Abstract
Biological experiments are most often performed with immortalized cell lines because they are readily available and can be expanded without limitation. However, cell lines may differ from the in vivo situation in important aspects. Here we introduce a straightforward methodology to compare cell lines to their cognate primary cells and to derive a comparative functional phenotype. We used SILAC (stable isotope labeling by amino acids in cell culture) for quantitative, mass spectrometry-based comparison of the hepatoma cell line Hepa1–6 with primary hepatocytes. The resulting quantitative proteome of 4,063 proteins had an asymmetric distribution, with many proteins down-regulated in the cell line. Bioinformatic analysis of the quantitative proteomics phenotypes revealed that Hepa1–6 cells were deficient in mitochondria, reflecting re-arrangement of metabolic pathways, drastically up-regulate cell cycle-associated functions and largely shut down drug metabolizing enzymes characteristic for the liver. This quantitative knowledge of changes provides an important basis to adapt cell lines to more closely resemble physiological conditions.
The development of tissue culture techniques and establishment of cell lines has been indispensable for biological research for several decades (1). However, disadvantages of cell lines are that they are usually derived from tumors and have adapted to growth in culture. Although cell culture tries to create a close-to-physiology milieu by adding appropriate amounts of salt, glucose, amino acids, vitamins, and serum, the lack of tissue architecture and heterogeneous population of cell types often abolishes cell-cell interaction, secretion, and other functions based on tissue context. Cells in culture are prone to genotypic and phenotypic drifting. Thereby cell lines can lose tissue-specific functions and acquire a molecular phenotype quite different from cells in vivo. Acceptance of cell lines as model for biological function varies between fields. Cell biological studies on basic mechanisms, such as the cell cycle are routinely and overwhelmingly carried out in long-established cell lines (2). This is particularly the case for microscopy studies, including large-scale siRNA screens with imaging read out. In contrast, there is substantial controversy of how well cell lines, which are often established from late stage cancer, preserve aspects of the disease and whether or not they should be used in cancer drug development (3–5). Thus animal experiments or studies in primary cell lines are often preferred despite their added complexity. Accurate molecular phenotypes to determine whether the function to be investigated is preserved in cell lines would enable a rational choice of the most appropriate experimental system (6). In biotechnology and the pharmaceutical industry this goal obtains added urgency in the light of efforts to reduce animal experimentation to a minimum.
In this work we ask how primary cells and cell lines differ in their functions. This question has been addressed by comparing gene expression profiles at the transcriptome level in a substantial body of literature (for recent examples, see Refs. 7, 8). However, transcriptome studies are not quantitative with respect to changes at the proteome level. Ideally, the different molecular phenotypes should be assessed by quantitatively comparing the proteomes of the primary cells versus the cell lines. Here we report such a study and develop an algorithm to extract functional phenotypes from the resulting differential protein distributions.
EXPERIMENTAL PROCEDURES
Materials and Reagents—
Mouse hepatoma cell line Hepa1–6 was obtained from American Type Culture Collection (ATCC). l-arginine, l-lysine, l-13C615N4-arginine and l-13C615N2-lysine were purchased from Sigma-Aldrich. Chemicals for the “in solution” and “in gel” digests were purchased from Sigma-Aldrich; endoproteinase Lys-C was obtained from Waco, and sequencing grade-modified trypsin was from Promega.
Isolation of Mouse Primary Hepatocytes—
Isolation and culture of mouse hepatocytes was performed according to standard operating procedures (9). For biological and analytical reproducibility, primary hepatocytes were isolated from two mice and processed separately. After cultivation for 14 h, the cells were placed on ice and the medium was removed. The cells were lysed in radioimmune precipitation assay (RIPA) buffer.
SILAC Labeling of Mouse Hepatoma Cell Line Hepa1–6—
Hepa1–6 cells were grown in SILAC “light” (l-arginine and l-lysine) and “heavy” (l-13C615N4-arginine and l-13C615N2-lysine) conditions for 8 passages before the experiment. This period lasted around 3 weeks. Unless stated, cell culture medium contained 4.5 g/liter glucose, following the standard culture condition from ATCC. Other cell culture conditions were essentially the same as described (10).
