Abstract
We integrated five sets of proteomics data profiling the constituents of cerebrospinal fluid (CSF) derived from Huntington disease (HD)-affected and -unaffected individuals with genomics data profiling various human and mouse tissues, including the human HD brain. Based on an integrated analysis, we found that brain-specific proteins are 1.8 times more likely to be observed in CSF than in plasma, that brain-specific proteins tend to decrease in HD CSF compared with unaffected CSF, and that 81% of brain-specific proteins have quantitative changes concordant with transcriptional changes identified in different regions of HD brain. The proteins found to increase in HD CSF tend to be liver-associated. These protein changes are consistent with neurodegeneration, microgliosis, and astrocytosis known to occur in HD. We also discuss concordance between laboratories and find that ratios of individual proteins can vary greatly, but the overall trends with respect to brain or liver specificity were consistent. Concordance is highest between the two laboratories observing the largest numbers of proteins.
Huntington disease (HD)1 is an inherited neurodegenerative disorder characterized by progressive cognitive decline and psychiatric and movement symptoms. The cause of the disease is the expansion of trinucleotide (CAG) repeats in the coding region of the htt gene that translates into a polyglutamine tract in the huntingtin protein (1). Currently no treatment has been shown to delay the onset of the disease or slow its progression in patients. To speed assessment of therapies in clinical trials, it is critical to identify biological markers that can accurately monitor disease progression.
Several genomics and proteomics approaches to identifying biomarkers for HD have been undertaken previously. Genomics studies have determined the molecular phenotype of human HD brain (2) and different tissues of HD mouse models at the mRNA level (3–6). Proteomics approaches have been applied to brain tissues of HD mouse models and humans to identify candidate markers (7–9). Blood plasma in particular has received considerable attention recently because of its ready accessibility clinically (10, 11). The candidate protein biomarkers identified in the blood proteomics studies are largely known inflammatory markers. Because HD is regarded primarily as a neurodegenerative disease, it is not entirely clear how directly general markers of neuroinflammation relate to the pathophysiology of HD, although astrocytosis and microgliosis (12) are prominent components of HD in its mid- to late stages (13). Another concern regarding markers discovered primarily in blood is that the blood-brain barrier may restrict brain proteins from entering plasma, and so plasma candidates may not directly reflect HD progression in the brain.
Cerebrospinal fluid (CSF) is a more relevant biomaterial for biomarker discovery because it is proximal to the brain; it occupies the subarachnoid space of the central nervous system and the ventricular system around and inside the brain. Changes in CSF proteins have been identified for several diseases (14–17), and oligoclonal bands in CSF have long been used to aid in diagnosis of multiple sclerosis and encephalitis (18–20). CSF is an ultrafiltrate of arterial blood produced by the choroid plexus in the lateral, third, and fourth ventricles. However, it has been estimated that about 20% of the proteins in CSF are derived from brain (21), making CSF an attractive source of potential disease biomarkers in neurodegenerative diseases such as Alzheimer and Parkinson diseases (16, 22, 23). We report here an integrated proteomics approach to characterize the constituents of CSF and identify potential markers in CSF for human Huntington disease.
In this study, we analyzed and interpreted human HD CSF proteomics data generated by four laboratories using different proteomics approaches, including separation strategies, pooling strategies, depletion of proteins, quantitation methods, and mass spectrometry instruments. Although acquired using different biochemical approaches, all data were interpreted using a common protein database, algorithms for database search (24), and peptide and protein identification (25, 26) and quantitation (27) methods to allow comparison across laboratories.
The preplanned primary analysis of these data includes deriving rankings for protein changes in HD based on the synthesized data from all laboratories and then assessing biological and statistical significance by interrogating the rankings with gene annotations derived from independent data sets (e.g. gene set enrichment style analyses (28)). Annotations include the tissue specificity of a gene (e.g. brain or liver) and whether a gene is significantly changed in human HD brain when compared with non-HD brain, both derived from previously published data sets profiling the transcripts of normal human tissues (29) and human HD and non-HD brains (2).
This analysis reveals that proteins that have specifically high expression in the brain (brain-specific) are 1.8 times more enriched in CSF than in human plasma. These brain-specific proteins overall have lower concentrations in HD than normal CSF, and 81% of them are concordant with previously identified mRNA changes in HD versus normal brain. Altogether these results suggest that measuring proteins in CSF may be a useful way to assess the health of the brain, track progression of the disease, and improve our understanding of the disease.
Secondary analysis was also performed to investigate the concordance of protein changes across laboratories. Overall at the protein (e.g. International Protein Index (IPI) sequence) level, there is a low concordance of the disease/control ratios among laboratories, meaning that each laboratory would report different highest (or lowest) ranking proteins. However, the laboratories are consistent with respect to the overall trends that brain-specific proteins decline and liver-specific proteins increase in HD samples. Concordance in protein ratios and overall trends is greatest between the two laboratories identifying the highest number of proteins. This supports an argument that future studies with resource limitations should emphasize the depth of protein coverage and include multiple laboratories only if their experimental methods complement each other in terms of the protein observations (30). Our study also suggests that data integration plays a central role in studies to identify biomarkers as statistical significance could not have been demonstrated without the ability to evaluate changes in predefined groups of proteins using the gene set enrichment analysis (GSEA) approach.
EXPERIMENTAL PROCEDURES
Sample Collection
CSF samples were collected and processed by a single study site described previously (10). Briefly CSF was obtained by lumbar puncture from 20 HD gene-positive patients and 10 gene-negative controls with matched ages recruited through the University of British Columbia HD Medical Clinic. On the day of the lumbar puncture, all subjects had a comprehensive clinical evaluation including assessment on the complete United Huntington Disease Rating Scale. Based on the United Huntington Disease Rating Scale independence score as defined by the Huntington Study Group (31), 10 of the 20 gene-positive individuals were categorized as early stage (independence score >80), and the other 10 were categorized as moderate stage (65 < independence score ≤ 80). About 5–7 ml of CSF samples were collected in four or five standard lumbar puncture kit tubes (CardinalHealth, safe-t-LP kit) (catalog number 4301CSDF). Each collected sample was placed on ice and then centrifuged at 2000 × g (4000 rpm) for 10 min to eliminate cells and other insoluble material. The collected CSF was examined by microscopy, aliquoted, and frozen immediately on dry ice in polypropylene tubes in 1- or 3-ml aliquots and stored at −80 °C. Tubes were filled to the top to minimize oxidation during storage. Average total processing time was 76 min from the start of collection to final storage. No anticoagulants, preservatives, and protease inhibitors were added. The lumbar punctures were atraumatic with CSF cell counts revealing red blood cells from 0 to 171 counts/μl and white blood cells from 0 to 17 counts/μl, indicating no significant blood cell contamination (supplemental Table S1). Samples were stored at −80 °C for various lengths of time ranging from 17 to 27 months before they were thawed and subdivided into aliquots of 0.5 ml to be shipped using dry ice to individual labs for analysis. The duration of shipment was between 1 and 3 days, and dry ice was replenished during the shipment to keep samples frozen. All 30 samples were stored at −80 °C before thawing once again for analysis. Therefore, except one (HDU-2) that was thawed three times, all samples were thawed twice before analysis.
Proteomics Platforms
Five different laboratories received aliquots of CSF collected from the same 30 individuals described above. The disease statuses of the 10 HD gene-negative controls, 10 HD gene-positive early stage samples, and 10 HD gene-positive mid-stage samples were blinded to the research laboratories and labeled as group B, C, and A, respectively, with identifiers provided to labs only after raw data from all labs were received by the bioinformatics data core analysis group.
Each lab designed experiments based on their preferred comprehensive proteomics platform(s) (Table I) for the purpose of discovering biomarkers that can classify Huntington disease. These approaches include four quantitative mass spectrometry approaches and one gel-based quantitative approach that used the mass spectrometer for protein identification. Experimental designs varied in many respects across the labs, including the use of pooled and non-pooled designs, depletion and non-depletion of abundant proteins in the samples, and label-free and isotopically labeled quantitation. After evaluation, data from four laboratories were reported in this study. One of the five laboratories reported quality control issues that were also detected in the data analyses (fewer than 80 total peptides were identified), and therefore this data set was not considered for further analysis. Detailed experimental designs of the other four laboratories are described in the supplemental text, Part A.
Table I.
Experimental designs of five proteomics laboratories
1D, one-dimensional; 2D, two-dimensional; iTRAQ, isobaric tags for relative and absolute quantitation.
| Group | Mass spectrometer | Quantitative method | Pooling | Immunodepletion | Separation |
|---|---|---|---|---|---|
| Lab 1 | HCT-Ultra | DIGE | No | No | 2D gels |
| Lab 2 | Q-ToF Micro | Label-free (spectral counts) | No | IgY-12 High Capacity LC2 | 1D SDS-PAGE |
| Lab 3 | LTQ-FT | d0/d3 acetylation | Yes | ProteoExtract Albumin/IgG Removal kit | 1D SDS-PAGE |
| Lab 4 | LTQ OrbiTrap XL | Acrylamide labeling and label-free (AMT) | Yes | Multiple Affinity Removal System | Anion exchange and reverse phase chromatography |
| Lab 5a | LC-MALDI-TOF/TOF | iTRAQ | Yes | Multiple Affinity Removal System | 1D SDS-PAGE |
Data from Lab 5 was dropped prior to data synthesis across labs because of quality control issues (see “Experimental Procedures”).
