Abstract
A systematic study of posttranslational modifications of the estrogen receptor isolated from the MCF-7 human breast cancer cell line is reported. Proteolysis with multiple enzymes, mass spectrometry, and tandem mass spectrometry achieved very high sequence coverage for the full-length 66-kDa endogenous protein from estradiol-treated cell cultures. Nine phosphorylated serine residues were identified, three of which were previously unreported and none of which were previously observed by mass spectrometry by any other laboratory. Two additional modified serine residues were identified in recombinant protein, one previously reported but not observed here in endogenous protein and the other previously unknown. Although major emphasis was placed on identifying new phosphorylation sites, N-terminal loss of methionine accompanied by amino acetylation and a lysine side chain acetylation (or possibly trimethylation) were also detected. The use of both HPLC-ESI and MALDI interfaced to different mass analyzers gave higher sequence coverage and identified more sites than could be achieved by either method alone. The estrogen receptor is critical in the development and progression of breast cancer. One previously unreported phosphorylation site identified here was shown to be strongly dependent on estradiol, confirming its potential significance to breast cancer. Greater knowledge of this array of posttranslational modifications of estrogen receptor, particularly phosphorylation, will increase our understanding of the processes that lead to estradiol-induced activation of this protein and may aid the development of therapeutic strategies for management of hormone-dependent breast cancer.
The α-isoform of the estrogen receptor (ERα)1 is a 66-kDa nuclear transcription factor that mediates transcriptional regulation of genes involved in cell proliferation and differentiation and plays a pivotal role in the development and progression of breast cancer (1–3). Consequently the development of therapies for management of hormone-dependent breast cancer has targeted signaling pathways based on modulation of ERα activity (4–6). Current knowledge and understanding of ERα activity is derived from over 30 years of accumulated scientific evidence that has sought to delineate ER-mediated mechanisms and signaling pathways under pathological conditions modeled in cultured breast cancer cell lines. Because the function or activity of a protein may depend strongly on the presence of posttranslational modifications (PTMs), significant research has focused on detection and quantitation of such modifications in ERα. The constellation of PTMs on a protein constitutes a molecular code that may dictate protein conformation, localization, and function. Thus biological inference based on ERα structure requires a comprehensive study of all possible PTMs on the constituent amino acids.
Modifications to ERα reported to date are listed in Table I. Two of the most common and important PTMs that modulate the activity of ERα are redox-based modifications of cysteine residues (7–12) and phosphorylation of serine, threonine, and tyrosine residues (13). Biochemical techniques used previously to map ERα phosphorylation sites utilized radioactive labeling with 32P (14–16), Edman degradation, deletion and/or point mutations, and Western blots (17–19). Mutational analysis in combination with estrogen response element-luciferase reporter assays can validate the functional relevance of phosphorylation sites (16, 20). Although these techniques have provided substantial information regarding PTMs in ERα, they have several disadvantages. Autoradiography using radioisotope labeling (32P) lacks specificity, Edman degradation requires large amounts of purified protein and is not applicable to proteins/peptides with N-terminal acetylation, and quantitation of PTMs by Western blot analysis relies on prior knowledge of the type and position of specific modifications and on the availability of high quality antibodies. Deletional and mutational analyses on transfected rather than endogenous protein are laborious and time-consuming. Moreover discrepancies in the literature concerning ERα phosphorylation may arise from the use of different promoters in the reporter constructs (13).
Table I.
Posttranslational modifications previously reported in ERα
| Residue | Modification | Ref. |
|---|---|---|
| Serine 10 | O-Linked N-acetylglucosamine | Cheng and Hart (33) |
| Threonine 50 | O-Linked N-acetylglucosamine | Cheng and Hart (33) |
| Serine 102 | Phosphorylation | Medunjanin et al. (18) |
| Serine 104 | Phosphorylation | Le Goff et al. (29) |
| Serine 106 | Phosphorylation | Le Goff et al. (29) |
| Serine 118 | Phosphorylation | Ali et al. (34) |
| Serine 154 | Phosphorylation | Britton et al. (27) |
| Serine 167 | Phosphorylation | Arnold et al. (14) |
| Serine 236 | Phosphorylation | Chen et al. (15) |
| Various cysteine residues | S-Nitrosylation | Garban et al. (35) |
| Cysteines 221, 227, 237, and 240 | Thiol oxidation | Whittal et al. (11) |
| Lysines between 251 and 305 | Sumoylation | Sentis et al. (36) |
| Lysine 266 and lysine 268 | Acetylation | Kim et al. (37) |
| Lysine 299, 302, and 303 | Acetylation | Wang et al. (16) |
| Lysine 302 | Methylation | Subramanian et al. (28) |
| Serine 305 | Phosphorylation | Wang et al. (30) |
| Threonine 311 | Phosphorylation | Lee and Bai (31) |
| Cysteine 447 | S-Palmitoylation (C16) | Marino et al. (38) |
| Tyrosine 537 | Phosphorylation | Arnold et al. (32) |
| Threonine 575 | O-Linked N-acetylglucosamine | Jiang and Hart (39) |
| Lysines between 302 and 534 | Ligand-independent ubiquitination | Tateishi et al. (40) |
| Residues 535–595 (lysine 581) | Ligand-dependent ubiquitination | Lonard et al. (41) |
Because of its selectivity and sensitivity, MS/MS has emerged as a powerful tool for the analysis of PTMs such as phosphorylation (21–23). The analysis of phosphopeptides benefits from the use of multiple ionization methods and instrumentation platforms as ESI and MALDI interfaced with different tandem mass analyzers provide complementary information (21). Orthogonal Q-TOF and linear quadrupole trap (LTQ) instruments are widely used for ESI analysis, and several tandem instruments have been developed for or adapted to MALDI methods, including the LTQ (24–26). We previously utilized vacuum (v) MALDI-LTQ to optimize quantitation of cysteine oxidation within the DNA-binding domain (DBD) of recombinant ERα (7). More recently we enriched and immunoaffinity-purified endogenous ERα derived from the human breast cancer cell line MCF-7 in quantities compatible with tandem MS analysis and identified a novel phosphorylation site, Ser154. vMALDI-MSn confirmed Ser154 phosphorylation in human breast cancer cells grown under both ligand-dependent and ligand-independent conditions (27). Coincidentally we also observed known phosphorylation sites Ser118 and Ser167 not previously confirmed by MS methods.
Here we report a comprehensive MS study of ERα to achieve very high sequence coverage for the full-length 66-kDa endogenous protein from estradiol-treated MCF-7 cell cultures with particular emphasis on identifying new phosphorylation sites. Using multiple reaction monitoring (MRM)/MS we demonstrated that one previously unreported phosphorylation site is strongly dependent on estradiol, confirming its potential significance to ER activation and breast cancer. This study is the first to describe extensive mass spectrometric sequencing and phosphopeptide mapping of full-length ERα derived from a human breast cancer cell line.
EXPERIMENTAL PROCEDURES
Reagents and Chemicals—
These were as described previously (27) except that human recombinant ERα (rERα) was from Invitrogen, and sequencing grade chymotrypsin, Lys-C, Asp-N, and Glu-C were from Roche Diagnostics.
Cell Culture and Sample Preparation—
The human breast cancer cell line MCF-7 was obtained from the American Type Culture Collection (Manassas, VA). Protocols for cell culture and extraction, immunopurification, and in-gel digestion of ERα from 20 15-cm plates of cells were as described previously (27) except that for each experiment in-gel digestion was with one of the following enzymes: chymotrypsin (300 ng; 37 °C), trypsin (150 ng; 37 °C), Glu-C (250 ng; 25 °C), Lys-C (400 ng; 37 °C), Asp-N (280 ng; 37 °C), or a combination of the last two. The resultant peptides were extracted by vortexing and sonication with 50% ACN, 5% formic acid; vacuum centrifuged to remove most of the solvent; reconstituted in 20 μl of 0.1% formic acid; and stored at −80 °C.