Fluorescence Microscopy—
Primary hepatocytes were isolated and seeded at a density of 2 × 105 cells per well in collagen I-coated 12-well plates. Hepa1–6 cells were grown to a density of 2 × 105 cells per well in 12-well plates. Cells were stained for 15 min with 15 nm Mitotracker Orange CMTMRos (Invitrogen) and 1:5000 Hoechst 333342 (Sigma-Alldrich) at 37 °C in the corresponding cultivation medium. After three washing steps in cultivation medium cells were viewed with a ZeissAxioVert 200 m fluorescence microscope using a 40× LD-Plan Neofluor objective (numerical aperture 1.5). Cells were viewed under visible light, or excited with 345 nm (Hoechst 333342) or 550 nm (Mitotracker).
Protein Harvest, Digestion—
Primary hepatocytes and Hepa1–6 cells were lysed in a buffer containing 1% Nonidet P-40, 0.1% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 50 mm Tris, pH 7.5, 1 mm sodium orthavanadate, 5 mm NaF, 5 mm beta-glycerophosphate, and protease inhibitors (Complete tablet, Roche Diagnostics). The lysates were centrifuged in cold with 17,000 × g for 15 min to pellet cellular debris. Supernatant was collected, and a Bradford method was used to determine the protein concentrations. Equal amount of the proteins from the primary hepatocyte sample and Hepa1–6 sample were mixed, resulting in 100-μg proteins in total.
Protein mixtures were added with four volumes of methanol, one volume of chloroform, and three volumes of distilled water in a sequential manner. The addition of each solvent was followed by a short vortex. After centrifugation of 20,000 × g for 1 min, proteins were focused between organic and inorganic phases. The aqueous phase was discarded. Four starting volumes of methanol were added to the protein pellet followed by a short vortex. After spinning at 20,000 × g for 2 min, methanol was removed, and the protein pellet was air-dried.
Precipitated proteins were redissolved in a buffer containing 6 m urea, 2 m thiourea, 10 mm Hepes, pH 7.5. Proteins were reduced with 1 mm dithiothreitol for 1 h, alkylated with 5.5 mm iodoacetamide for 45 min in dark, and digested for four hours with endoproteinase Lys-C (1/50 w/w). After diluting four times with 20 mm ammonium bicarbonate, samples were digested overnight with sequencing grade-modified trypsin (1/50 w/w). The digestion was quenched by adding trifluoroacetic acid to reach pH <3.
Peptide Preparation for Mass Spectrometry—
Peptides were separated based on their isoelectric points in the Agilent 3100 OFFGEL fractionator (Agilent, G3100AA) in combination with commercially available IPG DryStrips, 13 cm, pH 3–10 (GE Healthcare, 17-6002-44) and IPG buffer, pH 3–10 (GE Healthcare, 17-6000-87) diluted 1:50 in 5% glycerol. Peptides were focused for 20 kVh at maximum current of 50 μA and maximum power of 200 milliwatt. Each peptide fraction was mixed with 10 μl of solvent containing 30% acetonitrile, 5% acetic acid, and 10% trifluoroacetic acid. The resulting solution was loaded into C18 reverse-phase StageTips (11).
Peptides were eluted from the StageTips by applying 80% acetonitrile, 0.5% acetic acid. Samples were dried down to 3 μl and mixed with equal volume of solvent containing 2% acetonitrile and 1% trifluoroacetic acid. 5-μl samples were applied for LC-MS/MS analysis.
Mass Spectrometry and Data Analysis—
Samples were injected via autosampler into a 15-cm fused silica emitter (75-μm inner diameter; Proxeon Biosystems) packed in-house with reverse-phase ReproSil-Pur C18-AQ 3-μm resin(12) and eluted with nanoflow in Agilent 1200 liquid chromatography system (Agilent Technologies, Waldbronn, Germany). The gradient induced a linear increase of 4–40% acetonitrile in 0.5% acetic acid over 90 min.