Individual Lab Data Processing and Analysis
All data files were transmitted to the bioinformatics data-processing lab for analysis after converting to the standard mzXML format (32, 33) and then searched with X! Tandem (January 1, 2007 release) configured with a scoring function (24) compatible with PeptideProphet (25) and ProteinProphet (26). The same database (human IPI version 3.20 consisting of 61,225 IPI numbers) was used to search all data. PeptideProphet (version 3.0) and ProteinProphet (version 3.0) were used to assign identified peptide and protein confidence scores for all but Lab 1 (details described below) where the complexity of samples interrogated was not sufficient to estimate the PeptideProphet error model. Quantitation was performed following tryptic search and protein inference. Finally multiple experiments within a lab were then aligned to create analytic data sets sufficient to determine relative disease status.
The following common criteria were applied to all searches. A ±2.0-Da error from the calculated peptide monoisotopic mass was allowed to determine whether a particular peptide sequence is to be considered as a possible model for a spectrum. Mass tolerance for fragment ions was 1 Da (24). The maximal number of missed cleavages permitted was 2. A static modification on cysteines of +57.021 Da was used for all labs except Lab 4, which performed acrylamide labeling on cysteines. A potential modification on methionines of +15.9949 Da was used. A weighted average mass was used to calculate the masses of the fragment ions in a tandem mass spectrum. A minimum number of one ion was required for a peptide to be scored. A default minimum PeptideProphet probability of 0.2 was used to calculate the protein group probability. Only peptides with probability ≥0.75 and mass error <20 ppm were selected for quantitation. Specific search parameters for the different labeling schemes by different labs are specified in the supplemental text, Part B. The following are descriptions of the software used for each lab's data and quantitation methods (summarized in supplemental Table S2) during data processing.
Lab 1—
Lab 1 used a DIGE method and quantitated a large number of fluorescent spots with two commercial software algorithms (DeCyder 6.5 (GE Healthcare) and SameSpots 2.0 (Nonlinear Dynamics)). A t test was performed on the log ratios using Statistica for Windows (StatSoft, Inc.) version 7 to estimate the significant difference of a protein. Only those spots found to have significant changes, based on fluorescence, between the HD and the control were selected for tandem MS analysis. Peak lists of MS/MS spectra acquired on the HCT-Ultra ion trap instrument (Bruker Daltonics) were generated using the software tool DataAnalysis 3.4.179 (Bruker Daltonics) with default parameters. The built-in algorithm version 2.0 was used, and neither smoothing nor any signal-to-noise filter was applied for compound detection. A maximum charge state of 3 was considered for deconvolution. Data were then converted to mzXML files using CompassXport (version 1.2.3). Peptides were identified using decoy database methods with an approach described by Elias and Gygi (34). After proteins were identified for selected spots using false discovery rate (FDR) and ProteinProphet (see details in the supplemental text, Part B), quantitation of -fold changes among different disease statuses was processed based on the following rules. 1) If a spot was quantitated by both DeCyder 6.5 and SameSpots methods, -fold changes by DeCyder 6.5 were selected because the differential expression resulting from this method is more significant on average. On the other hand, if only one quantitation method was used, results from that method were selected. 2) When multiple spots have the same protein identification, -fold changes were averaged for that protein. 3) When a spot resulted in several protein identifications, the same -fold changes were assigned to all proteins.
Lab 2—
Lab 2 performed label-free analysis. Peak lists were generated using MassLynx (4.0) based on signals obtained by Q-ToF Micro spectrometer from Waters Micromass. The MS duty cycle was set at 1,1,4 as described in detail in the supplemental text, Part A. These data were converted to mzXML files using MassWolf 1.02 with the Waters Datafile Access Component (DAC) library. Following the database search using the common criteria, quantitation was performed using a spectral count approach (35), which sums the number of total spectra assigned to the protein group in that sample. Only peptide spectra with PeptideProphet probability greater than 0.75 or an error rate of 5% were counted for each IPI entry identified. Because individual level variation can be determined for this design, we used a straightforward procedure that tracks all proteins that are members of a single ProteinProphet group within the experiment to associate groups across multiple experiments and to flag groups that are not directly comparable (supplemental text, Part B). After summing up to master protein groups across samples, the average spectral count of all IPI entries within a “master group” was assigned to that group. For each master group, total spectral counts from 10 HD-mid samples, 10 HD-early samples, and 10 control samples were summed up and used to calculate HD-mid/control, HD-early/control, and HD-mid/HD-early ratios. Intensity-dependent ratio plots (MA plots) and histograms of light/heavy ratios were examined to ensure the quality of data for the labeled experiments. M is the y-axis and A is the x-axis, where in this paper, M = Log2 (Heavy) − Log2 (Light); A = [½] (Log2 (Heavy) + Log2 (Light)).
Lab 3—
Data from Lab 3 were acquired from the LTQ-FT instrument (Thermo Finnigan). Peak lists were generated by Xcalibur (version 1.1) and converted to mzXML files using ReAdW 1.1 with XRawfile library. Default parameters were used. Database search was carried out using the common parameters and the designated specific modifications (supplemental text, Part B). Protein groups with probability score >0.9 (corresponding to an overall error rate of 0.01) were considered confident proteins for downstream analysis. The Q3 algorithm (27), developed to accommodate a 3-dalton mass shift in heavy and light peptides, was used to compute the ratios between the light and heavy isotopic pairs using peak areas. More specifically, only confidently identified peptides (PeptideProphet probability >0.75 and mass error <20 ppm) were selected for further quantitation at the protein level. In three pairwise comparisons, the internal standard (IS) containing equal amounts of 30 samples was labeled with light acetyl group, and each of the three disease statuses (A, B, or C) was labeled with heavy acetyl group (for details, see supplemental text, Part A). Preliminary analysis found that light/heavy ratios were skewed for both IS versus HD-early and IS versus HD-mid experiments. Because the same amount of protein was loaded into the MS instrument and, in theory, most proteins in the disease and control should remain unchanged, we normalized these ratios. Normalization at the peptide level was performed by median centering the log ratios. Experiments were then aligned to infer the protein changes of different disease status comparisons. Protein inference was performed using the ProteinProphet analysis tool at the lab level, and light/heavy ratios of these protein groups were inferred from ratios at the experiment level by comparing IPI numbers. HD-mid versus control, HD-early versus control, and HD-mid versus HD-early ratios for each protein group were calculated using IS/control, IS/HD-mid, and IS/HD-early ratios from the three experiments.
Lab 4—
LTQ OrbiTrap XL mass spectrometer from Thermo Finnigan was used, and Xcalibur (version 2.2) was applied to generate peak lists. Data were converted to mzXML files using ReAdW 1.1 with XRawfile library. Default parameters were used. Data were then searched with the common criteria plus specific modifications. Protein groups with probability score >0.9 (corresponding to an overall error rate of 0.01) were considered confident proteins for downstream analysis. For the labeled analysis, as with Lab 3, the Q3 algorithm (27) was used to compute ratios between light and heavy isotopic pairs. And similarly to the methods used for Lab 3, peptides with PeptideProphet scores greater than 0.75 and a mass error of less than 20 ppm were selected for the protein level quantitation. Histograms of light/heavy ratios based on the number of cysteines in the peptides revealed that peptides with one cysteine have better normal distributions than peptides with more than one cysteine. Because 80% of peptides contain only one cysteine, only peptides with one cysteine were selected for protein level analysis. To ensure a high confidence of quantitation at the protein level, only those proteins with at least three quantitated peptides were used for further analysis. For the label-free analysis, accurate mass and time (AMT) methods were used to identify peptides in LC-MS data using a single AMT database containing all high quality (PeptideProphet probability ≥0.95) peptide identifications from all labeled (fractionated) and unlabeled (unfractionated) data from Lab 4. The LC-MS peptide features from each unlabeled sample were matched against the combined AMT database to provide peptide assignments. Each match was assigned a probability value based on mass error and normalized retention time error between the MS1 feature and the AMT peptide entry, and only matches with probability ≥0.95 or a false assignment rate ≤0.05 were kept. LC-MS peptide intensity values for the peptides were normalized across runs, and peptide ratios were calculated. The AMT database and matching were performed using the msInspect/AMT software platform (36, 37). Protein inference was performed using ProteinProphet, and protein ratios were calculated as well. As with the Lab 3 analysis, experiments were aligned using the ProteinProphet analysis tool at the lab level to infer the protein changes of different disease status comparisons for the labeled and unlabeled methods. Ratios of these protein groups were inferred from ratios at the experiment level by IPI numbers.
Gene Name and Group Assignments
Proteins, identified by their IPI sequence, were assigned to gene symbols by IPI protein cross-reference. Because the peptide level evidence cannot uniquely identify all IPI sequences, proteins were assembled by ProteinProphet into groups (26). Some of these protein groups contain unique protein sequences and gene symbols, and some contain multiple sequences that may result from the same gene symbol or from multiple genes within the same family (e.g. a protein group is assigned with HBG2, HBE1, HBG1, HBB, and HBD, all of which belong to the hemoglobin gene family) or multiple incompatible genes.
Deriving a List of Consensus Proteins
To facilitate comparison across labs, a comprehensive list of protein groups consisting of all proteomics data reported in this study was generated by running ProteinProphet on all data. A minimum probability of 0.9 was used to generate the confident protein group list, resulting in an overall error rate of 0.01. A total of 1574 protein groups were identified, corresponding to 2012 gene symbols (supplemental Table S3). Only 34 of the protein groups were based on single peptides. Scores, sequences, and spectra for these single peptide-based proteins are provided in supplemental Table S4. HD-early/control, HD-mid/control, and HD-mid/HD-early ratios for each protein group were inferred from lab level analysis by comparing IPI numbers. For a few cases, multiple ratios were found for a protein group in HD-early versus control, HD-mid versus control, or HD-mid versus HD-early comparisons, and in those cases we took the geometric mean of these ratios. Detailed results of this search are available upon request. We have also deposited the comprehensive list of proteins at the PRoteomics IDentifications (PRIDE) database (accession number 3701).
