “Experimental Procedures”), the total number of unique proteins detected in feeder cell-dependent (MEF-CM) and independent (hESC-CM) microenvironments was 550 and 2493, respectively (Fig. 5d and supplemental Tables 1 and 2). This is significantly greater than the 136 mouse and 102 human proteins identified previously in related studies (9–11). Moreover, in analyzing CM from independent hESC cell lines (H1 and H9), we were able to observe an ∼80% overlap in the proteins identified (supplemental Table 2).
However, the objective of the analysis was not simply to increase the total proteins detected, but to identify growth regulating proteins secreted into the extracellular space. To avoid counting protein artifacts from cell lysis, we used gene ontology (vide supra) to sort the MEF- and hESC-CM datasets by cellular component. In doing so, we found 196 and 245 extracellular proteins in the MEF-CM and hESC-CM samples, respectively. Once again, the results presented here represent an approximate 10-fold enhancement over the 16 mouse and 31 human extracellular proteins identified in previously related datasets (9–11). These results also mark a significant improvement over other attempts to characterize cellular secretomes (12–17).
All of the proteins identified in each MEF- and hESC-CM samples were categorized based on their gene ontology, first filtering by those associated with the extracellular compartment and then by biological function (supplemental Fig. 3, a and b). The extracellular proteins identified in both the feeder cell-dependent (MEF-CM) and feeder cell-free (hESC-CM) conditions displayed a near identical distribution of inferred functions (supplemental Fig. 3, a and b). In both samples, proteins involved in the extracellular structure (ECM, collagen, basement membrane) had the highest representation, followed closely by those linked to growth factor (cytokine, chemokine, and hormone) function (supplemental Fig. 3, a and b). Interestingly, even though they represent two distinct components of the hESC microenvironment (one dependent and one free of feeder cells), the extracellular proteins found in both datasets possessed similar cellular functions based on the distribution of their biological roles.
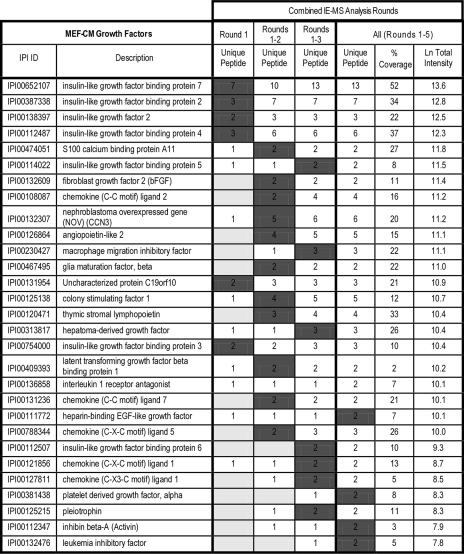
IE-MS Analysis Extends Protein Growth Factor Identification—
The benefits of the IE-MS analysis were best illustrated by considering the protein growth factors that were identified (Fig. 6 and Fig. 7). As a direct result of using the IE-MS analysis approach (Figs. 1 and 2), we report over 40 new potential growth factors (Figs. 6 and 7), as well as a large number of other proteins involved in extracellular structure, proteolysis, and development (supplemental Tables 3–8) as candidate hESC regulators. Specifically, we found 29 and 43 unique growth factor-like proteins in MEF and hESC-CM, respectively. This represents a far more comprehensive analysis than previously related studies (9–11) and the most successful proteomic analysis of growth factors to date. More importantly, we have used these data to establish that insulin-like growth factor II (IGF-II) in cooperation with bFGF and via one or more transforming growth factor β (TGFβ) signals establishes the regulatory niche of hESCs (30).
Fig. 6.
Table of extracellular protein growth factors identified in MEF-CM. High confidence protein growth factors identified in hESC feeder cell conditioned medium (MEF-CM). Based on combined analysis of replicates utilizing all pre-fractionation approaches, all factors were identified with > 2 unique peptides. Entries are listed according to total spectral intensity of unique peptides with the total unique peptides numbers identified for each analysis round. To illustrate IE analysis accessing species of lower spectral abundance each entry was highlighted in the round in which it was identified with two or more peptides.
Fig. 7.