Eluted peptides were sprayed into a 7-T LTQ-FT or LTQ-Orbitrap mass spectrometer (Thermo Electron, Bremen, Germany) via a nanoelectrospray ion source (Proxeon Biosystems, Odense, Denmark) and analyzed as described previously (12). Raw MS1 spectra were processed using in-house developed software MaxQuant (version 1.0.7.4) (13–15), which performed peak list generation, SILAC- and extracted ion current-based quantitation, calculated posterior error probability, and false discovery rate based on search engine results, peptide to protein group assembly, and data filtration and presentation (14). The derived peak list was searched with the Mascot search engine (version 2.1.04; Matrix Science, London, UK) against a concatenated database combining 52,326 proteins from International Protein Index mouse protein database version 3.24, 27 commonly observed contaminants (forward database), and the reversed sequences of all proteins (reverse database). Carbamidomethylation was set as fixed modification. Variable modifications included oxidation (M), N-acetylation (protein), pyro (N-term QC). Full tryptic specificity was required, i.e. enzyme specificity was set to trypsin, allowing for cleavage N-terminal to proline and between aspartic acid and proline (12). Up to three missed cleavages and three labeled amino acids (arginine and lysine) were allowed. Initial mass deviation of precursor ion and fragment ions were up to 10 ppm and 0.5 Da, respectively. The minimum required peptide length was set to 6 amino acids. To pass statistical evaluation, posterior error probability for peptide identification (MS/MS spectra) must be below or equal to 0.1; the maximum false discovery rate of proteins was set at 0.01.This ensures that in the worst case each MS/MS spectrum has a maximal false identification probability of 0.1, and the overall peptide identification list contains maximal 1% false identification hits. Posterior error probability for peptides was calculated by recording Mascot score and peptide sequence length-dependent histograms of forward and reverse hits separately and then, using Bayes theorem, deriving the probability of a false identification for a given top scoring peptide. False discovery rate was calculated by successively including best scoring peptide hits until the list contained 1% reverse hits. For protein identification, two peptides were required, among which at least one peptide was required to be unique in the database. False discovery rate of proteins was the product of the posterior error probability of the contained peptides where only peptides with distinct sequences were taken into account. If a group of identified peptide sequences belongs to multiple proteins and these proteins cannot be distinguished, i.e. no unique peptide reported, these proteins are then reported as a protein group in MaxQuant. Proteins were quantified if at least one MaxQuant-quantifiable SILAC pair was present.
Quantitation is explained in detail in Ref. 15 and was based on two-dimensional centroid of the isotope clusters within each SILAC pair. Ratios of the corresponding isotope forms in the SILAC pair were calculated, and linear line fitting to these intensities ratios gave the slope as the desired peptide ratio. To represent the ratio of a peptide being quantified several times, the median value was chosen. To minimize the effect of outliers, protein ratios were calculated as the median of all SILAC pair ratios that belonged to peptides contained in this protein. The % variability of the quantitation (supplemental Tables 1–4) was defined as the standard deviation of the natural logarithm of all ratios used for obtaining the protein ratio multiplied by a constant factor 100.
Gene Ontology and KEGG Enrichment Analysis-based Hierarchical Clustering—
In the primary against cell-line study the quantified proteome was divided into five quantiles corresponding to percentage cutoffs of 0, 15%, 25%, 75%, 85%, and 100%. The enrichment analysis for gene ontology (GO) biological process and cellular component were done separately for these quantiles with respect to the whole quantified proteome by conditional hypergeometric test available in the GO stats package (16) in the R statistical environment (17). For hierarchical clustering we first collated all the categories obtained after enrichment along with their p values and then filtered for those categories which were at least enriched in one of the quantiles with p value < 0.05. Categories which did not have a p value after collation in any quantile were provided a very conservative p value of 1. This filtered p value matrix was transformed by the function x = − log10 (p value). Finally these x values were transformed to z-score for each GO category by using the transformation [x-mean(x)]/sd(x). These z-scores were then clustered by one-way hierarchical clustering using “Euclidean distance” as distance function and “Average Linkage Clustering” method available in Genesis (18). KEGG pathway enrichment analysis was done in the same way, except that the hypergeometric test was employed, and the reference set was complete mouse KEGG annotation.
RESULTS
Quantitative Analysis of Hepa1–6 Against Primary Hepatocytes—
To characterize phenotypic differences between mouse liver cell lines and primary cells, we SILAC-labeled (19, 20) a murine hepatoma cell line, Hepa1–6 (21), and compared its proteome to that of primary hepatocytes prepared according to standard operating procedures established by the German systems biology competence network HepatoSys (9) (Fig. 1). We used high resolution MS to identify 3,400 proteins while quantifying more than 3,350 of them (see “Experimental Procedures”, supplemental Fig. 1A, and supplemental Table 1). We repeated the experiment with hepatocytes from another mouse and obtained excellent quantitative reproducibility (Pearson correlation coefficient 0.95; supplemental Fig. 1B). We then combined the two datasets and analyzed them together using stringent and unified criteria. At a false positive rate of less than one percent, a total of 4,063 proteins were identified and quantified between the two cell populations. The primary and cell line proteomes overlap qualitatively but are very different quantitatively, with more than half of the proteome changing at least 2-fold between the two conditions (Fig. 2, A and B; supplemental Tables 1 and 2; see Data deposition notes for access to raw data). Many proteins are expressed at much lower levels in the immortalized cell line than in the primary cells whereas comparatively few were up-regulated in Hepa1–6. This is surprising because cancer cells are thought to be de-differentiated and to express many genes inappropriately.