Deriving the Rank of Proteins Changes by Combining Ratios across Laboratories
Because various methods report protein changes on different scales, to make an effective and meaningful comparison (38) we integrated protein ratios across laboratories using the following meta-analysis procedures that combine the scale-free effect size measurements (z-scores). We first transformed the ratios to logarithm scale so that all the data were in a similar range, and then we standardized the scores by centering and scaling log ratios in each lab to have mean 0 and variance 1. The resulting z-score represents the number of standard deviations above or below the mean ratios within each lab. To combine z-scores, we chose to sum them across laboratories. Although one might have considered averaging the z-scores because not all proteins are quantified in the same number of experiments, we chose to sum them so that a protein quantified by only one laboratory must have a higher ratio to achieve the same rank as proteins observed across all laboratories with consistent and modest changes. For the purpose of finding proteins that classify HD, we combined the two HD groups and summed together the sum of z-scores of HD-mid/control and HD-early/control (sums of z-scores). Proteins that are most altered in HD are those with the highest and lowest sums of z-scores.
Annotating Proteins for Tissue Specificity and for Changes in HD Brain
We next annotated each protein in the synthesized protein list based on its behavior found in human transcriptional profiles of HD and normal brain (2) and other tissues (29). Specifically we annotated proteins based on 1) tissue specificity score: the relative expression of transcript in human brain, liver, and 25 other normal human tissues; and 2) changes in HD brain: the ratio of transcript abundance between HD and non-HD brains. All annotations were made by comparing gene symbols. IPI numbers identified in our proteomics study were associated with gene symbols by reference to the data (ipi.HUMAN.v3.20.dat) provided by the International Protein Index managed by the European Bioinformatics Institute.
Annotation of Protein Changes in CSF with Changes in the HD Brain by Microarray Analysis—
We compared protein changes from this proteomics study with the log2 -fold changes of mRNA in HD versus normal brain in caudate nucleus, cerebellum, and motor cortex (Brodmann area 4 (BA4)) based on a previously published study by Hodges et al. (2). Probe sets with significant changes (p values <0.001) were selected and collapsed to gene level based on gene symbols. When a gene has multiple probe sets, the median log2 -fold change of that gene was selected as the estimate of the mRNA change. As a result, 6183 genes in caudate nucleus, 1143 genes in BA4 cortex, and 440 in cerebellum have significant mRNA changes in HD brains from the normal.
Signal Processing of Tissue Transcriptional Expression Data—
Because a significant fraction of plasma proteins is derived from various tissues and so are CSF proteins, which are filtered from plasma, to annotate proteins for their source tissues, it is critical that the transcriptional data set includes tissues of human major organs. The normal human tissue expression data set was acquired from published data provided by Ge et al. (29) that includes a total of 36 types of normal human tissue, covering the complexity of human tissues. Because all CSF samples are from adult human, the annotation will focus on the adult tissue expression pattern. Therefore, data from three fetal tissues were removed. Ge et al. (29) examined a “whole brain” and six subregions of the brain (amygdala, corpus callosum, caudate nucleus, cerebellum, hippocampus, and thalamus). However, the cortex region that is noted to have severe cell loss in Huntington disease patients was not included. We removed the “whole-brain” data because it is not obvious to us how this RNA sample was prepared. We added the cortex signals and substituted cerebellum and caudate nucleus data with data derived from an extensive study that was carried out on four parts of human normal brain (caudate nucleus, cerebellum, and BA4 and BA9 cortex) using the same microarray chips (2). After these brain data were obtained from Gene Expression Omnibus (GEO) DataSets, signals detected using Affymetrix microarray suite version 5 software (MAS5) for each probe were averaged over 21 caudate nucleus, 21 cerebellum, and 24 cortex (12 BA4 and 12 BA9) arrays. We plotted log2 MAS5 signal of the caudate nucleus and cerebellum from Ge et al. (29) versus those from Hodges et al. (2) and found that the correlations are both 0.90 (supplemental Fig. S1). These high correlations suggest that data from the two studies may be combined. So far, we have data for seven subregions of the brain. Because we want to annotate a gene as being active in the brain if it is active in any part of the brain, we summarized the brain expression data by taking the maximum across all seven subregions. As a result, transcriptional data of “brain tissues” and 26 other types of human tissues were included in our analysis. These tissues are from brain, heart, thymus, spleen, ovary, kidney, muscle, pancreas, prostate, intestine, colon, placenta, bladder, breast, uterus, thyroid, skin, salivary, trachea, adrenal, bone marrow, pituitary, spinal cord, testis, liver, stomach, and lung (supplemental Table S5). Finally many genes have multiple probes; one can choose to use the average signal or the maximum signal for each gene. In our analysis, the one with maximum signal among all tissues was selected because we considered that the maximum signal is the highest above the noise level. As a result, we observed 1941 tissue markers based on human array data (supplemental Table S6). The definition of tissue-specific genes/proteins may vary with tissues included in the study and when the thresholds change.
Defining Tissue Specificity of a Gene—
Tissue specificity was derived from the human transcriptional data set profiling seven brain tissues plus 26 other normal tissues based on publicly available resources after processing as described above. The tissue specificity score for a gene is determined by the relative intensity of its probe on the array across tissues. We defined a gene as tissue-specific if the maximum intensity of its probe was highest in that tissue and the maximum intensity in every other tissue was at least 2.5 times lower. These tissue-specific genes were used to annotate the CSF proteome.
Verification of Protein Tissue-specific Annotation—
The definition of tissue-specific genes/proteins may vary with tissues included in the study, with detailed processing methods, and when thresholds are changed. To validate our definition of tissue-specific genes, we checked the description and functions of some genes chosen at random. For instance, muscle-specific genes include myosin, actinin, troponin, creatine kinase, and calcium channels. And testis-specific genes are associated with terms such as spermatogenic, sperm-specific, or male-enhanced antigen. In addition, normal mouse gene expression data from Zapala et al. (39) covering 20 subregions of the brain plus 14 tissues from other parts of the adult mouse was used to confirm the human array-based analysis. Similar to human tissue data processing, we combined data from 20 subregions of the brain including striatum, cortex, cerebellum, etc. into one data set called “brain” to simplify the analysis using the maximum MAS5 signal of the 20 brain subregions computed for each probe set. As a result, 15 mouse tissues were considered for further analysis: adrenal, pituitary, testis, thymus, spinal cord, choroid plexus, retina, brown adipose tissue, white adipose tissue, kidney, liver, heart, muscle, spleen, and brain (supplemental Table S7). Corresponding human orthologous genes were inferred by human-mouse ortholog data provided by the Human Gene Organisation Gene Nomenclature Committee. Using the same tissue specificity-defining methods, 1333 tissue markers with human orthologous genes were observed (supplemental Table S8).
Summary of Statistical Analysis Procedures Used to Interrogate the Protein List
Statistical procedures were used to interrogate the synthesized protein list for three hypotheses that the experiments were designed to address: 1) that CSF is enriched for brain-specific proteins compared with plasma, 2) that brain-specific proteins change in CSF with HD development, and 3) that brain-specific protein changes in CSF are concordant with transcriptional changes in the brain.
To evaluate hypothesis 1, we performed Pearson's χ2 tests on an over-representation analysis to compare the fraction of proteins annotated as being brain-specific in CSF and the Human Proteome Organization Plasma Proteome Project plasma data (results summarized in Table IV). To evaluate hypothesis 2, we used GSEA, which uses a non-parametric Wilcoxon test (40, 41) to compare the distribution of ratios between brain-specific and non-brain-specific proteins (see “Results”). For hypothesis 3, we coded all brain-derived proteins as up or down based on the sign of their sums of z-scores and all transcripts as up or down based on the sign of their log2 -fold changes of mRNA from the array and then applied Pearson's χ2 tests to evaluate the association (results are summarized in Table V).
Table IV.
Comparisons of brain- and liver-specific proteins in CSF and plasma
| Tissue | Marker genes detected in
|
-Fold of enrichment (CSF/plasma) | p valuea | |||
|---|---|---|---|---|---|---|
| CSF (all = 298)
|
Plasma (all = 414)
|
|||||
| Number | Percentage | Number | Percentage | |||
| Brain | 88 | 0.3 (88/298) | 71 | 0.17 (71/414) | 1.8 | 0.0001 |
| Liver | 97 | 0.33 (97/298) | 113 | 0.53 (113/414) | 0.6 | 0.15 |
p value is based on Pearson's χ2 test to evaluate the significance of differences of representativeness of brain (liver) proteins in CSF and plasma. Numbers of brain (88) versus non-brain (210) marker genes in CSF were tested against those numbers in plasma (71 versus 340). Likewise for liver proteins, numbers of liver (97) versus non-liver (201) proteins in CSF were tested against those numbers in plasma (113 versus 301).
Table V.
Concordance of proteomics and genomics data for brain-specific proteins, non-brain specific proteins, and proteins from unknown tissue origins
NA, not applicable.
| Proteins | Concordant | Discordant | Total | Percentage of concordance | p value |
|---|---|---|---|---|---|
| Brain-specific | 13 | 3 | 16 | 0.81 | NA |
| Non-brain specific | 13 | 18 | 31 | 0.42 | 0.024a |
| Unknown origin | 85 | 95 | 180 | 0.47 | 0.019b |
| All | 111 | 116 | 227 | 0.49 | NA |
This p value is from the Pearson's χ2 test of brain-specific proteins versus non-brain specific proteins. Numbers of concordant and discordant brain-specific proteins versus the two numbers for non-brain-specific proteins were used for the test.
This p value is from the Pearson's χ2 test of brain-specific proteins versus proteins of unknown sources. Numbers of concordant and discordant brain-specific proteins versus the two numbers for proteins with unknown tissue origin were used for the test.