Table of extracellular protein growth factors identified in hESC-CM. High confidence protein growth factors identified in hESC conditioned medium (hESC-CM) in the absence of MEF-CM. Based on combined analysis of replicates (H1 and H9 hESCs) utilizing all pre-fractionation approaches, all factors were identified with >2 unique peptides and were observed in both biological replicates. Entries are listed according to total spectral intensity of unique peptides with the total unique peptides numbers identified for each analysis round. To illustrate IE analysis accessing species of lower spectral abundance each entry was highlighted in the round in which it was identified with two or more peptides.
The IE-MS strategy was key in the detection of protein growth factors. The majority of growth factors were found in later rounds of IE-MS analysis in both the MEF-CM and hESC-CM (Figs. 6 and 7). Again, those growth factors identified in later rounds correlated strongly with decreasing spectral abundance, illustrating the ability for IE method to probe deeper into the proteome. Although protein growth factors best demonstrated the value of our IE-MS strategy for increasing depth and coverage, the same trend was also observed for those extracellular proteins identified in MEF-CM and hESC-CM with functions involved in extracellular structure (supplemental Tables 3 and 4), proteolysis (supplemental Tables 5 and 6), and development (supplemental Tables 7 and 8).
Finally, to quantify the success of our analysis, we performed immunoassays to validate two of the pertinent growth factors, IGF-II and TGFβ-1. The concentration of mouse IGF-II was determined to be 0.3–1.0 × 10−9 g/ml in MEF-CM and human IGF-II 2–20 × 10−9 g/ml in hESC-CM, as previously shown (30). To prevent artifacts in this analysis, mouse and human IGF-II-specific ELISA assays were used with mock CM as a control. In accordance with the MS analysis, TGFβ-1 was not detected by immunoassay in MEF-CM but was found to range between 8–30 × 10−11 g/ml in hESC-CM (feeder cell-free conditions). Additionally, the presence of bFGF and IGF-II has also been reconfirmed by Western blot analysis in both hESC- and MEF-CM (data not shown).
Although Matrigel (from mouse) used here in hESC culture has been shown to contain TGFβ-1, three of its four peptides identified by MS were human-specific (supplemental Fig. 4). The mock CM on Matrigel-coated plates did not produce a signal in the TGFβ-1 ELISA either. These observations further confirm that hESCs are producing factors independently to maintain a microenvironment where pluripotency and self-renewal are supported (30). Interestingly, in line with our previous findings for other growth factors, peptides corresponding to TGFβ-1 (supplemental Table 2 and supplemental Fig. 4) were not identified until the third round of IE-MS analysis. Taken together, these data demonstrate that the target limits of detection estimated for our MS-based analyses (1–10 × 10−9 g/ml) was not only met, but also exceeded, in the case of TGFβ-1. The complementary protein pre-fractionation combined with the IE-MS method was critical to achieve this level of sensitivity.
DISCUSSION
Our experimental approach was designed strategically to identify low-abundance stem cell regulatory proteins by circumventing limitations common to MS-based studies of complex samples, such as medium conditioned by heterogeneous cell types. The benefit of repeat analysis has already been demonstrated (31) where repeat injections of the same sample result in a 10–30% improvement in protein/peptide coverage by virtue of new peptides being randomly selected for MS/MS upon each successive round of analysis. In conjunction with what we show here, most of the abundant peptide ions are continually selected ultimately stifling the identification of lower spectral abundance peptides by MS/MS.
In an effort to increase the total coverage, MS-based exclusion has been described in previously LC-MALDI experiments using a single repeat analysis (32,33). With LC-ESI-MS platforms, more extensive exclusion experiments have recently been reported, but these studies are based only on using only identified peptides and predicted charge states (34). Generally, greater than 50% of all ions selected in an MS-based proteomic experiment are never identified. Consequently, this deficiency represents a significant inefficiency in the MS duty cycle when using a repeat analysis strategy. Moreover, excluding unselected ions of different charge state (independently or based on identified peptides) may not only require manual intervention via database searching, but also may exclude unrelated ions with similar m/z and retention time. Other instrument-based approaches have also been presented previously to account for peptide ions missed in LC-MS/MS analysis, the most popular of which is gas phase fractionation (24). Much like sample pre-fractionation, gas phase fractionation acts to simplify the peptide mixture seen by the MS instrumentation and has still been rooted in a single-pass analysis mentality, albeit with smaller m/z ranges. Consequently, gas phase fractionation does not account for low-abundance peptide ions not selected in complex mixtures, though it would most likely be complementary when combined with an IE-MS approach.