Fig. 1.
Strategy for comparing primary cells with immortalized cell lines. Primary hepatocytes were isolated and grown for 14 h. The Hepa1–6 cell line was completely SILAC-labeled with 13C615N4-arginine and 13C615N2-lysine. Cell extracts were combined and by online high-resolution MS on a linear ion trap Fourier transform instrument (LTQ-FT) or LTQ-Orbitrap. The bottom part of the figure exemplifies peptides from proteins mostly expressed in primary cells, equally expressed in both or mostly expressed in Hepa1–6.
Fig. 2.
Fold-change distributions of the proteome. A, quantitative comparison of the primary against the Hepa1–6 cell line proteome. The distribution was divided into five quantiles as follows. High relative expression in primary cells (0–15%, at least 4-fold down-regulation), mostly expressed in primary cells (15–25%, −4 to −1.5-fold regulation), not highly regulated proteins (25–75%; −1.5 to +2.8), mostly expressed in Hepa1–6 (75–85; 2.8 to 3.6-fold), highly expressed in Hepa1–6 (85–100%, more than 3.6-fold change). Color coding of these categories is indicated at the top of the panel. B, biological replicate of the experiment showing excellent reproducibility (see also supplemental Fig. 2). C, quantitative comparison of the Hepa1–6 proteome cultured in high glucose (28 nm) and physiological glucose concentration (8 nm).
It is usually recommended to cultivate Hepa1–6 cells in high glucose medium (38 mm). Therefore we asked whether some of the observed phenotypic changes were attributable to this circumstance. To address this experimentally, we performed another SILAC experiment comparing cells cultured in high glucose against physiological glucose levels in mice (8 mm) for 3 weeks (Fig. 2C and supplemental Table 3). In this experiment, there were hardly any overall changes in the proteome, and 96% of the proteins were of constant abundance within a factor of two. This was also confirmed in a replicate experiment (supplemental Fig. 2 and supplemental Table 4). These results rule out a dominant role of the superphysiological glucose level in the proteome differences between primary cells and cell lines. Furthermore, they demonstrate excellent quantitative accuracy of our experiment on a proteome-wide basis.
A Method for Proteomic Phenotyping—
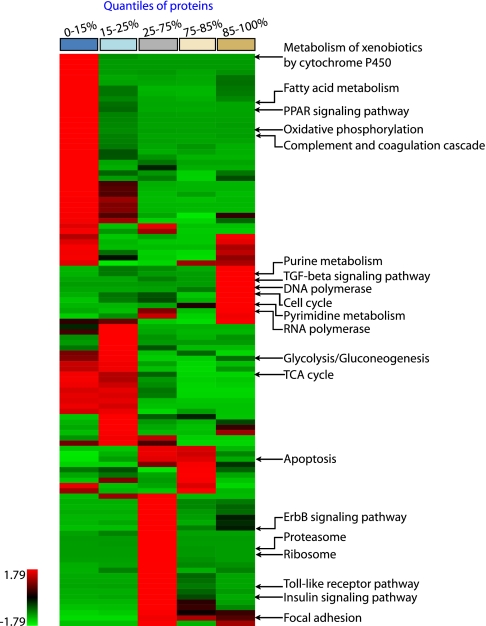
To functionally understand the differences between the two cell populations, we divided the fold-change distribution between primary hepatocytes and the Hepa1–6 cell line into five quantiles according to relative protein expression (Fig. 2, A and B). Each quantile was assessed separately for over-represented pathways, biological processes, and cellular components with GO and KEGG pathway analysis (22, 23) (see “Experimental Procedures”). We retained each functional category that reached at least 95% statistical significance in one of the quantiles and then performed one-way unsupervised clustering of the p values of the resulting categories (Fig. 3, supplemental Fig. 3, and supplemental Table 5). This analysis differs from the more familiar clustering of over-represented genes themselves, which is frequently employed in microarray-based experiments. It integrates the strength of statistical testing (taking p values as input for clustering) with the intuitive simplicity of hierarchical clustering. By automatically classifying related processes and pathways based on their up or down-regulated protein measurements, it provides an unbiased global portrait of representative biological functions, enabling visual interpretation of the phenotype in terms of aggregate functional modules on a systems level. We verified the robustness of these functional assignments by comparing the shared p value matrix of the replicate experiments against each other (supplemental Fig. 4). This correlation was 0.86 for KEGG, 0.85 for GO biological process, and 0.92 for GO cellular compartment.