RESULTS
General Work Flow of the Primary Data Analysis—
All proteomics data acquired by four laboratories utilizing different proteomics platforms were interpreted using a common protein database as well as the same search engine and peptide and protein validation methods to allow comparison across labs. The flow of data analysis is as follows. 1) Data sets generated based on each experimental design were searched with protein database independently. 2) Multiple data sets from the same experiment were aligned to determine the protein changes between different disease statuses. 3) Data were aligned across labs, and protein ratios were synthesized for the consistency of protein changes. 4) Proteins identified were assessed for their dominant expression tissues based on published human and mouse tissue expression data. The abundances of brain and liver proteins in CSF were compared with those in plasma. 5) Finally protein changes were integrated with the genomics profiling of mRNA changes in normal versus HD brain. Results from each step of analysis are presented sequentially.
Analysis of Proteomics Data—
Search engine performance and PeptideProphet details (25) were inspected (supplemental Fig. S2A) to assure that sensitivity and error distributions were sufficient to determine correct and incorrect identifications from Labs 2, 3, and 4. The quality of data quantitation was determined by examining MA plots, histograms of light labeled/heavy labeled ratios at the peptide level, and histograms of the HD-early/control, HD-mid/control, and HD-mid/HD-early ratios at the protein level (supplemental Fig. S2, B–D). We observed that the distributions of logarithms of ratios are around 0 before normalization for all experiments except two interrogations from Lab 3. The log ratios for these two data sets were normalized to have a median of 0.
Lab 1 used a DIGE method (see “Experimental Procedures”) and quantitated a large number of fluorescent spots. Only those spots found to have significantly different changes based on fluorescence between the HD and control CSF were selected for tandem MS analysis. As a result, a total of 19 unique proteins for 42 spots were identified based on the MS/MS data (Table II), each of which is a putative biomarker candidate. As a verification of these protein identifications (“Experimental Procedures”), we compared them with the results provided by Lab 1 using the Mascot search engine (supplemental Table S9) and found that there is a high consistency between the two results. In addition, proteins from more spots have been identified in this study. Lab 2 is the only lab that performed individual (non-pooled) interrogation. 335 confident protein groups (with ProteinProphet probability >0.9) were found and of these, 319 were quantitated using spectral counting (35) of highly confident peptide spectra (“Experimental Procedures”). Lab 3 pooled samples by disease status and analyzed by d0/d3 acetylation of the N terminus of the peptides. 263 confident protein groups were found after aligning the three experiments using ProteinProphet. After protein ratios were inferred from individual experiments, 161 were confidently quantitated. Lab 4 pooled samples by disease status and gender. In three pairwise comparisons, proteins in pooled CSF of two disease statuses were differentially labeled with light and heavy acrylamide on cysteine residues. Because a more extensive prefractionation strategy was used, these data resulted in identification of the majority of proteins (1179 groups) reported in this study of which 377 were confidently quantitated by the Q3 algorithm (27) (“Experimental Procedures”). In addition, Lab 4's label-free analysis using AMT methods (37) identified and quantitated 277 protein groups (Table II), ∼100 of which are not quantitated by the labeled approach (Table III).
Table II.
Numbers of peptides and protein groups identified with different methods
| Methods | Total peptidesa | Unique peptidesa | Unique protein groups | Quantitated unique protein groups |
|---|---|---|---|---|
| Lab 1 | 2,041 | 403 | 19b | 19b |
| Lab 2 | 122,353 | 3,556 | 335 | 319 |
| Lab 3 | 19,563 | 1,696 | 263 | 161 |
| Lab 4 (labeled) | 143,405 | 9,397 | 1,179 | 377 |
| Lab 4 (unlabeled) | 2,502 | 1,357 | 277 | 277 |
Number of peptides with FDR <0.1 for Lab 1 and PeptideProphet probability >0.75 for Labs 2–4.
These are number of unique proteins, not protein groups.
Table III.
Numbers of overlapping protein groups identified and quantitated among five methods
| Methods | Lab 1 | Lab 2 | Lab 3 | Lab 4 labeled | Lab 4 unlabeled |
|---|---|---|---|---|---|
| Lab 1 | 23 (23)a | 22 (22)b | 20 (19) | 23 (20) | 20 (20) |
| Lab 2 | 339 (327) | 190 (144) | 300 (197) | 187 (183) | |
| Lab 3 | 267 (170) | 235 (137) | 164 (135) | ||
| Lab 4 labeled | 1158 (376) | 293 (199) | |||
| Lab 4 unlabeled | 298 (298) |
The underlined numbers are the total of protein groups identified and quantified by each laboratory. These numbers are a little different with those in Table II because of the regrouping of proteins when combining all data across laboratories.
The numbers of overlapping protein groups quantitated are shown in parentheses.
Overall the number of proteins identified is highly related to the number of sample fractions and to the number of MS/MS spectra obtained. Because of the limited publication space and the large amount of data, all the confident protein groups identified and their quantitation results from each lab are provided in supplemental spreadsheets (supplemental Table S3). For each protein group (probability >0.9) identified, the number of total and unique peptide identifications are provided in the spreadsheets. p values (Lab 1 and Lab 2) and standard deviations (Lab 3 and Lab 4) are also included to indicate the significant changes and accuracy of the quantitations. In practice, one can use these values as filters to generate lists by lab for protein groups that significantly change with disease status. Because of limitations in the accuracy and comprehensiveness of any one data set due to the complex nature of the samples (19, 30) and current technology (42), a combination of these results across labs using completely different experimental designs and proteomics platforms will improve the accuracy, consistency, and comprehensiveness of protein candidate lists. Here we used a method described under “Experimental Procedures” of first performing a comprehensive search on all applicable data and then synthesizing the quantitation results.
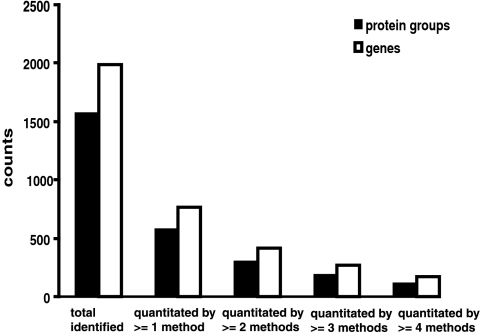
The integrated analysis of all proteomics data was performed by a comprehensive search combining Lab 2, Lab 3, and Lab 4 labeled and unlabeled data. This resulted in a total of 12,430 peptides and 1574 high confidence protein groups identified (supplemental Table S3). The greatest overlaps in both proteins identified and proteins quantitated are between Lab 2 and Lab 4 labeled methods (Table III). From the across-lab comparison, 577 protein groups (corresponding to 762 genes) have been quantitated by at least one experimental method, and 301 protein groups (419 genes) have been quantitated by more than one method (Fig. 1). We synthesized protein ratios across laboratories by methods described under “Experimental Procedures.” The resulting score (sum of z-scores) estimates the relative protein change in the HD versus normal CSF. Specifically a negative sum of z-scores indicates that the protein declines in the HD CSF when compared with normal, and a positive sum of z-scores indicates that the protein inclines. This score for each identified protein is shown in supplemental Table S3.
Fig. 1.
Number of total protein groups and genes identified and quantitated by different numbers of methods. Protein groups are in solid bars, and genes are in empty bars.
Annotation of HD CSF Proteins with Human and Mouse Tissue Expression Data—
The general perception of HD is that the most important clinical signs and symptoms can be traced to neurodegeneration in the brain. Furthermore by definition, the most powerful and useful biomarkers are intimately related to the etiology of a disease. This raises the question of the precise source of the proteins detected in CSF and whether they can be traced to brain and substructures within the brain or they arise from other sources. Formally one cannot definitively identify the source of each particular CSF protein. Direct evidence must come from some type of tracer experiment. But we can begin to make a circumstantial argument that a substantial fraction of our identified CSF proteins are based upon mRNA expression patterns.
Several human brain gene expression profiling data have been published in recent years (29, 43). We selected an expression data set that was generated by Ge et al. (29) using a total of 36 types of normal human tissues. Data on four regions of brain (amygdala, corpus callosum, hippocampus, and thalamus) and other adult tissues were selected and combined with data from a comprehensive study carried out on four parts of normal human brain (caudate, cerebellum, and BA4 and BA9 cortex) (2) (“Experimental Procedures”). Based on the algorithm described under “Experimental Procedures,” 1941 tissue-specific proteins are identified: 445 are brain- and 225 are liver-specific (supplemental Table S6). Integration with the CSF proteomics data found that 298 proteins/genes of 1574 are tissue markers, among which two major species are brain-specific (∼30%) and liver-specific (∼33%) (Table IV). One intriguing question is how representative these proteins are compared with plasma. Starting from a list of 3020 proteins identified with two or more peptides provided by the Human Proteome Organization Plasma Proteome Project, we aligned the 1941 tissue markers and found that of 414 proteins that are annotated as tissue-specific proteins 17% are brain- and 53% are liver-specific (Table IV). Therefore, brain-specific proteins are 1.8-fold enriched in CSF over plasma, whereas liver-specific proteins are about half as represented in CSF as in plasma. The Pearson's χ2 test shows that brain-specific proteins significantly predominate in CSF compared with plasma (Table IV). This observation can also be confirmed by performing the same analysis using the normal mouse gene expression data from Zapala et al. (39) that covered 20 neural tissues from the adult mouse central nervous system plus 14 tissues from other parts of the body (data not shown).
Next we examined whether the brain-specific proteins are specific to any regions of the brain. Seven regions of the brain were included in this human tissue array data: amygdala, corpus callosum, hippocampus, thalamus, caudate, cerebellum, and cortex. Among 88 CSF proteins considered to be brain-specific, 29 are cerebellum-specific, 26 are cortex-specific, 12 are amygdala-specific, eight are caudate-specific, and 13 belong to the other regions. This suggests that more than 60% of these brain-specific proteins are specifically expressed in cerebellum and frontal cortex.