The IE-MS method presented here circumvents many of these previous pitfalls by systematically excluding previously selected ions (identified or not) using optimized m/z and RT windows. As such, it is the only strategy that targets low abundant peptide ions in a de novo analysis, working toward a comprehensive profile of any sample type. Currently, the manual preparation of an IE-MS exclusion list takes only a few seconds and requires no special software, computer, data analysis, or interpretation (a copy of the MS Excel script can be found on our website). As a result, it could be completely automated in any MS platform using only the instrument's control software. Moreover, for the first time, we demonstrate the utility of combining a targeted repeat analysis strategy (IE-MS) with complex sample pre-fractionation (MuD-PIT and gel-enhanced separation). The results of this approach provide an unprecedented description of the hESC protein microenvironment in vitro.
Previous attempts to characterize supportive CM from both mouse embryonic and human fibroblasts identified only a few proteins as potential regulators of hESCs. In terms of potential growth regulators, Lim and Bodnar (9) detected insulin-like growth factor-binding protein 4 and pigment epithelium derived factor in MEF-CM. In two separate studies of different hESC supportive fibroblasts, Prowse et al. (10,11) identified gremlin, insulin-like growth factor-binding protein 3,6,7, follistatin, DKK3, TGFβ-binding protein, pigment epithelium derived factor, inhibin β A (activin), and slit 2 homologue. Even though the biological systems analyzed here are not identical, of all these factors, only the bone morphogenic protein agonist gremlin was not identified in this study. More significantly, the majority of these previously reported factors were detected in the first round of analysis in our study, reflecting their higher abundance, where the host of newly identified factors reported here for the first time were detected in later IE-MS rounds of analysis (Figs. 6 and 7 and supplemental Tables 3–8).
The identification of low-level components of the hESC microenvironment was accomplished by combining complementary pre-fractionation techniques with the IE-MS method. This offered a significant improvement over earlier studies on similar extracellular environments (8–11), as well as on general high-throughput proteomics strategies (31). As such, implementation of the methodology outlined here is well suited for the characterization of the secretome from a multitude of tissues and cell types in culture. The availability of more reproducible and sensitive LC-MS/MS instrumentation will improve the performance of this IE-MS approach even further. The IE-MS method should be straightforward to automate, and it will find applications in a wide range of future proteomic practices.
Acknowledgments
We are grateful for the help of D. George for technical work and L. Gallacher and L. Li for cell culture assistance and histology.
Footnotes
Published, MCP Papers in Press, October 20, 2008, DOI 10.1074/mcp.M800190-MCP200
The abbreviations used are: hESC, human embryonic stem cell; DDA, data-dependent acquisition; ACN, acetonitrile; bFGF, basic fibroblast growth factor; CM, conditioned medium; MuD-PIT, multi-dimensional protein identification technology; FA, formic acid; IE, iterative exclusion; IGF, insulin-like growth factor; LC, liquid chromatography; ELISA, enzyme-linked immunosorbent assay; MEF, mouse embryonic fibroblast; MS, mass spectrometry; MS/MS, tandem mass spectrometry; Q-ToF, quadrupole time of flight; RT, retention time; SCX, strong cation exchange; TGF, transforming growth factor.