Fig. 3.
Functional phenotyping of the proteome. The quantiles resulting from quantitative proteome comparison in Fig. 2 were separately analyzed for enriched KEGG pathways and clustered for the z-transformed p values. The color bar on top represents the quantiles. Representative pathways enriched in the protein population of each quantile are annotated. For complete listing of significant categories see supplemental Fig. 3.
Proteomic Differences between Hepa1–6 and Primary Hepatocytes—
The most prominent cluster of proteins expressed at higher levels in Hepa1–6 relates to cell division and encompasses categories such as cell cycle (p < 10−9) (supplemental Fig. 5), DNA synthesis (p < 10−4), and RNA polymerase (p < 10−3). This cluster consists of 10 enriched pathways, of which at least five relate to increased cell proliferation. Biologically, this is not surprising because hepatocytes in the liver and in our primary culture are largely arrested in the GO phase of the cell cycle, whereas Hepa1–6 cells double every 18 h. Nevertheless, the fact that this phenotypic trait is so clearly grouped in the cluster analysis makes it an excellent positive control.
One of the most enriched categories in the quantile most expressed in primary cells is the P450 family of enzymes (p < 10−16). These enzymes are mainly involved in metabolizing endogenous substances and xenobiotics (24), a prototypical function of the liver. We identified 32 different P450 proteins, and 25 of them were down-regulated at least 10-fold in the cell line. Furthermore, the flavin monooxygenase (FMO), UDP-glucuronosyltransferase (UGT), sulfotransferase (SULT), and glutathione S-transferase (GST), additional prominent drug metabolizing enzyme families (DMEs), were also severely down-regulated in Hepa1–6 (supplemental Table 6). Only three P450s were up-regulated. Two of them (CYP1A1 and CYP2S1) are known to be regulated by the aryl hydrogen receptor (25, 26). This receptor was also more highly expressed in Hepa1–6, providing a ready explanation for the up-regulation. The third up-regulated P450 protein (RIKEN clone E130013F06) has only been characterized on the basis of sequence homology and may have functions different from traditional P450 enzymes.
Reduction of DME activity is a notorious difficulty in toxicological assays in cell lines. Toxicologists therefore attempt to stimulate liver cell lines with the aim of boosting DME activity (27). Quantitative knowledge of the changes in the profile of DME could provide a rational basis to adapt cell systems to more closely mimic hepatocytes in vivo.
Another prominent and cell-specific function of hepatocytes is production of plasma proteins. Fig. 3 reveals that “complement and coagulation cascade” is specific for the primary cells (p < 10−2). Inspection of the pathway involved (supplemental Fig. 5) shows that major liver-produced factors, such as C3, C4, MBP-C, F2, F5, A2M, Serpin A1/C1 and apolipoproteins are down-regulated more than 5-fold in Hepa1–6. Thus, loss of tissue context allows the cell line to shut down this function, which is nonessential for propagation in culture.
The cellular compartments most over-represented in the primary cells are mitochondria (p < 10−62) (Fig. 4A) and extracellular matrix (p < 10−18) (supplemental Fig. 3). Apparently, the cell line under-expresses proteins related to communication with stroma and with tissue maintenance. Our proteome contained a total of 452 proteins annotated as mitochondrial in GO. Of these, 67% were in the asymmetric tail of the distribution, indicating they were expressed several-fold lower in Hepa1–6 cells than in primary hepatocytes. We independently confirmed this observation by 4′,6-diamidino-2-phenylindole (DAPI) and Mitotracker staining (Fig. 4B). Indeed, primary hepatocyte nuclei were smaller whereas in these cells mitochondria were more abundant with respect to Hepa1–6. Concurrent with this, fatty acid metabolism was drastically down-regulated according to enrichment analysis of KEGG pathways (Fig. 5A). Likewise, “oxidative phosphorylation” (p < 10−29), “urea cycle” (p < 10−4), and “steroid biosynthesis” (p < 10−2) were statistically significantly enriched in the quantile most expressed in primary hepatocytes. These down-regulated metabolic functions at least partially take place in mitochondria. Conversely, parts of the glycolysis pathway were up-regulated in Hepa1–6 (supplemental Fig. 5). Together, our results portray a drastic metabolic rearrangement, away from oxidative metabolism in the mitochondria and toward less efficient anerobic metabolism. These findings provide evidence for the Warburg hypothesis, that cancer cells shift toward glycolytic metabolic pathways (28, 29).
Fig. 4.