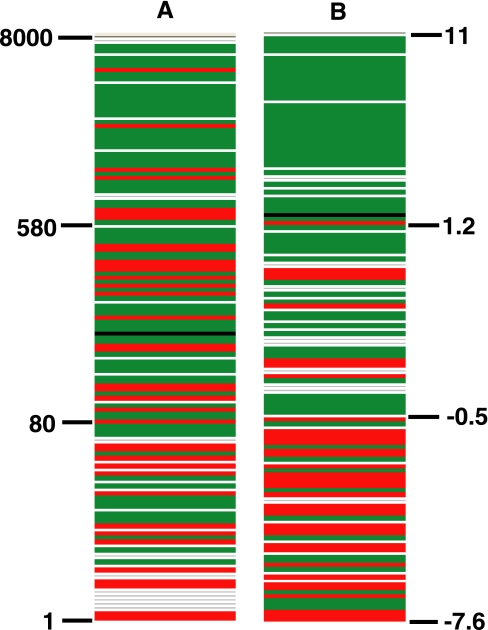
To have an overview of the relative abundance of these tissue-specific proteins/genes in CSF, we used the overall spectral count for each protein as a surrogate for the concentration. When the spectral counts were sorted in descending order, most of the liver-specific proteins (colored in green in Fig. 2A) show up at the top of the list. The brain-specific proteins (red) are distributed from upper middle to the bottom. The observation that liver-specific proteins are abundant makes sense because most CSF proteins come from plasma in which liver-derived proteins are highly represented.
Fig. 2.
Patterns of tissue-specific proteins in CSF. A, proteins were sorted in descending order by spectral counts that reflect the relative concentration in CSF with highest concentrations at the top. B, proteins were sorted in descending order by sums of z-scores that indicate the trends of changes in HD-affected individuals relative to control with the most increasing one at the top. Red, brain-specific proteins; green, liver-specific proteins; black, muscle- and heart-specific proteins; white, other tissue-specific proteins.
Finally we evaluated the trends of changes for these tissue-specific proteins in normal versus HD patients. When 150 tissue-specific proteins were sorted by sums of z-scores (Fig. 2B), there is a clear bias of liver proteins (colored in green) at higher scores and brain proteins at lower scores, indicating that liver-specific proteins tend to increase and brain-specific proteins decrease in HD CSF. The p value based on Wilcoxon test on sums of z-scores of brain-specific proteins versus other proteins is 6.3 × 10−9, and that on sums of z-scores of liver-specific proteins versus others is 3.3 × 10−8. This result indicates that the trends of changes in HD CSF from the normal are significant for brain- and liver-specific proteins.
Comparison of Protein Changes with Human HD Brain Transcriptional Profiling Data—
Gene expression changes in four brain regions of Huntington disease patients have been studied by Hodges et al. (2). Their results revealed that 21, 1, and 3% of probe sets were significantly differentially expressed in HD caudate, cerebellum, and BA4, respectively, and that no significant changes were found for BA9. An immediate question raised is what the concordance of changes in HD patients is between the proteomics and microarray studies. Among the genes that are significantly differentially expressed in HD caudate, cerebellum, and BA4 cortex, ∼665, 57, and 165 of their products are identified in CSF, respectively. Because the most significant mRNA changes occur in HD caudate and the expression profile of HD BA4 is strikingly similar to that of HD caudate (2), we compared all CSF protein changes with HD caudate data and additionally looked at cerebellum- and cortex-specific proteins when mRNA expression data were available.
To examine the concordance of changes, we used a sign test that compares the negative or positive signs of sums of z-scores from our proteomics study with the log2 -fold changes from the microarray study because in both data sets positive values indicate an increasing trend of proteins/genes in HD status and negative values indicate a decreasing trend. Therefore, proteins/genes with these two values in the same signs were considered concordant. Overall about half (111 of 227) of protein groups that have both sums of z-scores and significant mRNA changes are concordant (Table V). Among the 227 proteins, 47 have tissue annotations, and 16 are brain-specific. We found that 13 of 16 (81%) brain-specific proteins are concordant. However, only 42 and 47% of the proteins are concordant for non-brain tissue-specific proteins and proteins with unknown tissue origin, respectively. The χ2 tests on the numbers of concordant and discordant for 1) brain-specific versus other tissue-specific proteins and 2) brain-specific versus those that are not tissue markers both gave a p value ≤0.024, indicating that the consistency of expression changes in HD status measured by a proteomics and genomics approach are significant for brain-specific proteins compared with other proteins (Table V). This concordance suggests that these proteins might be derived from neurons or glial cells in the brain. Moreover 11 of the 13 brain-specific genes that have concordant mRNA and protein changes show the trend of declining in HD, consistent with the above observation that brain-specific proteins tend to decline in HD samples.
The Most Significantly Changed Proteins in HD CSF Based on Proteomics Data—
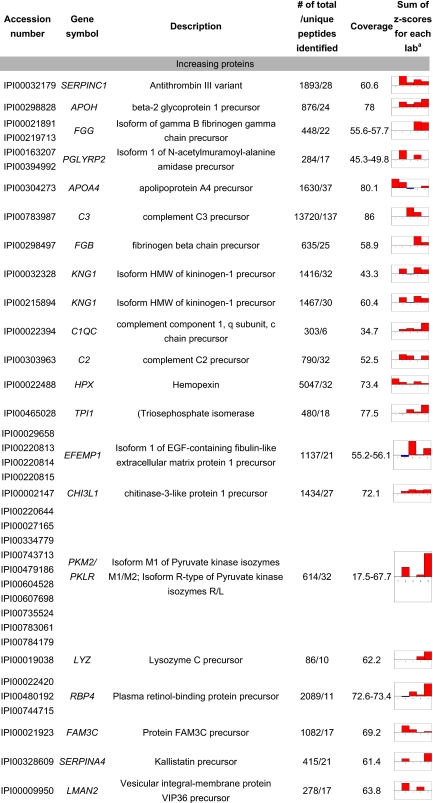
With sums of z-scores that estimate protein changes between disease states across labs (Table VI), we were able to select 20 most increasing and 20 most decreasing proteins in HD CSF (relative to controls). This selection is naturally biased toward proteins that are observed by many labs and that have consistent trends of changes in HD CSF.
Table VI.
The most significantly changed proteins in CSF of HD-affected individuals compared with unaffected ones
Each bar in this column represents the sums of z-scores based on logarithm of early/control and mid/control ratios for each lab. The negative scores are plotted in blue, and the positive scores are in red.
Among the identified proteins, 12 of them (CHGB, SIAE, IDS, NRXN3, GSN, ENDOD1, GRIA4, GGH, GC, C4B, and PRNP; see Table VI for the full protein names) have a trend of declining with disease progression (control > HD-early > HD-mid). Among the most increasing proteins, seven of them (C1QC, HPX, TPI1, PKM2/PKLR, LYZ, FAM3C, and LMAN2) follow the trend consistent with elevating as disease progresses (control < HD-early < HD-mid). C1QC, C2, and C3 are complement factors. PGLYRP2 and APOA4 are also associated with the inflammatory pathway. SERPINC1, APOH, FGG, FGB, and KNG1 are related to the coagulation system that cross-talks to the immune system (44). The immune system is activated in Huntington disease patients (45), and recently Dalrymple et al. (10), who used a proteomics approach to profile plasma rather than CSF in Huntington disease, found several inflammatory proteins. Because CSF is a filtration of plasma, our observations are consistent with their findings.
Integration of these most altered proteins in HD CSF with the tissue expression data shows that although most increasing proteins are liver-specific only three of the decreasing proteins are brain-specific (Table VII). This result is expected given that the method we used to generate this list of 40 proteins is biased toward more abundant proteins and given the above result suggesting that most liver-specific proteins are abundant and increased in HD CSF, whereas most brain-specific proteins are decreased, but not all of them are abundant enough to be selected. However, some of the decreased proteins may come from other substructures of the brain. For example, CHGB has the highest mRNA level in the mouse. TTR, ENPP2, and GGH are choroid plexus-specific genes according to the mouse array data and Allen Brain Atlas data. Moreover although not exclusively expressed in the brain, MEGF8, ALDOC, ENPP2, ENDOD1, and PRNP have the highest mRNA expression level in the brain. In addition, TTR, CHGB, and PAM have high mRNA expressions in the brain when compared with the median expression level. It is possible that a majority of these proteins found in CSF were derived from the brain.
Table VII.