P. Thibault, personal communication.
This work was supported by a grant from the Ontario Research and Development Challenge Fund (to G. A. L.), a Canadian Institutes of Health Research (CIHR) multi resource (to P. T.), and by CIHR and Stem Cell Network (to M. B.).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Reubinoff, B. E., Pera, M. F., Fong, C. Y., Trounson, A., and Bongso, A. ( 2000) Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat. Biotechnol. 18, 399–404 [DOI] [PubMed] [Google Scholar]
- 2.Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., and Jones, J. M. ( 1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 3.Menendez, P., Wang, L., and Bhatia, M. ( 2005) Genetic manipulation of human embryonic stem cells: a system to study early human development and potential therapeutic applications. Curr. Gene Ther. 5, 375–385 [DOI] [PubMed] [Google Scholar]
- 4.Evans, M. J., and Kaufman, M. H. ( 1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- 5.Hoffman, L. M., and Carpenter, M. K. ( 2005) Characterization and culture of human embryonic stem cells. Nat. Biotechnol. 23, 699–708 [DOI] [PubMed] [Google Scholar]
- 6.Van Hoof, D., Mummery, C. L., Heck, A. J., and Krijgsveld, J. ( 2006) Embryonic stem cell proteomics. Expert Rev. Proteomics 3, 427–437 [DOI] [PubMed] [Google Scholar]
- 7.Boyer, L. A., Mathur, D., and Jaenisch, R. ( 2006) Molecular control of pluripotency. Curr. Opin. Genet. Dev. 16, 455–462 [DOI] [PubMed] [Google Scholar]
- 8.Chin, A. C., Fong, W. J., Goh, L. T., Philp, R., Oh, S. K., and Choo, A. B. ( 2007) Identification of proteins from feeder conditioned medium that support human embryonic stem cells. J. Biotechnol. 130, 320–328 [DOI] [PubMed] [Google Scholar]
- 9.Lim, J. W., and Bodnar, A. ( 2002) Proteome analysis of conditioned medium from mouse embryonic fibroblast feeder layers which support the growth of human embryonic stem cells. Proteomics 2, 1187–1203 [DOI] [PubMed] [Google Scholar]
- 10.Prowse, A. B., McQuade, L. R., Bryant, K. J., Marcal, H., and Gray, P. P. ( 2007) Identification of potential pluripotency determinants for human embryonic stem cells following proteomic analysis of human and mouse fibroblast conditioned media. J. Proteome Res. 6, 3796–3807 [DOI] [PubMed] [Google Scholar]
- 11.Prowse, A. B., McQuade, L. R., Bryant, K. J., Van Dyk, D. D., Tuch, B. E., and Gray, P. P. ( 2005) A proteome analysis of conditioned media from human neonatal fibroblasts used in the maintenance of human embryonic stem cells. Proteomics 5, 978–989 [DOI] [PubMed] [Google Scholar]
- 12.Alvarez-Llamas, G., Szalowska, E., de Vries, M. P., Weening, D., Landman, K., Hoek, A., Wolffenbuttel, B. H., Roelofsen, H., and Vonk, R. J. ( 2007) Characterization of the human visceral adipose tissue secretome. Mol. Cell. Proteomics 6, 589–600 [DOI] [PubMed] [Google Scholar]
- 13.Khwaja, F. W., Svoboda, P., Reed, M., Pohl, J., Pyrzynska, B., and Van Meir, E. G. ( 2006) Proteomic identification of the wt-p53-regulated tumor cell secretome. Oncogene 25, 7650–7661 [DOI] [PubMed] [Google Scholar]
- 14.Mbeunkui, F., Fodstad, O., and Pannell, L. K. ( 2006) Secretory protein enrichment and analysis: an optimized approach applied on cancer cell lines using 2D LC-MS/MS. J. Proteome Res. 5, 899–906 [DOI] [PubMed] [Google Scholar]
- 15.Pellitteri-Hahn, M. C., Warren, M. C., Didier, D. N., Winkler, E. L., Mirza, S. P., Greene, A. S., and Olivier, M. ( 2006) Improved mass spectrometric proteomic profiling of the secretome of rat vascular endothelial cells. J. Proteome Res. 5, 2861–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thouvenot, E., Lafon-Cazal, M., Demettre, E., Jouin, P., Bockaert, J., and Marin, P. ( 2006) The proteomic analysis of mouse choroid plexus secretome reveals a high protein secretion capacity of choroidal epithelial cells. Proteomics 6, 5941–5952 [DOI] [PubMed] [Google Scholar]