On-bead Proteolytic Digestion—
After immunoprecipitation and PBS washes the immune complex was transferred to a siliconized 1.5-ml Eppendorf tube and washed twice in 100 mm Tris, pH 7, by gentle rotation for 2 min at room temperature and three times in 25 mm NH4HCO3 with centrifugation between washes. The pelleted beads were suspended in a further 150 μl of NH4HCO3 and 170 ng of sequence grade trypsin (10 μl of 17 ng/μl stock solution) and then incubated overnight at 37 °C with shaking at 1,200 rpm. Eppendorf tubes were centrifuged at 12,000 rpm, and the supernatant was transferred to low binding polymer technology 0.65-ml microcentrifuge tubes (PGC Scientifics). Ten microliters of acetonitrile and 1 μl of 10% formic acid were added to the peptide solution, which was then evaporated down to ∼15 μl. Each sample was divided into three 5-μl aliquots and stored at −80 °C until used for mass spectrometry.
Peptide Mass Fingerprinting by MALDI-TOF—
Peptides to be analyzed by MALDI were desalted with C18 ZipTips (Millipore), and then 1 μl of peptide solution and 1 μl of 2,5-dihydroxybenzoic acid or α-cyano-4-hydroxycinnamic acid matrix (50 mg/ml in 4:1 ACN, 0.6% phosphoric acid) were mixed, deposited on a stainless steel sample plate, and air-dried. Alternatively 1 μl of peptide solution was mixed with 1 μl of 50% ACN, 0.5% TFA; spotted on the MALDI plate; and dried. This addition of solvent and drying was repeated three more times, and then 1 μl of matrix solution was added and dried. 1 μl of 0.1% TFA was added and aspirated off by applying a tissue. Mass spectra were recorded on a Voyager-DE STR Plus instrument (Applied Biosystems, Framingham, MA) with 337 nm laser radiation, positive ion delayed extraction, reflector mode, and 20-kV accelerating voltage.
Peptide Identification by Tandem Mass Spectrometry—
Peptide mixtures were further analyzed on a vMALDI-LTQ linear ion trap (Thermo Fisher, San Jose, CA) using both manual and automated data acquisition. Both methods used “Tune plus” using the “crystal-positioning system” and low threshold settings for “auto spectrum filter.” MS2 spectra were typically collected using threshold values of 500 counts for MS and 150 counts for MS2 with “auto gain control” turned on, limiting the number of ions admitted to the trap. A parent mass isolation width of 3 m/z units and fragmentation settings of activation Q = 0.25, activation time of 30 ms, and relative collision energy of 35% were used. For phosphopeptide mapping, potential peptide peaks were interrogated by MS/MS for the neutral loss of phosphoric acid, and (MH − 98)+ peaks were selected for MS3. Loss of phosphoric acid converted serine residues to dehydroalanine (dS). Reversed phase nano-HPLC-MS/MS (LC-MS/MS) was performed using an Ultimate HPLC instrument (Dionex, Sunnyvale, CA) with a C18 analytical column directly connected to a Q-STAR Pulsar I quadrupole orthogonal TOF mass spectrometer (MDS Sciex, Concorde, Canada) as described previously (27).
LC-MS/MS and vMALDI-LTQ fragment ion data were searched against a database constructed for human full-length ERα using Mascot in-house licensed version 2.2 (Matrix Sciences, London, UK) with the “no enzyme” option. LC-MS/MS data were also searched with Paragon (Protein Pilot 2.0, Applied Biosystems).
LC-MRM/MS—
Samples were analyzed by nano-LC-MRM/MS on a 4000 QTRAP hybrid triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems). Chromatography was performed using an Eksigent (Dublin, CA) NanoLC-2D LC system with buffer A (0.1% formic acid) and buffer B (90% acetonitrile in 0.1% formic acid). Digests were loaded at 20 μl/min in buffer A onto a 5-mm × 300-μm reversed phase C18 column (5 μm, 100 Å; Dionex) and eluted at 300 nl/min with a 75-μm-inner diameter Integrafrit column (New Objective, Woburn, MA) packed in house with 10–12 cm of ReproSil-Pur C18-AQ 3-μm reversed phase resin (Dr. Maisch, GmbH) with a gradient of 2–70% B over 32 min. Peptides were ionized using a PicoTip emitter (75 μm, 15-μm tip; New Objective). Data acquisition was performed with ion spray voltage at 2450 V, curtain gas pressure at 10 p.s.i., nebulizer gas at 20 p.s.i., and interface heater temperature at 150 °C. Collision energy, declustering potential, and collision cell exit potential were optimized using recombinant ER for maximum sensitivity. MRM transitions were monitored and acquired at unit resolution both in the first and third quadrupoles (Q1 and Q3) to maximize specificity. The y7 transition was monitored at Q1 670.9 m/z and Q3 785.5 m/z for the unmodified peptide and at Q1 710.9 m/z and Q3 865.4 m/z for the phosphorylated peptide with dwell times of 10 and 320 ms, respectively. Furthermore although the y7 transition gave the most intense signal, MRM transitions y8 and y7 − phosphoric acid were also assayed to confirm the retention time of the Ser294 peptides. A minimum of nine data points were collected per peak.
RESULTS
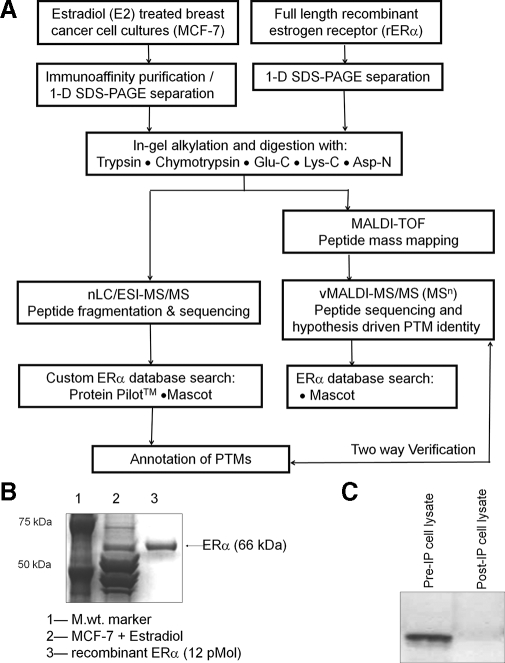
The overall analytical strategy (Fig. 1A) required optimizing immunoaffinity enrichment of human ERα from estradiol-treated MCF-7 cells for peptide analysis by tandem mass spectrometry. Estradiol treatment of confluent MCF-7 cells for 30 min, overnight incubation of cell lysates with agarose-coupled ERα antibody, and one-dimensional gel electrophoresis separation yielded endogenous ERα protein at 66 kDa (Fig. 1B). Recombinant protein (rERα) was used to confirm this identification and to model in-gel digestions. Comparison with 13 pmol of rERα indicates a yield of ∼8–10 pmol of protein/20 plates of cultured MCF-7 cells. Western blots of cell lysates before and after immunoprecipitation indicate the ERα immunocapture to be as much as 98% efficient (Fig. 1C).
Fig. 1.
A, analytical strategy for affinity purification, digestion, and sequence mapping of PTMs in immunopurified endogenous ERα. rERα was used as a standard for optimization of experimental conditions. B, Coomassie-stained one-dimensional (1-D) gel electrophoresis of affinity-purified MCF-7 ERα showing co-migration with rERα at 66 kDa. C, Western blots of estradiol (E2)-treated MCF-7 cell lysates before and after immunopurification (IP) demonstrate the effectiveness of the enrichment protocol.