Phenotypic proteome comparison at the cellular component level. A, the quantiles resulting from quantitative proteome comparison in Fig. 2 were separately analyzed for enriched gene ontology cellular components and clustered for the z-transformed p values. The color bar on top represents the quantiles. Representative categories enriched in the protein population of each quantile are annotated. For complete listing of significant categories and p values, see supplemental Fig. 3. Prominent mitochondria-related categories for the primary cells are highlighted in red and prominent nucleus related categories in blue. B, nuclear (DAPI) and mitochondrial (Mitotracker) staining of primary hepatocytes and Hepa1–6 cells. Most primary hepatocytes are binuclear (34). Magnification factor is ×400. DAPI, 4′,6-diamidino-2-phenylindole.
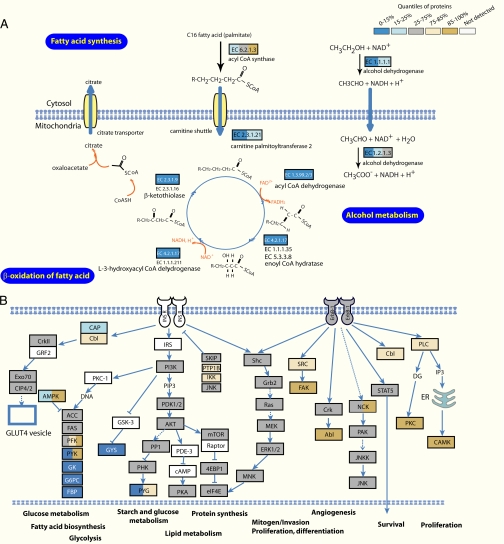
Fig. 5.
Phenotypic proteome comparison at the pathway level. A, mapping of protein ratios on the fatty acid metabolism pathway reveals that almost the entire module is down-regulated several-fold in Hepa1–6. Proteins are color-coded according to their relative expression in the two cell types according to the scheme in Fig. 2. B, KEGG pathway mapping of ErbB and PI3K signaling pathway shows that they are equally present in primary cells and the cell line. PI3K, phosphatidylinositol 3-kinase.
In the category containing the 50% of proteins with the least change, many household functions and organelles including ribosome (p < 10−2), proteasome (p < 10−3), splicing (p < 10−4), and Golgi apparatus (p < 10−3) are significantly enriched. Interestingly, several signaling pathways are also preferentially located in this quantile. These include the ErbB and phosphatidylinositol 3-kinase signaling pathways (Fig. 5B). This finding is in agreement with the requirement of growth factor containing serum for the maintenance of most cell lines. Conversely, TGFβ-mediated signaling is higher represented in the Hepa1–6 cell line and the canonical members TGFβ R1, Smad2/3, Smad4, p107, and p15 are all up-regulated significantly (supplemental Fig. 5). This was unexpected because TGFβ is usually associated with growth inhibition whereas Hepa1–6 has increased proliferation rate compared with primary hepatocytes. However, the biological actions of TGFβ are complex and it is thought to shift from a growth inhibitory to a growth-promoting role during cancer development (30). Thus up-regulation of this pathway suggests that in the Hepa1–6 tumor cells, TGFβ may have growth promoting effects. Taken together, our data indicate that biological functions related to many important signaling pathways are well preserved in Hepa1–6.
Some categories shared by both cell types represent non-liver functions (such as “long term potentiation”) or even non-animal functions (such as “CO2 fixation”). However, the enzymes found in these categories function both in liver tissue as well as in neurons or plants. Therefore, over-representation of these categories reflects the still evolving state of annotation of pathway databases rather than a limitation of our technology.
DISCUSSION
Taking advantage of the ability of SILAC to compare the levels of thousands of proteins in different cellular states (13, 14) and a novel bioinformatic approach, we have, for the first time, compared the proteomes of primary cells to cell lines. The overall picture that emerges is that Hepa1–6 has lost many of the specific functions typical of hepatocytes in vivo. Examples are the DMEs, complement production, and synthesis of extracellular matrix. Conversely, the cell line shifts more of its resources into functions associated with proliferation, but maintains important cell signaling pathways. This phenotype is “functionally beneficial” for rapidly dividing and not nutrient limited cells and may partly reflect Darwinian selection of cell clones. Note that we have only analyzed a specific time point of primary cells and a specific cell type. Although we believe many of the findings will be general for different methods to prepare primary cells and for different cell types, this will need to be established in the future.