Integration of HD CSF most significantly changed proteins with tissue expression data and HD-brain transcriptional data
NA, not available.
| Protein | mRNA changes (HD vs. control) | Specific tissue | Highest mRNA expression tissue | Expression ratioa
|
|
|---|---|---|---|---|---|
| Brain/median | Liver/median | ||||
| A. Decreasing proteins | |||||
| EPHA4 | Decrease | Brain | Brain | 41.2 | 0.7 |
| CHGB | Decrease | NA | Pituitary | 23.9 | 0.8 |
| TTR | No change | NA | Liver | 145.7 | 218.9 |
| SIAE | NA | NA | NA | NA | NA |
| MEGF8 | NA | NA | Brain | 5.2 | 2.1 |
| CTSD | Increase | NA | Lung | 1.1 | 1.8 |
| IDS | Decrease | Brain | Brain | 10.2 | 0.5 |
| ALDOC | No change | NA | Brain | 12.2 | 1.5 |
| ZNF503 | Decrease | NA | NA | NA | NA |
| NRXN3 | Decrease | Brain | Brain | 142.6 | 0.41 |
| PAM | Decrease | NA | Salivary | 1.9 | 0.11 |
| PGCP | Increase | NA | Thyroid | 1.2 | 1.5 |
| ENPP2 | Decrease | NA | Brain | 10 | 0.5 |
| GSN | Increase | NA | Bladder | 1.2 | 0.2 |
| ENDOD1 | NA | NA | Brain | 4.1 | 0.6 |
| GRIA4 | No change | NA | Pancreas | 1.8 | 1.4 |
| GGH | Decrease | NA | Liver | 2.7 | 8.1 |
| GC | No change | Liver | Liver | 4.2 | 154.7 |
| C4B | Increase | Liver | Liver | 1.3 | 15.1 |
| PRNP | Decrease | NA | Brain | 2.82 | 0.58 |
| B. Increasing proteins | |||||
| SERPINC1 | No change | Liver | Liver | 10.6 | 688.9 |
| APOH | No change | Liver | Liver | 16.2 | 603.9 |
| FGG | No change | Liver | Liver | 6 | 231.4 |
| PGLYRP2 | No change | NA | NA | NA | NA |
| APOA4 | No change | Intestine | Intestine | 3.7 | 15 |
| C3 | Increase | NA | NA | NA | NA |
| FGB | No change | Liver | Liver | 7.5 | 352.7 |
| KNG1 | No change | Liver | Liver | 6.7 | 581.3 |
| C1QC | No change | NA | NA | NA | NA |
| C2 | NA | Liver | Liver | NA | NA |
| HPX | No change | Liver | Liver | 2.1 | 190 |
| TPI1 | Decrease | NA | Adrenal | 1.9 | 1 |
| EFEMP1 | Increase | NA | Placenta | 1.8 | 0.15 |
| CHI3L1 | Increase | Liver | Liver | 2.1 | 26.8 |
| PKM2 and PKLR | Increase | NA | Brain | 2.5 | 0.1 |
| LYZ | No change | NA | NA | NA | NA |
| RBP4 | Decrease | Liver | Liver | 10.7 | 242.9 |
| SERPINA4 | No change | Liver | Liver | 1 | 13.5 |
| FAM3C | Decrease | NA | Intestine | 2.7 | 0.9 |
| LMAN2 | No change | NA | Thyroid | 0.6 | 1.2 |
Expression ratios are calculated from the human tissue transcription data by dividing the brain or liver MAS5 signal by the median MAS5 signal of each tissue. This value represents how many -fold the mRNA expression level of the brain or liver tissue is above the average.
The changes of three brain-specific proteins (EPHA4, IDS, and NRXN3) in HD CSF agree with their transcriptional changes in HD brain. In addition, CHGB, ZNF503, PAM, ENPP2, GGH, and PRNP have the same trend at both the protein and mRNA levels. These proteins are potentially interesting biomarker candidates once validated. Many liver-specific proteins shown to be increased in HD CSF according to the proteomics analysis are not significantly changed based on transcriptional profiling of the brain. However, these proteins can provide additional information on the peripheral manifestations of Huntington disease and may be used in combination with the brain-specific CSF proteins as biomarkers.
Assessing Cross-lab Comparability—
As shown above, the labeled methods of Lab 2 and Lab 4 have the greatest overlap in both protein identification and quantitation (Table III), and 301 protein groups have been quantitated by more than one method (Fig. 1). Questions we addressed include the concordance of protein ratios and the concordance of each laboratory with the overall trends of brain- and liver-specific protein changes identified by the integrated analysis.
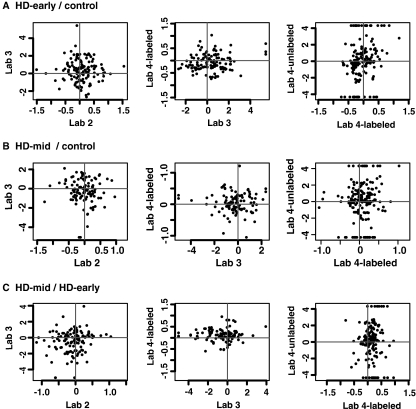
Scatter plots of relative protein abundance ratios between two disease states across different proteomics methods are shown in Fig. 3. The apparent low proteome-wide correlation of protein ratios should be expected given the nature of data we were interrogating. Specifically whenever high dimensional analyses such as proteomics (or genomics) are used for comparisons, as in our example, one expects that most proteins do not change between the two conditions (HD versus control). For these proteins the sources of variation are random (from experiments) and so will not correlate across laboratories. Only proteins that systematically changed as a result of differences in the case and the control are concordant. Thus, instead of inspecting the correlation of all data points in Fig. 3, one should focus on those ratios with higher magnitude and determine the concordance among them as these should be enriched for proteins having the systematic change. We can see that this trend of changes is rather consistent among laboratories for the 40 most altered proteins, especially for those increasing proteins that are more abundant and quantified by more laboratories (Table VI). Concordance was strongest among laboratories identifying the largest number of proteins.
Fig. 3.
Correlations of relative protein abundance ratios of two disease statuses among different proteomics methods. Both axes are the following three ratios on a logarithm scale based on different quantitation methods: HD-early/control (A), HD-mid/control (B), and HD-mid/HD-early (C).
We also evaluated whether single laboratories could demonstrate the overall trend of changes in HD versus normal for brain- versus liver-specific proteins. As shown in Table VIII, each lab's sum of z-scores are negative for brain-specific proteins and positive for liver-specific proteins, and the differences are statistically significant based on Wilcoxon test. This suggests that all laboratories are concordant in observing the trend that brain-specific proteins tend to decrease and liver-specific proteins tend to increase in HD CSF.
Table VIII.
The significant differences of protein changes for brain-specific and liver-specific proteins observed by each laboratory
| Methods | Brain-specific
|
Liver-specific
|
p valuec | ||
|---|---|---|---|---|---|
| Counta | Meanb | Counta | Meanb | ||
| Lab 2 | 9 | −1.28 | 27 | 0.96 | 0.0039 |
| Lab 3 | 6 | −1.15 | 21 | 0.25 | 0.025 |
| Lab 4 (labeled) | 9 | −0.94 | 31 | 1.64 | 0.0002 |
| Lab 4 (unlabeled) | 8 | −1.26 | 30 | 1.26 | 0.0009 |
Numbers of proteins that are counted in the test.
Mean sum of z-scores of the individual lab.
p value is derived by performing Wilcoxon test on the sum of z-scores of brain- and liver-specific proteins for each individual lab.
DISCUSSION
In this study, we report an extensive list of proteins identified in CSF with a high degree of confidence and provide their concentrations in human HD relative to control CSF. Because of the complexity of the CSF proteome and the current shotgun proteomics technology (42), there is a limitation in the accuracy and comprehensiveness of a single proteomics experiment. It was suggested that, similarly to gene expression profiling, which cannot draw meaningful conclusions from a single quantitative gene expression profile, multiple profiles from related samples allow extraction of signature patterns containing diagnostic or functional information (42, 46). For example, to comprehensively characterize the human CSF proteome, Pan et al. (30) have used several different separation strategies and proteomics platforms. The analysis of the same samples by four different proteomics platforms provides a rather in-depth characterization of HD CSF.
From a practical standpoint, the protein changes identified in CSF of Huntington disease patients are candidate biomarkers that may be useful for tracking the HD progression or as surrogate end points in clinical trials. Integrating results between laboratories provided confirmation of both protein identification and quantitation. The universality of our findings will require further validation with additional CSF samples using specific technologies such as multiple reaction monitoring and ELISA rather than shotgun proteomics.
Before a candidate protein can serve as a biomarker, it is important to understand its role in the pathophysiology of the disease process. In the case of Huntington disease this is not yet possible because the exact sequence of pathological events downstream from expression of mutant huntingtin protein remains elusive. However, the predominant view is that the most clinically important signs and symptoms of HD relate to neurodegeneration and dysfunction in the brain. The hallmark neuropathology of HD is degeneration of medium spiny neurons in the striatum accompanied by extensive astrocytosis (47) and microgliosis (12). Gene expression profiling of postmortem human HD brain has been performed using striatal tissue as well as cerebellum and two cortical areas (2, 6). The known pathology and gene expression changes provide a perspective from which to view the proteomic changes.
To identify the most probable origin of the proteins detected in CSF, we queried a published microarray survey of gene expression in human tissues. Although the assumptions and methods used were somewhat crude and only constitute circumstantial evidence for the source tissue, this created a very biologically plausible list of “tissue markers.” Integrating the expression data tissue markers with the CSF proteomics results revealed that many of the most abundant CSF proteins were probably derived from the liver. This is consistent with the known origin of CSF, which is a complex filtrate of blood produced by the choroid plexus. Importantly many proteins likely to have been derived from the brain were also detected in CSF. This was not seen in a re-examination of a protein component list for blood plasma. In plasma, liver-specific proteins are also highly over-represented, whereas very few brain-specific proteins are detected. The over-representation of brain-specific proteins in CSF supports the hypothesis that it is feasible to monitor some aspects of the health of the brain using CSF. This has important implications for biomarker discovery in neurological disease. However, proving that these proteins are derived from brain tissue is difficult and requires some type of labeling experiment.
The overall trends between changes in concentrations of brain-specific proteins and their corresponding mRNAs in HD brain tissues were consistent. This provides further support for the hypothesis that the concentration of some proteins in HD CSF may provide a window into the health of the brain and that CSF may be a fruitful source of HD biomarkers. However, to the extent that the candidate brain-specific proteins can be traced to a particular region of the brain, most appear to be cerebellum- and cortex-specific rather than specific to the striatum. This may be because the greater mass and surface area of the cortex and cerebellum provide more exposure to the CSF.
Two noticeable trends in the data were for the brain-specific proteins to decrease in concentration in HD CSF, whereas most proteins we detected with higher concentrations in HD CSF were functionally associated with the immune system. The latter may relate to astrocytosis, microgliosis, and neuroinflammation. Neuroinflammation is a common component of many neurodegenerative diseases, and these changes are unlikely to be specific to Huntington disease. Thus, although we did not detect a large number of protein changes that can be directly linked to striatal degeneration in HD, we did detect a general trend for brain-related proteins to decrease in HD CSF, and we detected increases in proteins that may reflect neuroinflammatory processes. Both trends are consistent with the known neuropathology of HD and bolster the biological relevance of our findings.