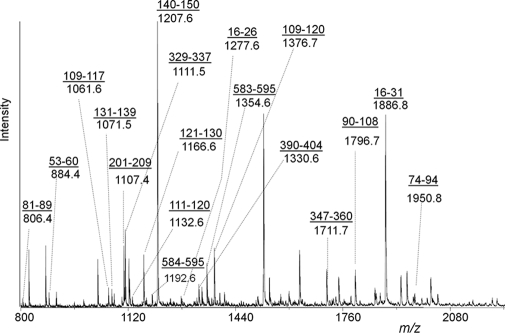
Peptidecutter (ExPASy) was used to predict peptide fragments of a size and nature suitable for ionization and sequencing by LC-MS/MS and/or vMALDI-MSn, showing that multiple enzymes would be required to achieve comprehensive coverage of the primary sequence. Digests were prepared using trypsin, chymotrypsin, Glu-C, Lys-C, Asp-N, and the combination of Lys-C and Asp-N and analyzed as unseparated mixtures by MALDI-TOF. Fig. 2 shows a typical peptide mass fingerprint derived from a chymotryptic digest of immunopurified MCF-7 ERα. Internally calibrated peak lists were searched against the ERα database on Swiss-Prot using Protein Prospector Version 4.5.1. Peptide sequence information was then obtained by LC-MS/MS and vMALDI-MSn. All spectra were interrogated by database searching, and sequence identifications were confirmed by visual inspection. All identified ERα peptides are listed in supplemental Table 1 and summarized in Fig. 3 with combined sequence coverage of 95%. Peptides assigned by peptide mass fingerprinting were not classified as identified unless confirmed by tandem MS. LC-MS/MS analysis yielded more peptides than vMALDI-MSn of unseparated mixtures. Chymotrypsin gave the highest sequence coverage by both LC-MS/MS and vMALDI-MS/MS (73% combined) and the highest coverage for detection of critical residues: serine (43 of 45), threonine (20 of 25), tyrosine (19 of 23), and lysine (17 of 29) but not for monitoring another important residue, cysteine. Spectra presented in the text represent previously unreported PTMs in endogenous ERα from MCF-7 cells. Spectra confirming previous reports or observations made only in recombinant protein are presented as supplemental data. Data for Ser(P)118, Ser(P)154, and Ser(P)167 have been reported previously and are not repeated here except for a Ser(P)154 peptide that models an unexpected MS/MS fragmentation.
Fig. 2.
MALDI-TOF peptide mass fingerprint of chymotrypsin-digested endogenous ERα. All underlined sequences were confirmed by MS/MS.
Fig. 3.
Coverage map for MS/MS identification of amino acid residues within MCF-7 ERα digested with various enzymes. chym, chymotrypsin; tryp, trypsin; LyAs, Lys-C/Asp-N.
Monitoring of Cysteine Residues
To facilitate detection of the 13 ERα cysteine residues, protein samples were reduced and alkylated. For the important DBD, consecutive digestion by Lys-C and Asp-N gave four major peptides, each containing two cysteine residues. Cys381 and Cys417 were not identified in Lys-C/Asp-N digests of MCF-7-derived ERα, although Cys417 was identified in rERα following chymotryptic digestion. Cys530 was observed only in Glu-C digests, and Cys381 was not observed at all. Our studies on the oxidation of cysteine residues in the DBD have been described elsewhere (8, 10, 11).
Observation of Acetylated Amino Acids
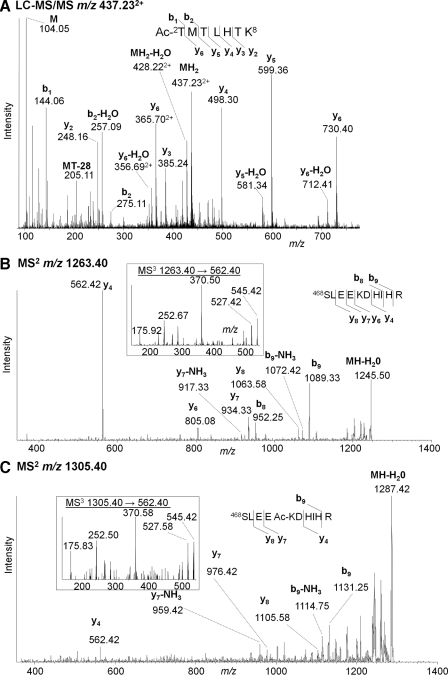
No identified peptides contained the N-terminal methionine residue predicted by the gene sequence. Broadening the search criteria to allow for loss of this residue together with N-terminal acetylation led to the identification of Ac-2TMTLHTK in tryptic and Lys-C/Asp-N digests. The LC-MS/MS spectrum of the MH22+ is illustrated in Fig. 4A: b1 and b2 ions confirm N-terminal acetylation, whereas y2 and y3 exclude the possibility of the acetyl group being located on Lys8. Ac-2TMTLHTKASGMALLHQIQGNE was also identified with a Mascot score of 72 (Glu-C digest; spectrum not shown). Thus there is strong evidence that endogenous MCF-7 ERα undergoes this common modification, which would render it blocked for Edman sequencing.
Fig. 4.
Acetylation at ERα N terminus and acetylation (or trimethylation) of Lys472. A, LC-MS/MS of MH22+ (m/z 437.23) of peptide Ac2TMTLHTK from endogenous ERα. B, vMALDI-MS/MS of 468SLEEKDHIHR (m/z 1263.4). C, the corresponding spectrum of a modified peptide (m/z 1305.4). Inset to B and C, MS3 of m/z 562.4 common to both MS/MS spectra.
Two tryptic peptides having similar MS/MS spectra differed in mass by 42 Da, suggesting another acetyl group. Alternatively this mass difference could correspond to trimethylation, which could not be differentiated with the instruments used here. Several ERα acetylation sites have been reported (Table I), but a methylation was also detected recently (28). For the current discussion, it is assumed the modification is an acetylation. vMALDI-LTQ fragmentation of the unmodified MH+ (m/z 1263.4) identified it as 468SLEEKDHIHR with sequence ions y4, y6, y7, y7−NH3, y8, b8, b8−NH3, and b9 (Fig. 4B). The putative acetylated form (m/z 1305.4) showed the same fragments (Fig. 4C), all but y4 (m/z 562.4) were shifted by 42 Da, consistent with a centrally located modification. MS3 of both y4 ions gave identical spectra for both the non-acetylated and acetylated forms (inset). Thus the acetyl group is within EKD, placing it on the side chain of Lys472.
Phosphopeptide Identification by MS/MS
Several phosphopeptides were identified in this study (Table II), some confirming previously known sites as well as several that were previously unreported. As a potential aid to phosphopeptide analysis, three Web-based algorithms, Scansite, NetPhos, and Disphos, were used to predict phosphorylation sites, but the correlation between prediction and experiment proved weak (results not shown). MS/MS analysis was performed on digested ERα peptides without any prior phosphopeptide enrichment step. Consequently it was often possible to observe both the phosphorylated and unphosphorylated species. In LC-MS/MS the phosphopeptides were consistently eluted later than the unphosphorylated forms, providing additional chromatographic confirmation of their existence. Phosphopeptides usually resulted in pairs of peptides separated by 80 Da plus yn − 98 ions for the higher mass species. Intact phosphate-containing yn ions were seen infrequently.
Table II.