Our technology is accurate, relatively rapid and should now allow selection of the appropriate cell system based on a global and unbiased profile according to desired biological function. Furthermore, it can be used to manipulate the cell line system to better reflect the in vivo situation at the proteome level. Although we have based our analysis on protein expression levels, it could just as well be applied to assess fidelity of signaling pathways in cell lines using SILAC-based quantitative and global phosphoproteomics (31). This work complements other approaches dedicated to infer pathway information from quantitative proteomics data (32).
Our analysis differs in important points from the more familiar measurement of mRNA levels by microarray and its associated bioinformatics (33). Even though reproducibility of microarray chips has become much better during recent years, the data is not quantitative with respect to the final, desired parameter, the global change in protein levels. Furthermore, results of any specific transcript on the chip generally have to be validated by RT-PCR and then by quantitative immunoblotting. This is impractical for large numbers of proteins. In contrast, quantitative proteomics inherently contains the fold-change for each protein and increasingly also that of specific isoforms. The quantitative nature of our results also made it possible to directly group over-represented functions and processes instead of the genes themselves.
Here we have analyzed interesting but relatively general phenotypic traits of two cell populations. Although many of the resulting observations can be immediately rationalized in terms of biological function, they have never been quantified in a global and unbiased way. Our data furthermore contains a wealth of functional leads that could not be explored in depth here. The combination of very high quantitative accuracy at the proteome level with increasingly accurate pathway databases should allow even richer assessment of the phenotypic state of any cell population in the future.
Supplementary Material
Acknowledgments
We thank other members of the department for proteomics and signal transduction, the department of molecular medicine, and the division of systems biology of signal transduction for sharing insights. We also thank Tobias Walther for critical reading and help with the manuscript.
Footnotes
Published, MCP Papers in Press, October 23, 2008, DOI 10.1074/mcp.M800258-MCP200
The abbreviations used are: MS, mass spectrometry; MS/MS, tandem mass spectrometry; LC, liquid chromatography; FT, Fourier transform; KEGG, Kyoto encyclopedia of genes and genomes; GO, gene ontology; SILAC, stable isotope labeling by amino acids in cell culture; DMEs, drug metabolizing enzyme families.
This work was supported by HepatoSys, by Interaction Proteome, a 6th framework program of the European Union, and by the Max-Planck Society.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Freshney, R. I. ( 2005) Culture of Animal Cells: A Manual of Basic Technique, 5th Ed., Wiley
- 2.Masters, J. R. ( 2002) HeLa cells 50 years on: the good, the bad and the ugly. Nat. Rev. Cancer 2, 315–319 [DOI] [PubMed] [Google Scholar]
- 3.Masters, J. R. ( 2000) Human cancer cell lines: fact and fantasy. Nat. Rev. Mol. Cell. Biol. 1, 233–236 [DOI] [PubMed] [Google Scholar]
- 4.Burdall, S. E., Hanby, A. M., Lansdown, M. R., and Speirs, V. ( 2003) Breast cancer cell lines: friend or foe? Breast Cancer Res. 5, 89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nolan, G. P. ( 2007) What's wrong with drug screening today. Nat. Chem. Biol. 3, 187–191 [DOI] [PubMed] [Google Scholar]
- 6.Kamb, A. ( 2005) What's wrong with our cancer models? Nat. Rev. Drug Discov. 4, 161–165 [DOI] [PubMed] [Google Scholar]
- 7.Sandberg, R., and Ernberg, I. ( 2005) The molecular portrait of in vitro growth by meta-analysis of gene-expression profiles. Genome Biol. 6, R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsavsky, K. M., Page, J. L., Johnson, M. C., Zarbl, H., Strom, S. C., and Omiecinski, C. J. ( 2007) Gene expression profiling and differentiation assessment in primary human hepatocyte cultures, established hepatoma cell lines, and human liver tissues. Toxicol. Appl. Pharmacol. 222, 42–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klingmuller, U., Bauer, A., Bohl, S., Nickel, P. J., Breitkopf, K., Dooley, S., Zellmer, S., Kern, C., Merfort, I., Sparna, T., Donauer, J., Walz, G., Geyer, M., Kreutz, C., Hermes, M., Gotschel, F., Hecht, A., Walter, D., Egger, L., Neubert, K., Borner, C., Brulport, M., Schormann, W., Sauer, C., Baumann, F., Preiss, R., MacNelly, S., Godoy, P., Wiercinska, E., Ciuclan, L., Edelmann, J., Zeilinger, K., Heinrich, M., Zanger, U. M., Gebhardt, R., Maiwald, T., Heinrich, R., Timmer, J., von Weizsacker, F., and Hengstler, J. G. ( 2006) Primary mouse hepatocytes for systems biology approaches: a standardized in vitro system for modeling of signal transduction pathways. Syst. Biol. ( Stevenage) 153, 433–447 [DOI] [PubMed] [Google Scholar]