- 17.Zvonic, S., Lefevre, M., Kilroy, G., Floyd, Z. E., DeLany, J. P., Kheterpal, I., Gravois, A., Dow, R., White, A., Wu, X., and Gimble, J. M. ( 2007) Secretome of primary cultures of human adipose-derived stem cells: modulation of serpins by adipogenesis. Mol. Cell. Proteomics 6, 18–28 [DOI] [PubMed] [Google Scholar]
- 18.Au, C. E., Bell, A. W., Gilchrist, A., Hiding, J., Nilsson, T., and Bergeron, J. J. ( 2007) Organellar proteomics to create the cell map. Curr. Opin. Cell Biol. 19, 376–385 [DOI] [PubMed] [Google Scholar]
- 19.Aebersold, R., and Mann, M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 20.Hui, J. P. M., Tessier, S., Butler, H., Badger, J., Kearney, P., Carrier, A., and Thibault, P. ( 2003) Iterative exclusion and directed MS-MS from trace-level protein digests of differentiated cell extracts. In 53rd ASMS Conference on Mass Spectrometry and Allied Topics, American Society for Mass Spectrometry, Nashville, TN
- 21.Xu, C., Inokuma, M. S., Denham, J., Golds, K., Kundu, P., Gold, J. D., and Carpenter, M. K. ( 2001) Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 19, 971–974 [DOI] [PubMed] [Google Scholar]
- 22.Wang, L., Li, L., Menendez, P., Cerdan, C., and Bhatia, M. ( 2005) Human embryonic stem cells maintained in the absence of mouse embryonic fibroblasts or conditioned media are capable of hematopoietic development. Blood 105, 4598–4603 [DOI] [PubMed] [Google Scholar]
- 23.Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V., and Mann, M. ( 2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 24.Yi, E. C., Marelli, M., Lee, H., Purvine, S. O., Aebersold, R., Aitchison, J. D., and Goodlett, D. R. ( 2002) Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis 23, 3205–3216 [DOI] [PubMed] [Google Scholar]
- 25.Rao, B. M., and Zandstra, P. W. ( 2005) Culture development for human embryonic stem cell propagation: molecular aspects and challenges. Curr. Opin. Biotechnol. 16, 568–576 [DOI] [PubMed] [Google Scholar]
- 26.Anderson, N. L., and Anderson, N. G. ( 2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 [DOI] [PubMed] [Google Scholar]
- 27.Blagoev, B., Kratchmarova, I., Ong, S. E., Nielsen, M., Foster, L. J., and Mann, M. ( 2003) A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21, 315–318 [DOI] [PubMed] [Google Scholar]
- 28.Washburn, M. P., Wolters, D., and Yates, J. R., 3rd ( 2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 29.Barnea, E., Sorkin, R., Ziv, T., Beer, I., and Admon, A. ( 2005) Evaluation of prefractionation methods as a preparatory step for multidimensional based chromatography of serum proteins. Proteomics 5, 3367–3375 [DOI] [PubMed] [Google Scholar]
- 30.Bendall, S. C., Stewart, M. H., Menendez, P., George, D., Vijayaragavan, K., Werbowetski-Ogilvie, T., Ramos-Mejia, V., Rouleau, A., Yang, J., Bosse, M., Lajoie, G., and Bhatia, M. ( 2007) IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature 448, 1015–1021 [DOI] [PubMed] [Google Scholar]
- 31.Elias, J. E., Haas, W., Faherty, B. K., and Gygi, S. P. ( 2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2, 667–675 [DOI] [PubMed] [Google Scholar]
- 32.Chen, H. S., Rejtar, T., Andreev, V., Moskovets, E., and Karger, B. L. ( 2005) Enhanced characterization of complex proteomic samples using LC-MALDI MS/MS: exclusion of redundant peptides from MS/MS analysis in replicate runs. Anal. Chem. 77, 7816–7825 [DOI] [PubMed] [Google Scholar]
- 33.Scherl, A., Francois, P., Converset, V., Bento, M., Burgess, J. A., Sanchez, J. C., Hochstrasser, D. F., Schrenzel, J., and Corthals, G. L. ( 2004) Nonredundant mass spectrometry: a strategy to integrate mass spectrometry acquisition and analysis. Proteomics 4, 917–927 [DOI] [PubMed] [Google Scholar]
- 34.Seo, J., Jeong, J., Kim, Y. M., Hwang, N., Paek, E., and Lee, K. J. ( 2008) Strategy for comprehensive identification of post-translational modifications in cellular proteins, including low abundant modifications: application to glyceraldehyde-3-phosphate dehydrogenase. J. Proteome Res. 7, 587–602 [DOI] [PubMed] [Google Scholar]