Modifications identified by tandem mass spectrometry in this study in endogenous ERα from MCF-7 cells and recombinant ERα
All peptide sequences were confirmed by MS/MS. Modification sites are bold in the sequences, and lowercase m in the sequences represents oxidized methionine. Mod., modification; Msct, Mascot; Chymo., chymotrypsin; tr, retention time.
| Mod. site | Protein source | Enzyme | Peptide | Sequence | vMALDI
|
LC-MS/MS
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MH+ | Msct score | Precursor ion | Error | Msct score | tr
|
||||||
| Modified | Unmodified | ||||||||||
| ppm | min | ||||||||||
| Thr(Ac)2 | Both | Trypsin | 2–8 | M↓AcTMTLHTK↓A | 437.232+ | 33 | 25 | ||||
| Both | Glu-C | 2–22 | M↓AcTMTLHTKASGMALLHQIQGNE↓L | 775.073+ | 13 | 72 | |||||
| Ser(P)102 | MCF7 | Chymo. | 90–108 | F↓GSNGLGGFPPLNpSVSPSPL↓M | 1876.9 | 27 | |||||
| Ser(P)104 | Both | Chymo. | 90–108 | F↓GSNGLGGFPPLNSVpSPSPL↓M | 1876.9 | 25 | 938.97+2 | 21 | 9 | 28.6 | 24.8 |
| Ser(P)106 | Both | Chymo. | 90–108 | F↓GSNGLGGFPPLNSVSPpSPL↓M | 1876.9 | 24 | 938.94+2 | 1 | 17 | 26.6 | 24.8 |
| Ser(P)118 | Both | Chymo. | 109–120 | L↓mLLHPPPQLpSPF↓L | 1472.7 | 10 | 736.85+3 | 9 | 16 | 37.6 | 36.72 |
| Both | Chymo. | 109–120 | L↓MLLHPPPQLpSPF↓L | 1456.4 | 11 | 728.88+2 | 39 | 18 | 48.5 | 47.06 | |
| rER | Chymo. | 109–130 | L↓MLLHPPPQLpSPFLQPHGQQVPY↓Y | 868.82+3 | 67 | 15 | 49.8 | ||||
| rER | Glu-C | 109–130 | L↓MLLHPPPQLpSPFLQPHGQQVPY↓Y | 868.85+3 | 99 | 18 | 50.1 | ||||
| Both | Chymo. | 111–120 | L↓LHPPPQLpSPF↓L | 1212.4 | 7 | 606.77+2 | 27 | 35 | 39.8 | 37.59 | |
| MCF7 | Asp-N | 118–132 | L↓pSPFLQPHGQQVPYYL↓E | 927.54+2 | 119 | 61 | 33.1 | ||||
| Ser(P)154 | Both | Trypsin | 143–158 | R↓EAGPPAFYRPNpSDNRR↓Q | 1927.0 | 6 | 642.97+3 | 33 | 10 | 27.8 | 26.97 |
| Both | Asp-N | 143–154 | R↓EAGPPAFYRPNpS↓D | 1385.7 | |||||||
| Ser(P)167 | MCF7 | Asp-N | 155–169 | S↓DNRRQGGRERLApSTN↓D | 453.25+4 | 81 | 17.3 | 16.9 | |||
| MCF7 | Glu-C | 164–177 | E↓RLApSTNDKGSmAmE↓S | 541.61+3 | 105 | 36 | 17.9 | 17.15 | |||
| MCF7 | Trypsin | 165–180 | R↓LApSTNDKGSMAMESAK↓E | 574.29+3 | 81 | 35 | 15.2 | 13.93 | |||
| MCF7 | Trypsin | 165–180 | R↓LApSTNDKGSMAMESAK↓E (one m) | 579.57+3 | 3 | 14 | 13.5 | 12.94 | |||
| MCF7 | Trypsin | 165–180 | R↓LApSTNDKGSmAmESAK↓E | 584.90+3 | 11 | 33 | 12.7 | 12.37 | |||
| MCF7 | Chymo. | 165–177 | R↓LApSTNDKGSmAME↓S | 484.22+3 | 50 | 12 | 25.7 | ||||
| Ser(P)212 | Both | Asp-N | 203–217 | C↓EGCKAFFKRpSIQGHN↓D | 1778.9 | ||||||
| Ser(P)236 | rER | Lys-C | 236–244 | K↓pSCQACRLRK↓C | 420.19 | 6.2 | 5 | 16.1 | 15.5 | ||
| Ser(P)294 | Both | Lys-C/Asp-N | 285–299 | G↓DMRAANLWPpSPLMIK↓R | 608.15+3 | 80 | 22 | 40.1 | 38.93 | ||
| MCF7 | Chymo. | 287–296 | M↓RAANLWPpSPL↓M | 1204.8 | |||||||
| MCF7 | Trypsin | 288–299 | R↓AANLWPpSPLMIK↓R | 1420.7 | |||||||
| MCF7 | Trypsin | 288–299 | R↓AANLWPpSPLmIK↓R | 1436.7 | 718.88+2 | 35 | 20 | 23.4 | 22.44 | ||
| Lys(Ac)472 | Both | Trypsin | 468–477 | K↓SLEEAcKDHIHR↓V | 1305.7 | ||||||
| Ser(P)554 | rER | Trypsin | 549–555 | R↓LHAPTpSR↓G | 861.4 | 5 | 431.21+2 | 23 | 21 | 18.1 | 14.83 |
| rER | Chymo. | 550–568 | L↓HAPTpSRGGASVEETDQSHL↓A | 686.96+3 | 0.38 | 15 | 13.6 | 12.69 | |||
| Ser(P)559 | Both | Trypsin | 556–581 | R↓GGApSVEETDQSHLATAGSTSSHSLQK↓Y | 2665.4 | 8 | 667.10+4 | 77 | 52 | 27.1 | 26.4 |
| rER | Chymo. | 556–577 | R↓GGApSVEETDQSHLATAGSTSSH↓S | 552.9+4 | −148 | 12 | 25 | ||||
Complementary peptide information was obtained by automated vMALDI-LTQ-MS/MS experiments, initially based on MALDI-TOF-generated inclusion lists. Manual data acquisition was also used to sequence peptides with low ion counts not initially observed by MS and to isolate weak precursor ions and monitor peptides not detected by LC-MS/MS analysis. vMALDI-MSn also allowed repeated analysis of the same sample as well as extensive MSn interrogation of ions, whereas LC analysis was limited to the brief time during which a peptide eluted.
All ERα peptides containing unmodified serine, threonine, or tyrosine residues were interrogated by vMALDI-LTQ-MS/MS at 80 m/z units above the observed molecular ion even if no ions were seen at this mass. Additionally hypothetical phosphopeptide m/z values were generated by in silico proteolysis of ERα and probed for putative phospho-Ser, -Thr, and -Tyr residues in both recombinant and endogenous ERα by multiple stage vMALDI-MSn. The loss of phosphoric acid (98 Da) in the linear ion trap provided a sensitive readout for the presence of a phosphopeptide; thus m/z regions that might correspond to (MH − 98)+ ions were examined for indications of phosphorylated residues using MS3 to probe for fragment ions that correlated with the MS2 spectra derived from the corresponding unphosphorylated parent ion.
Systematic Search for Phosphorylation Sites
The N-terminal Domain—
Serine phosphorylation within the N-terminal domain containing activation factor-1 (AF-1) contributes to ERα activation and may lead to drug resistance in breast cancer treatment. We recently reported Ser154 as a new phosphorylation site within the N-terminal domain of ERα from cultured human breast cancer cells. Here we found no other new sites but observed known sites at Ser102/104/106, Ser118, and Ser167. Data for Ser(P)118 and Ser(P)167 were reported previously (27) and are not presented here.
We sought to confirm whether each of the previously reported sites Ser102/104/106 can be phosphorylated; the data is presented in supplemental Fig. 1. The analysis was on an MCF-7 ERα chymotryptic peptide containing all three residues, 90GSNGLGGFPPLNSVSPSPL. LC-MS/MS spectra in supplemental Fig. 1, A and B, are from the MH22+ ions of unphosphorylated peptide (m/z 898.95) eluting at 24.80 min and a monophosphorylated moiety (m/z 938.94) eluting at 26.56 min, i.e. differing by 80 Da. A strong y2 ion of m/z 229.15 is common to both spectra, whereas y4 at m/z 413.26 for the unphosphorylated peptide is replaced by y4 − H3PO4 (m/z 395.22) in the phosphorylated case, localizing the modification on Ser106. All y ions above y2 show the loss of phosphoric acid, but y4 alone definitively locates the site of the modification. Supplemental Fig. 1C shows a weaker MS/MS spectrum for an additional isomer eluting later (28.59 min). The y3/y4 ions show no phosphorylation, whereas y5 has lost phosphoric acid, suggesting phosphorylation of Ser104.