- 10.Blagoev, B., Ong, S. E., Kratchmarova, I., and Mann, M. ( 2004) Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 11.Rappsilber, J., Ishihama, Y., and Mann, M. ( 2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 12.Olsen, J. V., Ong, S. E., and Mann, M. ( 2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics 3, 608–614 [DOI] [PubMed] [Google Scholar]
- 13.Cox, J., and Mann, M. ( 2007) Is proteomics the new genomics? Cell 130, 395–398 [DOI] [PubMed] [Google Scholar]
- 14.Graumann, J., Hubner, N. C., Kim, J. B., Ko, K., Moser, M., Kumar, C., Cox, J., Schoeler, H., and Mann, M. ( 2008) SILAC-labeling and proteome quantitation of mouse embryonic stem cells to a depth of 5111 proteins. Mol. Cell. Proteomics 7, 672–683 [DOI] [PubMed] [Google Scholar]
- 15.Cox, J., and Mann, M. ( 2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 16.Falcon, S., and Gentleman, R. ( 2007) Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 [DOI] [PubMed] [Google Scholar]
- 17.Team, R. D. C. ( 2008) R: a language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria
- 18.Sturn, A., Quackenbush, J., and Trajanoski, Z. ( 2002) Genesis: cluster analysis of microarray data. Bioinformatics 18, 207–208 [DOI] [PubMed] [Google Scholar]
- 19.Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A., and Mann, M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 20.Mann, M. ( 2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell. Biol. 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 21.Darlington, G. J., Bernhard, H. P., Miller, R. A., and Ruddle, F. H. ( 1980) Expression of liver phenotypes in cultured mouse hepatoma cells. J. Natl. Cancer Inst. 64, 809–819 [PubMed] [Google Scholar]
- 22.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. ( 2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. ( 2004) The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aitken, A. E., Richardson, T. A., and Morgan, E. T. ( 2006) Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu. Rev. Pharmacol. Toxicol. 46, 123–149 [DOI] [PubMed] [Google Scholar]
- 25.Kawajiri, K., and Fujii-Kuriyama, Y. ( 2007) Cytochrome P450 gene regulation and physiological functions mediated by the aryl hydrocarbon receptor. Arch. Biochem. Biophys. 464, 207–212 [DOI] [PubMed] [Google Scholar]
- 26.Rivera, S. P., Saarikoski, S. T., and Hankinson, O. ( 2002) Identification of a novel dioxin-inducible cytochrome P450. Mol. Pharmacol. 61, 255–259 [DOI] [PubMed] [Google Scholar]
- 27.Hewitt, N. J., Lechon, M. J., Houston, J. B., Hallifax, D., Brown, H. S., Maurel, P., Kenna, J. G., Gustavsson, L., Lohmann, C., Skonberg, C., Guillouzo, A., Tuschl, G., Li, A. P., LeCluyse, E., Groothuis, G. M., and Hengstler, J. G. ( 2007) Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab. Rev. 39, 159–234 [DOI] [PubMed] [Google Scholar]
- 28.Warburg, O., Posener, K., and Negelein, E. ( 1930) Ueber den Stoffwechsel der Tumoren. Biochemische Z. 152, 319–344 [Google Scholar]
- 29.Hsu, P. P., and Sabatini, D. M. ( 2008) Cancer cell metabolism: warburg and beyond. Cell 134, 703–707 [DOI] [PubMed] [Google Scholar]
- 30.Jakowlew, S. B. ( 2006) Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev. 25, 435–457 [DOI] [PubMed] [Google Scholar]
- 31.Olsen, J. V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P., and Mann, M. ( 2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 32.Zubarev, R. A., Nielsen, M. L., Fung, E. M., Savitski, M. M., Kel-Margoulis, O., Wingender, E., and Kel, A. ( 2008) Identification of dominant signaling pathways from proteomics expression data. J. Proteomics 71, 89–96 [DOI] [PubMed] [Google Scholar]
- 33.Butte, A. ( 2002) The use and analysis of microarray data. Nat. Rev. Drug Discov. 1, 951–960 [DOI] [PubMed] [Google Scholar]
- 34.Guidotti, J. E., Bregerie, O., Robert, A., Debey, P., Brechot, C., and Desdouets, C. ( 2003) Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J. Biol. Chem. 278, 19095–19101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.