However, an interesting alternative mechanism may cause or contribute to the increase of blood-derived proteins and decrease of brain-derived proteins in HD CSF. Disruption of the blood-brain barrier is widely accepted in inflammatory conditions such as neurosystemic lupus erythematosus (48, 49) and multiple sclerosis (50, 51) and increasingly in conditions traditionally seen as degenerative with secondary neuroinflammation, like Alzheimer disease (52). Interestingly microglial activation and the presence of inflammatory cytokines could alter the properties of brain microvascular endothelial cells and the tight junctions that link them (53, 54), raising the possibility that the blood-brain barrier is also disturbed in Huntington disease patients. This hypothesis is consistent with the observed differences of brain- and blood-derived proteins in HD CSF. The integrity of the blood-brain barrier in HD can be examined by detecting changes of brain proteins in HD versus normal plasma or more directly using magnetic resonance imaging.
Because of the large source of variation among multiple laboratories, no general consensus can be made with regard to the specific ranking of proteins or their magnitude of change. In our experiments the statistical significance was not derived from establishing the significance of the top ranked individual proteins by traditional FDR (55) but rather based on interrogating the rankings based on protein sets using GSEA (28) methods where the gene sets were derived from external transcriptional data. We also found that results in this study are most strongly supported by laboratories that obtained the greatest depth of protein coverage. Our results suggest that additional biomarker studies should focus on designs that obtain the greatest depth of coverage and that interrogate the data analysis with externally derived hypotheses, perhaps from data integration. Our study could not have led to a positive finding without taking these advantages.
In summary, we provide a comprehensive profiling of the human HD CSF proteome. The integration of the proteomics data with various genomics data supports the idea of CSF as a rich source of biomarkers for neurological diseases. For Huntington disease in particular we derived a list of proteins that are altered in HD CSF and that have the potential to be used as a specific signature of HD progression.
Supplementary Material
Acknowledgments
We thank Allan Tobin from the High Q Foundation for the suggestions and encouragement in this project and Sara Lynn Zriny for project administration. Yan Liu, Jimmy K. Eng, Ted Holzman, and Lynn Amon each provided technical help in various aspects during the data analysis. Peter Hussey, from LabKey, assisted in developing the Web site.
Footnotes
Published, MCP Papers in Press, November 4, 2008, DOI 10.1074/mcp.M800231-MCP200
The comprehensive list of proteins has been deposited in the PRoteomics IDentifications (PRIDE) database under accession number 3701.
The abbreviations used are: HD, Huntington disease; CSF, cerebrospinal fluid; IPI, International Protein Index; AMT, accurate mass and time; LTQ-FT, hybrid linear ion trap-Fourier transform ICR mass spectrometer designed by Thermo Finnigan; LTQ OrbiTrap XL, mass spectrometer by Thermo Finnigan that is based on LTQ XL™ linear ion trap and the patented Orbitrap™ technology; HCT-Ultra ion trap, High-Capacity Trap (HCT™) mass spectrometer system by Bruker Daltonics; mzXML, XML (extensible markup language)-based common file format for proteomics mass spectrometric data; FDR, false discovery rate; MAS5, Affymetrix microarray suite version 5 software; GSEA, gene set enrichment analysis; IS, internal standard; BA, Brodmann area; MA plots, intensity-dependent ratio plots.
This work was supported by the High Q Foundation, Inc. The Canary Foundation provided financial support for publishing and disseminating data.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.The Huntington's Disease Collaborative Research Group ( 1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 2.Hodges, A., Strand, A. D., Aragaki, A. K., Kuhn, A., Sengstag, T., Hughes, G., Elliston, L. A., Hartog, C., Goldstein, D. R., Thu, D., Hollingsworth, Z. R., Collin, F., Synek, B., Holmans, P. A., Young, A. B., Wexler, N. S., Delorenzi, M., Kooperberg, C., Augood, S. J., Faull, R. L., Olson, J. M., Jones, L., and Luthi-Carter, R. ( 2006) Regional and cellular gene expression changes in human Huntington's disease brain. Hum. Mol. Genet. 15, 965–977 [DOI] [PubMed] [Google Scholar]
- 3.Chan, E. Y., Luthi-Carter, R., Strand, A., Solano, S. M., Hanson, S. A., DeJohn, M. M., Kooperberg, C., Chase, K. O., DiFiglia, M., Young, A. B., Leavitt, B. R., Cha, J. H., Aronin, N., Hayden, M. R., and Olson, J. M. ( 2002) Increased huntingtin protein length reduces the number of polyglutamine-induced gene expression changes in mouse models of Huntington's disease. Hum. Mol. Genet. 11, 1939–1951 [DOI] [PubMed] [Google Scholar]
- 4.Strand, A. D., Aragaki, A. K., Shaw, D., Bird, T., Holton, J., Turner, C., Tapscott, S. J., Tabrizi, S. J., Schapira, A. H., Kooperberg, C., and Olson, J. M. ( 2005) Gene expression in Huntington's disease skeletal muscle: a potential biomarker. Hum. Mol. Genet. 14, 1863–1876 [DOI] [PubMed] [Google Scholar]
- 5.Luthi-Carter, R., Hanson, S. A., Strand, A. D., Bergstrom, D. A., Chun, W., Peters, N. L., Woods, A. M., Chan, E. Y., Kooperberg, C., Krainc, D., Young, A. B., Tapscott, S. J., and Olson, J. M. ( 2002) Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum. Mol. Genet. 11, 1911–1926 [DOI] [PubMed] [Google Scholar]
- 6.Luthi-Carter, R., Strand, A., Peters, N. L., Solano, S. M., Hollingsworth, Z. R., Menon, A. S., Frey, A. S., Spektor, B. S., Penney, E. B., Schilling, G., Ross, C. A., Borchelt, D. R., Tapscott, S. J., Young, A. B., Cha, J. H., and Olson, J. M. ( 2000) Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum. Mol. Genet. 9, 1259–1271 [DOI] [PubMed] [Google Scholar]
- 7.Zabel, C., Chamrad, D. C., Priller, J., Woodman, B., Meyer, H. E., Bates, G. P., and Klose, J. ( 2002) Alterations in the mouse and human proteome caused by Huntington's disease. Mol. Cell. Proteomics 1, 366–375 [DOI] [PubMed] [Google Scholar]
- 8.Zabel, C., and Klose, J. ( 2004) Influence of Huntington's disease on the human and mouse proteome. Int. Rev. Neurobiol. 61, 241–283 [DOI] [PubMed] [Google Scholar]
- 9.Kaltenbach, L. S., Romero, E., Becklin, R. R., Chettier, R., Bell, R., Phansalkar, A., Strand, A., Torcassi, C., Savage, J., Hurlburt, A., Cha, G. H., Ukani, L., Chepanoske, C. L., Zhen, Y., Sahasrabudhe, S., Olson, J., Kurschner, C., Ellerby, L. M., Peltier, J. M., Botas, J., and Hughes, R. E. ( 2007) Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 3, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalrymple, A., Wild, E. J., Joubert, R., Sathasivam, K., Bjorkqvist, M., Petersen, A., Jackson, G. S., Isaacs, J. D., Kristiansen, M., Bates, G. P., Leavitt, B. R., Keir, G., Ward, M., and Tabrizi, S. J. ( 2007) Proteomic profiling of plasma in Huntington's disease reveals neuroinflammatory activation and biomarker candidates. J. Proteome Res. 6, 2833–2840 [DOI] [PubMed] [Google Scholar]
- 11.Wild, E. J., Petzold, A., Keir, G., and Tabrizi, S. J. ( 2007) Plasma neurofilament heavy chain levels in Huntington's disease. Neurosci. Lett. 417, 231–233 [DOI] [PubMed] [Google Scholar]
- 12.Sapp, E., Kegel, K. B., Aronin, N., Hashikawa, T., Uchiyama, Y., Tohyama, K., Bhide, P. G., Vonsattel, J. P., and DiFiglia, M. ( 2001) Early and progressive accumulation of reactive microglia in the Huntington disease brain. J. Neuropathol. Exp. Neurol. 60, 161–172 [DOI] [PubMed] [Google Scholar]
- 13.Myers, R. H., Vonsattel, J. P., Paskevich, P. A., Kiely, D. K., Stevens, T. J., Cupples, L. A., Richardson, E. P., Jr., and Bird, E. D. ( 1991) Decreased neuronal and increased oligodendroglial densities in Huntington's disease caudate nucleus. J. Neuropathol. Exp. Neurol. 50, 729–742 [DOI] [PubMed] [Google Scholar]
- 14.Strand, T., Alling, C., Karlsson, B., Karlsson, I., and Winblad, B. ( 1984) Brain and plasma proteins in spinal fluid as markers for brain damage and severity of stroke. Stroke 15, 138–144 [DOI] [PubMed] [Google Scholar]
- 15.Boesenberg-Grosse, C., Schulz-Schaeffer, W. J., Bodemer, M., Ciesielczyk, B., Meissner, B., Krasnianski, A., Bartl, M., Heinemann, U., Varges, D., Eigenbrod, S., Kretzschmar, H. A., Green, A., and Zerr, I. ( 2006) Brain-derived proteins in the CSF: do they correlate with brain pathology in CJD? BMC Neurol. 6, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verbeek, M. M., De Jong, D., and Kremer, H. P. ( 2003) Brain-specific proteins in cerebrospinal fluid for the diagnosis of neurodegenerative diseases. Ann. Clin. Biochem. 40, 25–40 [DOI] [PubMed] [Google Scholar]