Crystal heterogeneity of unseparated digests can cause some separation of isomers, and different regions of the MALDI spot may yield distinct spectra. Several spectra from phosphorylated 90GSNGLGGFPPLNSVSPSPL indicated multiple isomeric forms. Supplemental Fig. 1D presents a vMALDI-LTQ-MS/MS spectrum with (MH − 98)+ as the base peak. N-terminal fragments b12 and below show no evidence of phosphate, whereas b13 and above are all bn − 98, suggesting phosphorylation at Ser102. Another spectrum (not shown) gave similar fragments, but a distinctive b13 ion (m/z 1198.2) indicated some molecules not phosphorylated at Ser102, and a y6 − 98 ion (m/z 581.2) implied phosphorylation of Ser104.
Peptides possessing both two and three phosphate groups prove that multiple serine residues can be phosphorylated. Supplemental Fig. 1E shows vMALDI-MS/MS of a doubly phosphorylated peptide of m/z 1956.5. The y11 and y11 − 98 ions confirm two phosphates C-terminal to Phe97 not involving Ser91, whereas b15 − 98 and b16 − 98 − H2O ions suggest that Ser106 is not phosphorylated. Supplemental Fig. 1F shows a triply phosphorylated species of m/z 2036.6. The stoichiometry for triple phosphorylation is clearly low, but three consecutive neutral losses of 98 Da confirm all three serine residues can be phosphorylated simultaneously.
The DBD—
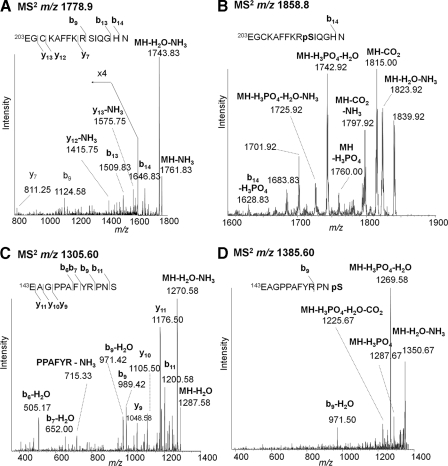
A new phosphorylation site was detected at Ser212 within the loop between the two zinc fingers of this domain. Two Asp-N peptides separated by 80 Da gave similar fragments in vMALDI-LTQ-MS/MS. The unphosphorylated MH+ (m/z 1778.9) showed losses of water and ammonia and weak backbone fragments even at increased collision energy (Fig. 5A). Fragment ions b9, b13, b14, y7, y12 − NH3, y13, and y13 − NH3 identified the peptide as 203EGCKAFFKRSQGHN with cysteine modified by iodoacetamide. The heavier mass MH+ (1858.8) gave a weaker spectrum (Fig. 5B) but showed loss of phosphoric acid (−98 Da) and additional losses of water and ammonia. An N-terminal fragment common to both spectra was identified as b14 − H2O for the unmodified peptide and b14 − H3PO4 for the phosphopeptide. The heavier molecular ion also retained phosphate but lost combinations of water, ammonia, and carbon dioxide. The peptide is clearly phosphorylated, and the only possible location for a phosphate group in this peptide is Ser212.
Fig. 5.
Identification of novel phosphorylation site Ser212 and comparison with a structurally similar phosphopeptide containing Ser(P)154. A, vMALDI-LTQ-MS/MS of MH+ for the Asp-N peptide 203EGCKAFFKRSIQGHN (m/z 1778.9) from endogenous ERα with expansion of weak fragment peaks at m/z 800–1600. B, the corresponding MH+ spectrum from the phosphorylated peptide (m/z 1858.8). C, vMALDI-LTQ-MS/MS of MH+ for the Asp-N peptide 143EAGPPAFYRPNS (m/z 1305.6) from endogenous ERα. D, the corresponding MH+ spectrum from the phosphorylated peptide (m/z 1385.6). pS, phosphoserine.
It was hypothesized that such a phosphopeptide with N-terminal glutamate might undergo the combined loss of phosphoric acid and water to form a new ionic moiety containing dehydroalanine and pyroglutamate. Identical fragmentation was observed for an analogous peptide incorporating the known Ser154 site. Fig. 5C shows MS/MS of the unmodified peptide of m/z 1305.6 with (MH − NH3 − H2O)+ as the base peak plus b11, b9, b9 − H2O, b7 − H2O, b6 − H2O, y11, y10, and y9 that confirm this peptide as 143EAGPPAFYRPNS. Fig. 5D shows the spectrum of the phosphopeptide at m/z 1385.6. Again the base peak corresponds to combined losses of phosphoric acid and water, and other prominent peaks combine losses of phosphoric acid, H2O, NH3, and CO2. The only significant backbone fragment (b9) also shows water loss; thus such fragmentation appears to characterize phosphoserine peptides with N-terminal glutamate.
A previously reported phosphorylation site falling within the second zinc finger of the DBD at Ser236 was confirmed here in rERα only. LC-MS/MS of a Lys-C digest of rERα identified the triply charged peptide 236SCQACRLRK (with iodoacetamide-derivatized cysteines) with and without serine phosphorylation; the modified form eluted half a minute later. Supplemental Fig. 2A represents the unmodified MH33+ (m/z 393.5) with fragment ions MH3 − NH33+, y62+, y72+, y82+, and y83+. Supplemental Fig. 2B shows the corresponding spectrum of the phosphorylated MH33+ (m/z 420.2) with base peak corresponding to the loss of phosphoric acid (triply charged) and y62+, y72+, and y82+ ions that locate the modification at Ser236.
The Hinge Region between the DBD and AF-2—
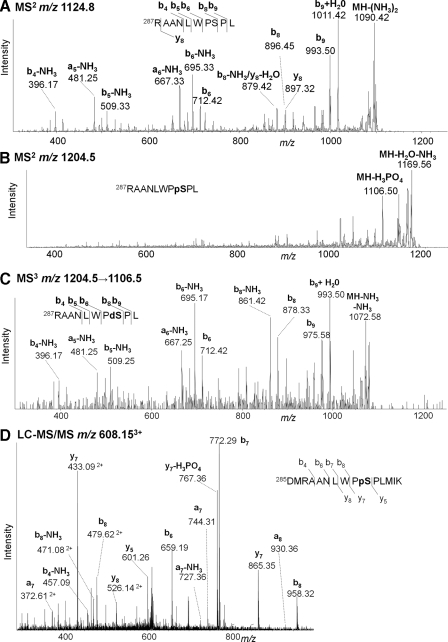
Although no phosphorylation has been reported for this region, residues 263–301, we identified Ser294 as a phosphorylation site in MCF-7 cells. Fig. 6A shows the vMALDI-LTQ-MS/MS of MH+ of a chymotryptic peptide (m/z 1124.8) for which b4 − NH3, a5 − NH3, b5 − NH3, a6 − NH3, b6, b6 − NH3, b8, b8 − NH3, b9, b9 + H2O, and MH+ − 2NH3 identify 287RAANLWPSPL. The N-terminal arginine is consistent with predominant b ions. Fig. 6B for the analogous species 80 Da heavier (m/z 1204.5) shows (MH − 98)+ at m/z 1106.5, which was selected for MS3 analysis (Fig. 6C). This gives identical fragments to MS2 of the unmodified peptide for ions b6 and below, but b8, b8 − NH3, b9, b9 + H2O, and (MH − 2NH3)+ are all 18 Da lower, confirming that the phosphate is situated between the b6 and b8 positions, i.e. Ser294. Further evidence for this new site came from a 15-residue Asp-N/Lys-C peptide, 285DMRAANLWPSPLMIK. The LC-MS/MS spectrum of MH33+ (m/z 608.15) is illustrated in Fig. 6D; Ser(P)294 is confirmed by the y5, y7, and y7 − 98 ions.
Fig. 6.
Identification of novel phosphorylation site Ser294 in endogenous MCF-7 ERα. A, vMALDI-LTQ-MS/MS of MH+ from chymotryptic peptide 287RAANLWPSPL (m/z 1124.8). B, the corresponding MH+ spectrum from the phosphorylated peptide (m/z 1204.5). C, MS3 spectrum of (MH − 98)+ (m/z 1106.5) from the phosphopeptide MH+. D, LC-MS/MS spectrum of MH33+ of phosphorylated Lys-C/Asp-N peptide 285DMRAANLWPSPLMIK (m/z 608.15). pS, phosphoserine; dS, dehydroalanine.
The Ligand-binding Domain—
The largest domain of ERα extends from residue 302 to the C terminus at residue 595 and contains AF-2. The extreme C-terminal region 552–595 (the F domain) may have a specific modulatory function that affects the agonist/antagonist effectiveness of antiestrogens and the transcriptional activity of ligand-bound ER in cells. No phosphorylation sites were identified within AF-2, but we found two novel phosphorylation sites in the F-domain at Ser554 and Ser559. The assignment of Ser554 was based only on observations made with recombinant protein, not with MCF-7 protein, so it is illustrated in supplemental Fig. 3. Panel A shows a vMALDI-MS/MS spectrum, and panel B shows an LC-MS/MS spectrum for a tryptic phosphopeptide of sequence 549LHAPTSR phosphorylated at either Thr553 or Ser554. Both spectra show strong fragment ions, including the loss of phosphoric acid. Only the vMALDI spectrum contains a strong b5 fragment ion that confirms the phosphate group to be on Ser554 rather than Thr553.
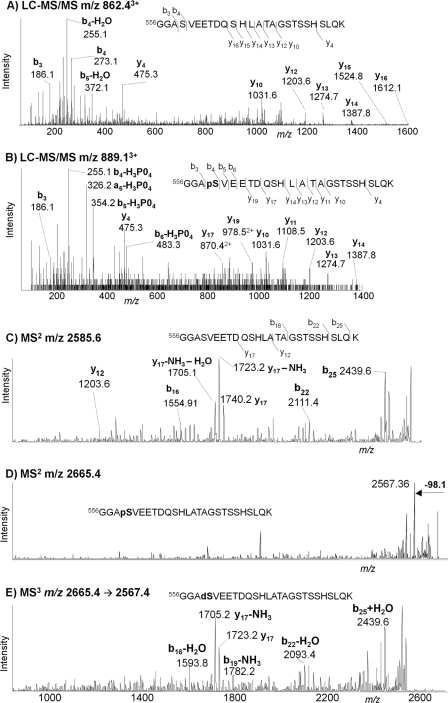
Fig. 7 shows MS/MS spectra that identify phospho-Ser559 in MCF-7-derived ERα digested with trypsin. Fig. 7A shows the LC-MS/MS spectrum of the unmodified peptide MH33+ (m/z 862.4) eluting at 26.4 min and identified as 556GGASVEETDQSHLATAGSTSSHSLQK. Fig. 7B shows the corresponding spectrum of the modified form (m/z 889.1) eluting at 27.1 min. This 26-residue peptide contains six serines and three threonines, any one of which could potentially be the phosphorylation site. Comparing the two spectra, the similarity of the y-series ions indicates that the modification is not in the C-terminal region, whereas strong b4–b7 ions show the loss of phosphoric acid, which unambiguously establishes Ser559 as the site of the modification. The same peptide was also observed by vMALDI. Fig. 7C shows the vMALDI-MS2 spectrum of the unmodified peptide at m/z 2585.3, Fig. 7D shows the species 80 Da heavier at m/z 2665.3, and Fig. 7E represents the MS3 spectrum of the (M − 98)+ at m/z 2567.2. Although these spectra show numerous fragment ions that clearly define the sequence, no unique ions identify the specific location of the phosphate group, unlike the LC-MS/MS data.
Fig. 7.
Identification of novel phosphorylation site Ser559 in endogenous MCF-7 ERα. A, LC-MS/MS spectrum of MH33+ of tryptic peptide 556GGASVEETDQSHLATAGSTSSHSLQK (m/z 889.1) eluting at 26.4 min. B, LC-MS/MS spectrum of MH33+ of the analogous phosphorylated peptide (m/z 889.1) eluting at 27.1 min. C, vMALDI-LTQ-MS/MS spectrum of MH+ of the unmodified peptide (m/z 2585.3). D, the corresponding spectrum of MH+ of the phosphorylated peptide (m/z 2665.3). E, MS3 spectrum of (MH − 98)+ (m/z 2567.2) from the phosphopeptide. pS, phosphoserine; dS, dehydroalanine.
Quantitation of Ser294 Phosphorylation by LC-MRM/MS
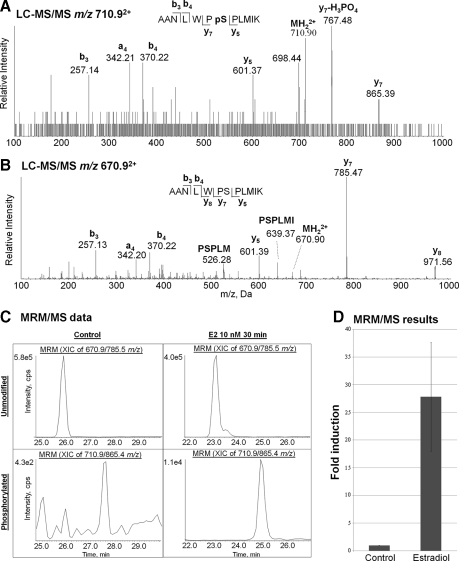
To determine the biological relevance of these previously unreported phosphorylation sites an MRM/MS assay was developed to monitor any change in Ser294 phosphorylation following estradiol treatment of MCF-7 cells. MRM is the most sensitive and robust quantitative mass spectrometric method because of (i) coincident detection of both the precursor ion (Q1) and a designated fragment ion (Q3) and (ii) rapid cycling that allows each transition to be sampled multiple times during chromatographic elution of the selected species. Fragment ions with m/z values greater than that of the precursor ion lead to lower noise levels, which requires that the fragment ion has a lower charge state than the precursor. The LC-MS/MS spectra in Fig. 8, A and B, of tryptic peptide 288AANLWPSPLMIK with and without phosphorylation of Ser294 show that the most abundant fragment ions that fit this criterion are y7 − phosphoric acid (m/z 767.48) from the doubly charged Ser294 phosphorylated peptide (m/z 710.9) and y7 (m/z 785.47) from the doubly charged unmodified peptide (m/z 670.9).
Fig. 8.
LC-MS/MS of tryptic peptides from endogenous MCF-7 ERα. A, spectrum of MH22+ (m/z 710.9) of peptide 288AANLWPpSPLMIK (where pS is phosphoserine). B, spectrum of MH22+ (m/z 670.9) of the analogous unphosphorylated peptide. C, MRM/MS analysis of Ser294 induction following estradiol (E2) treatment showing chromatograms of the y7 Q1/Q3 MRM transitions for the unmodified (670.9/785.5) and phosphorylated (710.9/865.4) peptide from control and estradiol-treated (10 nm; 30 min) cells. D, comparing peak areas for the phosphorylated with unmodified peptides demonstrates that estradiol induces Ser294 oxidation 28-fold. The error bar represents the standard deviation for three biological replicates. XIC, extracted ion chromatogram; cps, counts/s.
Because of the high specificity of MRM, gel isolation of the immunoprecipitated ERα from background antibody proteins, protein A, and other contaminants was not required before trypsin digestion. This eliminated the sample losses typical of in-gel digestion and also minimized methionine oxidation, significantly increasing the peptide signal strength as this was not split between the oxidized and non-oxidized forms. Fig. 8C illustrates MRM data for the doubly protonated molecular ion fragmenting to y7 for unmodified and phosphorylated Ser294 peptides from immunoprecipitated ERα obtained from control conditions and following 30-min incubation with 10 nm estradiol. As expected, the phosphopeptide eluted 1–2 min later than the unmodified peptide. The increased peak heights following estradiol treatment relative to control conditions clearly indicate significant induction of phospho-Ser294. The -fold induction shown in Fig. 8D was obtained by dividing the phospho-Ser294 integrated peak areas by the corresponding Ser294 integrated peak areas normalized to the value obtained for the control sample. Three biological replicates demonstrated a mean 27.8-fold induction of phosphorylation following estradiol treatment relative to control with a standard deviation of 9.8.