- 17.Davidsson, P., Ekman, R., and Blennow, K. ( 1997) A new procedure for detecting brain-specific proteins in cerebrospinal fluid. J. Neural Transm. 104, 711–720 [DOI] [PubMed] [Google Scholar]
- 18.Vandvik, B. ( 1977) Oligoclonal IgG and free light chains in the cerebrospinal fluid of patients with multiple sclerosis and infectious diseases of the central nervous system. Scand. J. Immunol. 6, 913–922 [DOI] [PubMed] [Google Scholar]
- 19.Pan, S., Wang, Y., Quinn, J. F., Peskind, E. R., Waichunas, D., Wimberger, J. T., Jin, J., Li, J. G., Zhu, D., Pan, C., and Zhang, J. ( 2006) Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J. Proteome Res. 5, 2769–2779 [DOI] [PubMed] [Google Scholar]
- 20.Verbeek, M. M., Willemsen, M. A., and Bloem, B. R. ( 2005) Diagnosis in cerebrospinal fluid: possible applications in neurological practice. Ned. Tijdschr. Geneeskd. 149, 1833–1838 [PubMed] [Google Scholar]
- 21.Thongboorkerd, V. ( 2007) Proteomics of human body fluids: principles methods and applications, p. 270, Humana Press, New Jersey
- 22.Zetterberg, H., Pedersen, M., Lind, K., Svensson, M., Rolstad, S., Eckerstrom, C., Syversen, S., Mattsson, U. B., Ysander, C., Mattsson, N., Nordlund, A., Vanderstichele, H., Vanmechelen, E., Jonsson, M., Edman, A., Blennow, K., and Wallin, A. ( 2007) Intra-Individual stability of CSF biomarkers for Alzheimer's disease over two years. J. Alzheimer's Dis. 12, 255–260 [DOI] [PubMed] [Google Scholar]
- 23.Finehout, E. J., Franck, Z., Choe, L. H., Relkin, N., and Lee, K. H. ( 2007) Cerebrospinal fluid proteomic biomarkers for Alzheimer's disease. Ann. Neurol. 61, 120–129 [DOI] [PubMed] [Google Scholar]
- 24.MacLean, B., Eng, J. K., Beavis, R. C., and McIntosh, M. ( 2006) General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics 22, 2830–2832 [DOI] [PubMed] [Google Scholar]
- 25.Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. ( 2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 26.Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. ( 2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 27.Faca, V., Coram, M., Phanstiel, D., Glukhova, V., Zhang, Q., Fitzgibbon, M., McIntosh, M., and Hanash, S. ( 2006) Quantitative analysis of acrylamide labeled serum proteins by LC-MS/MS. J. Proteome Res. 5, 2009–2018 [DOI] [PubMed] [Google Scholar]
- 28.Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., Paulovich, A., Pomeroy, S. L., Golub, T. R., Lander, E. S., and Mesirov, J. P. ( 2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge, X., Yamamoto, S., Tsutsumi, S., Midorikawa, Y., Ihara, S., Wang, S. M., and Aburatani, H. ( 2005) Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 86, 127–141 [DOI] [PubMed] [Google Scholar]
- 30.Pan, S., Zhu, D., Quinn, J. F., Peskind, E. R., Montine, T. J., Lin, B., Goodlett, D. R., Taylor, G., Eng, J., and Zhang, J. ( 2007) A combined dataset of human cerebrospinal fluid proteins identified by multi-dimensional chromatography and tandem mass spectrometry. Proteomics 7, 469–473 [DOI] [PubMed] [Google Scholar]
- 31.Huntington Study Group ( 1996) Unified Huntington's Disease Rating Scale: reliability and consistency. Mov. Disord. 11, 136–142 [DOI] [PubMed] [Google Scholar]
- 32.Turck, C. W., Maccarrone, G., Sayan-Ayata, E., Jacob, A. M., Ditzen, C., Kronsbein, H., Birg, I., Doertbudak, C. C., Haegler, K., Lebar, M., Teplytska, L., Kolb, N., Uwaje, N., and Zollinger, R. ( 2005) The quest for brain disorder biomarkers. J. Med. Investig. 52, (suppl.) 231–235 [DOI] [PubMed] [Google Scholar]
- 33.Pedrioli, P. G., Eng, J. K., Hubley, R., Vogelzang, M., Deutsch, E. W., Raught, B., Pratt, B., Nilsson, E., Angeletti, R. H., Apweiler, R., Cheung, K., Costello, C. E., Hermjakob, H., Huang, S., Julian, R. K., Kapp, E., McComb, M. E., Oliver, S. G., Omenn, G., Paton, N. W., Simpson, R., Smith, R., Taylor, C. F., Zhu, W., and Aebersold, R. ( 2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 34.Elias, J. E., and Gygi, S. P. ( 2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 35.Old, W. M., Meyer-Arendt, K., Aveline-Wolf, L., Pierce, K. G., Mendoza, A., Sevinsky, J. R., Resing, K. A., and Ahn, N. G. ( 2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 36.Bellew, M., Coram, M., Fitzgibbon, M., Igra, M., Randolph, T., Wang, P., May, D., Eng, J., Fang, R., Lin, C., Chen, J., Goodlett, D., Whiteaker, J., Paulovich, A., and McIntosh, M. ( 2006) A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics 22, 1902–1909 [DOI] [PubMed] [Google Scholar]
- 37.May, D., Fitzgibbon, M., Liu, Y., Holzman, T., Eng, J., Kemp, C. J., Whiteaker, J., Paulovich, A., and McIntosh, M. ( 2007) A platform for accurate mass and time analyses of mass spectrometry data. J. Proteome Res. 6, 2685–2694 [DOI] [PubMed] [Google Scholar]
- 38.Pepe, M. S., and Longton, G. ( 2005) Standardizing diagnostic markers to evaluate and compare their performance. Epidemiology 16, 598–603 [DOI] [PubMed] [Google Scholar]
- 39.Zapala, M. A., Hovatta, I., Ellison, J. A., Wodicka, L., Del Rio, J. A., Tennant, R., Tynan, W., Broide, R. S., Helton, R., Stoveken, B. S., Winrow, C., Lockhart, D. J., Reilly, J. F., Young, W. G., Bloom, F. E., Lockhart, D. J., and Barlow, C. ( 2005) Adult mouse brain gene expression patterns bear an embryologic imprint. Proc. Natl. Acad. Sci. U. S. A. 102, 10357–10362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilcoxon, F. ( 1945) Individual comparisons by ranking methods. Int. Biometric Soc. 1, 80–83 [Google Scholar]
- 41.Siegel, S. ( 1956) Nonparametric Statistics for the Behavioral Sciences, pp. 75–83, McGraw-Hill, New York
- 42.Aebersold, R., and Mann, M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 43.Jongeneel, C. V., Delorenzi, M., Iseli, C., Zhou, D., Haudenschild, C. D., Khrebtukova, I., Kuznetsov, D., Stevenson, B. J., Strausberg, R. L., Simpson, A. J., and Vasicek, T. J. ( 2005) An atlas of human gene expression from massively parallel signature sequencing (MPSS). Genome Res. 15, 1007–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi, G., Schultz, M. J., Levi, M., and van der Poll, T. ( 2006) The relationship between inflammation and the coagulation system. Swiss Med. Wkly. 136, 139–144 [DOI] [PubMed] [Google Scholar]
- 45.Leblhuber, F., Walli, J., Jellinger, K., Tilz, G. P., Widner, B., Laccone, F., and Fuchs, D. ( 1998) Activated immune system in patients with Huntington's disease. Clin. Chem. Lab. Med. 36, 747–750 [DOI] [PubMed] [Google Scholar]
- 46.Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. ( 1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graveland, G. A., Williams, R. S., and DiFiglia, M. ( 1985) Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington's disease. Science 227, 770–773 [DOI] [PubMed] [Google Scholar]
- 48.Bluestein, H. G., and Zvaifler, N. J. ( 1983) Antibodies reactive with central nervous system antigens. Hum. Pathol. 14, 424–428 [DOI] [PubMed] [Google Scholar]
- 49.Hoffman, S. A., Arbogast, D. N., Day, T. T., Shucard, D. W., and Harbeck, R. J. ( 1983) Permeability of the blood cerebrospinal fluid barrier during acute immune complex disease. J. Immunol. 130, 1695–1698 [PubMed] [Google Scholar]
- 50.Tourtellotte, W. W., and Ma, B. I. ( 1978) Multiple sclerosis: the blood-brain-barrier and the measurement of de novo central nervous system IgG synthesis. Neurology 28, 76–83 [DOI] [PubMed] [Google Scholar]
- 51.Correale, J., and Villa, A. ( 2007) The blood-brain-barrier in multiple sclerosis: functional roles and therapeutic targeting. Autoimmunity 40, 148–160 [DOI] [PubMed] [Google Scholar]
- 52.Rhodin, J. A., Thomas, T. N., Clark, L., Garces, A., and Bryant, M. ( 2003) In vivo cerebrovascular actions of amyloid beta-peptides and the protective effect of conjugated estrogens. J. Alzheimer's Dis. 5, 275–286 [DOI] [PubMed] [Google Scholar]
- 53.Han, H. S., and Suk, K. ( 2005) The function and integrity of the neurovascular unit rests upon the integration of the vascular and inflammatory cell systems. Curr. Neurovasc. Res. 2, 409–423 [DOI] [PubMed] [Google Scholar]
- 54.Jovanova-Nesic, K., and Shoenfeld, Y. ( 2007) Autoimmunity in the brain: the pathogenesis insight from cell biology. Ann. N. Y. Acad. Sci. 1107, 142–154 [DOI] [PubMed] [Google Scholar]
- 55.Storey, J. D., and Tibshirani, R. ( 2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.