DISCUSSION
Previous comprehensive mapping of ERα and its PTMs by mass spectrometry had been hampered by the inability to purify/enrich sufficient endogenous protein. The optimization of ERα immunoaffinity enrichment from MCF-7 cells for MS analysis reported here is a significant achievement in ERα biochemical studies as demonstrated by our recent observation of the novel phosphorylation site Ser154 in the N-terminal domain of ERα (27).
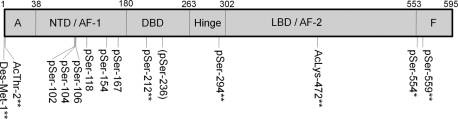
Several PTMs of ERα summarized in Table II and Fig. 9 were identified in this tandem mass spectrometry study, which focused on detection and characterization of phosphorylation sites. The majority of phosphorylation sites identified here occur in endogenous protein isolated from the estradiol-stimulated MCF-7 human breast cancer cell line (Ser102, Ser104, Ser106, Ser118, Ser154, Ser167, Ser212, Ser294, and Ser559), although two were observed only in recombinant protein (Ser236 and Ser554). Ser154 was a novel phosphorylation site previously detected by us using the same methods, but several other sites had not been reported previously: namely Ser212, Ser294, Ser554, and Ser559. Most or all of these serine phosphorylations are likely estradiol-dependent, although evaluation of the induction level and functional consequences for these various residues are not yet completed. However, in regard to Ser294, Fig. 8 demonstrates that estradiol stimulation of MCF-7 cells produced an approximate 28-fold phosphorylation enhancement of this residue, strongly implicating its functional relevance to ER activation. It is interesting to note that an early study examining the phosphorylation of ER expressed in COS-1 cells failed to detect Ser294 phosphorylation (29). We note that our studies differ from these initial studies in several key aspects including our inclusion of the potent phosphatase inhibitor okadaic acid in the immunoprecipitation buffer; the use of MCF-7 cells, which endogenously express ER instead of the more artificial system of ER expressed in COS-1 cells; and an estradiol exposure time of 30 min in our experiments versus the 4 h used in the earlier study.
Fig. 9.
Summary of ERα modifications observed in this study. ** signifies previously unreported modifications identified in ERα from MCF-7 cells. * signifies a previously unreported modification identified only in rERα. In parentheses previously known modification identified here only in rERα. All others are previously known modifications identified in ERα from MCF-7 cells. LBD, ligand-binding domain; NTD, N-terminal domain; pSer, phosphoserine.
Previously reported phosphorylation sites Ser305, Thr311, and Tyr537 were not confirmed here, and Ser236 phosphorylation was observed only in recombinant protein. However, Ser236 is phosphorylated by cAMP-dependent protein kinase (ligand-independent) so 30-min treatment with estradiol may not induce phosphorylation in MCF-7 cells, although 1–5-min exposure to estradiol might activate the cAMP-dependent protein kinase pathway through membrane ER activation (15). Residue 305 is in a region for which no peptide was observed so no data were obtained to support or refute its phosphorylation. However, it is phosphorylated by PAK1 (growth factors heregulin and epidermal growth factor induce PAK1 activation), which again is ligand-independent (30). Unmodified Thr311 was identified by LC-MS/MS in chymotryptic peptide 309SLTADQMVSALL with good mass accuracy (an error of 19 ppm) and a significant Mascot score of 35 and probably would be observed as phosphorylated if that actually occurred. This residue was reported phosphorylated by p38 kinase and was estradiol-dependent in cells overexpressing ER and tagged p38 using in vitro kinase assays (31). We observed unmodified Tyr537 peptides with trypsin (532–548), Glu-C (524–538), and Asp-N (527–537), so any phosphorylation of this residue would probably be observed, but phosphorylation may not be induced by 30-min estradiol treatment as it may be Src-dependent (32).
This study demonstrates the sensitivity of tandem mass spectrometric methods for detection of phosphorylation sites in low level proteins such as ERα, and it reiterates the advantages of using complementary methods of ionization and analysis as no single method was able to detect all the modifications reported here. Additional modifications detected in ERα from MCF-7 cells were N-terminal cleavage of methionine accompanied by acetylation of Thr2 and side chain acetylation or possibly trimethylation of Lys472.
Acknowledgments
The Mass Spectrometry Core at the Buck Institute is a Nathan Shock Center of Excellence for Basic Mechanisms of Aging and Age-related Diseases, which funded the development of methods for analyzing modified proteins through National Institutes of Health Grant P30-AG025708 (Proteomics Core E, BWG). The 4000 QTRAP mass spectrometer was purchased by NCRR shared instrumentation Grant 1S10RR024615 (BWG).
Footnotes
Published, MCP Papers in Press, November 3, 2008, DOI 10.1074/mcp.M800282-MCP200
The abbreviations used are: ERα, α-isoform of the estrogen receptor; PTM, posttranslational modification; LTQ, linear quadrupole trap; AF, activation factor; DBD, DNA-binding domain; vMALDI, vacuum MALDI; ER, estrogen receptor; rER, recombinant ER; MRM, multiple reaction monitoring.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-CA71468 and AG-020521. This work was also supported by California Breast Cancer Research Program Grant 10YB-0125.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Ciocca, D. R., and Fanelli, M. A. ( 1997) Estrogen receptors and cell proliferation in breast cancer. Trends Endocrinol. Metab. 8, 313–321 [DOI] [PubMed] [Google Scholar]
- 2.Conzen, S. D. ( 2008) Nuclear receptors and breast cancer. Mol. Endocrinol. 22, 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuske, B., Naughton, C., Moore, K., MacLeod, K. G., Miller, W. R., Clarke, R., Langdon, S. P., and Cameron, D. A. ( 2006) Endocrine therapy resistance can be associated with high estrogen receptor α (ERα) expression and reduced ERα phosphorylation in breast cancer models. Endocr.-Relat. Cancer 13, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann, J., Bohlmann, R., Heinrich, N., Hofmeister, H., Kroll, J., Kunzer, H., Lichtner, R. B., Nishino, Y., Parczyk, K., Sauer, G., Gieschen, H., Ulbrich, H.-F., and Schneider, M. R. ( 2004) Characterization of new estrogen receptor destabilizing compounds: effects on estrogen-sensitive and tamoxifen-resistant breast cancer. J. Natl. Cancer Inst. 96, 210–218 [DOI] [PubMed] [Google Scholar]
- 5.Howell, S. J., Johnston, S. R. D., and Howell, A. ( 2004) The use of selective estrogen receptor modulators and selective estrogen receptor down-regulators in breast cancer. Best Pract. Res. Clin. Endocrinol. Metab. 18, 47–66 [DOI] [PubMed] [Google Scholar]
- 6.Robertson, J. F. R. ( 2004) Selective oestrogen receptor modulators/new antioestrogens: a clinical perspective. Cancer Treat. Rev. 30, 695–706 [DOI] [PubMed] [Google Scholar]
- 7.Atsriku, C., Benz, C. C., Scott, G. K., Gibson, B. W., and Baldwin, M. A. ( 2007) Quantification of cysteine oxidation in human estrogen receptor by mass spectrometry. Anal. Chem. 79, 3083–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atsriku, C., Scott, G. K., Benz, C. C., and Baldwin, M. A. ( 2005) Reactivity of zinc finger cysteines: chemical modifications within labile zinc fingers in estrogen receptor. J. Am. Soc. Mass Spectrom. 16, 2017–2026 [DOI] [PubMed] [Google Scholar]
- 9.Baldwin, M. A., and Benz, C. C. ( 2002) Redox control of zinc finger proteins. Methods Enzymol. 353, 54–69 [DOI] [PubMed] [Google Scholar]
- 10.Meza, J. E., Scott, G. K., Benz, C. C., and Baldwin, M. A. ( 2003) Essential cysteine-alkylation strategies to monitor structurally altered estrogen receptor as found in oxidant-stressed breast cancers. Anal. Biochem. 320, 21–31 [DOI] [PubMed] [Google Scholar]
- 11.Whittal, R. M., Benz, C. C., Scott, G., Semyonov, J., Burlingame, A. L., and Baldwin, M. A. ( 2000) Preferential oxidation of zinc finger 2 in estrogen receptor DNA-binding domain prevents dimerization and, hence, DNA binding. Biochemistry 39, 8406–8417 [DOI] [PubMed] [Google Scholar]
- 12.Schilling, B., Yoo, C. B., Collins, C. J., and Gibson, B. W. ( 2004) Determining cysteine oxidation status using differential alkylation. Int. J. Mass Spectrom. 236, 117–127 [Google Scholar]
- 13.Lannigan, D. A. ( 2003) Estrogen receptor phosphorylation. Steroids 68, 1–9 [DOI] [PubMed] [Google Scholar]
- 14.Arnold, S., Obourn, J., Jaffe, H., and Notides, A. ( 1994) Serine 167 is the major estradiol-induced phosphorylation site on the human estrogen receptor. Mol. Endocrinol. 8, 1208–1214 [DOI] [PubMed] [Google Scholar]
- 15.Chen, D., Pace, P. E., Coombes, R. C., and Ali, S. ( 1999) Phosphorylation of human estrogen receptor α by protein kinase A regulates dimerization. Mol. Cell. Biol. 19, 1002–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, C., Fu, M., Angeletti, R. H., Siconolfi-Baez, L., Reutens, A. T., Albanese, C., Lisanti, M. P., Katzenellenbogen, B. S., Kato, S., Hopp, T., Fuqua, S. A. W., Lopez, G. N., Kushner, P. J., and Pestell, R. G. ( 2001) Direct acetylation of the estrogen receptor α hinge region by p300 regulates transactivation and hormone sensitivity. J. Biol. Chem. 276, 18375–18383 [DOI] [PubMed] [Google Scholar]
- 17.Buteau-Lozano, H., Ancelin, M., Lardeux, B., Milanini, J., and Perrot-Applanat, M. ( 2002) Transcriptional regulation of vascular endothelial growth factor by estradiol and tamoxifen in breast cancer cells: a complex interplay between estrogen receptors α and β. Cancer Res. 62, 4977–4984 [PubMed] [Google Scholar]
- 18.Medunjanin, S., Hermani, A., De Servi, B., Grisouard, J., Rincke, G., and Mayer, D. ( 2005) Glycogen synthase kinase-3 interacts with and phosphorylates estrogen receptor α and is involved in the regulation of receptor activity. J. Biol. Chem. 280, 33006–33014 [DOI] [PubMed] [Google Scholar]
- 19.Joel, P. B., Traish, A. M., and Lannigan, D. A. ( 1998) Estradiol-induced phosphorylation of serine 118 in the estrogen receptor is independent of p42/p44 mitogen-activated protein kinase. J. Biol. Chem. 273, 13317–13323 [DOI] [PubMed] [Google Scholar]
- 20.Rayala, S. K., Talukder, A. H., Balasenthil, S., Tharakan, R., Barnes, C. J., Wang, R.-A., Aldaz, M., Khan, S., and Kumar, R. ( 2006) p21-activated kinase 1 regulation of estrogen receptor-α activation involves serine 305 activation linked with serine 118 phosphorylation. Cancer Res. 66, 1694–1701 [DOI] [PubMed] [Google Scholar]
- 21.Mirza, S. P., and Olivier, M. ( 2008) Methods and approaches for the comprehensive characterization and quantification of cellular proteomes using mass spectrometry. Physiol. Genomics 33, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins, M. O., Yu, L., and Choudhary, J. S. ( 2007) Analysis of protein phosphorylation on a proteome-scale. Proteomics 7, 2751–2768 [DOI] [PubMed] [Google Scholar]
- 23.Zabrouskov, V., Senko, M. W., Du, Y., Leduc, R. D., and Kelleher, N. L. ( 2005) New and automated MSn approaches for top-down identification of modified proteins. J. Am. Soc. Mass Spectrom. 16, 2027–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang, E. J., Archambault, V., McLachlin, D. T., Krutchinsky, A. N., and Chait, B. T. ( 2004) Analysis of protein phosphorylation by hypothesis-driven multiple-stage mass spectrometry. Anal. Chem. 76, 4472–4483 [DOI] [PubMed] [Google Scholar]
- 25.BoeriErba, E., Matthiesen, R., Bunkenborg, J., Schulze, W. X., DiStefano, P., Cabodi, S., Tarone, G., Defilippi, P., and Jensen, O. N. ( 2007) Quantitation of multisite EGF receptor phosphorylation using mass spectrometry and a novel normalization approach. J. Proteome Res. 6, 2768–2785 [DOI] [PubMed] [Google Scholar]
- 26.Boeri Erba, E., Bergatto, E., Cabodi, S., Silengo, L., Tarone, G., Defilippi, P., and Jensen, O. N. ( 2005) Systematic analysis of the epidermal growth factor receptor by mass spectrometry reveals stimulation-dependent multisite phosphorylation. Mol. Cell. Proteomics 4, 1107–1121 [DOI] [PubMed] [Google Scholar]
- 27.Britton, D. J., Scott, G. K., Schilling, B., Atsriku, C., Held, J. M., Gibson, B. W., Benz, C. C., and Baldwin, M. A. ( 2008) A novel serine phosphorylation site detected in the N-terminal domain of estrogen receptor isolated from human breast cancer cells. J. Am. Soc. Mass Spectrom. 19, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian, K., Jia, D., Kapoor-Vazirani, P., Powell, D. R., Collins, R. E., Sharma, D., Peng, J., Cheng, X., and Vertino, P. M. ( 2008) Regulation of estrogen receptor α by the SET7 lysine methyltransferase. Mol. Cell 30, 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Goff, P., Montano, M. M., Schodin, D. J., and Katzenellenbogen, B. S. ( 1994) Phosphorylation of the human estrogen receptor. J. Biol. Chem. 269, 4458–4466 [PubMed] [Google Scholar]
- 30.Wang, R. A., Mazumda, A., Vadlamudi, R. K., and Kumar, R. ( 2002) p21-activated kinase-1 phosphorylates and transactivates estrogen receptor-α and promotes hyperplasia in mammary epithelium. EMBO J. 21, 5437–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, H., and Bai, W. ( 2002) Regulation of estrogen receptor nuclear export by ligand-induced and p38-mediated receptor phosphorylation. Mol. Cell. Biol. 22, 5835–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold, S. F., Vorojeikina, D. P., and Notides, A. C. ( 1995) Phosphorylation of tyrosine 537 on the human estrogen receptor is required for binding to an estrogen response element. J. Biol. Chem. 270, 30205–30212 [DOI] [PubMed] [Google Scholar]
- 33.Cheng, X., and Hart, G. W. ( 2000) Glycosylation of the murine estrogen receptor-α. J. Steroid Biochem. Mol. Biol. 75, 147–158 [DOI] [PubMed] [Google Scholar]
- 34.Ali, S., Metzger, D., Bornert, J. M., and Chambon, P. ( 1993) Modulation of transcriptional activation by ligand-dependent phosphorylation of the human estrogen receptor A/B region. EMBO J. 12, 1153–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garbán, H. J., Márquez-Garbán, D. C., Pietras, R. J., and Ignarro, L. J. ( 2005) Rapid nitric oxide-mediated S-nitrosylation of estrogen receptor: regulation of estrogen-dependent gene transcription. Proc. Natl. Acad. Sci. U. S. A. 102, 2632–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sentis, S., Le Romancer, M., Bianchin, C., Rostan, M. C., and Corbo, L. ( 2005) Sumoylation of the estrogen receptor α hinge region regulates its transcriptional activity. Mol. Endocrinol. 19, 2671–2684 [DOI] [PubMed] [Google Scholar]
- 37.Kim, M. Y., Woo, E. M., Chong, Y. T., Homenko, D. R., and Kraus, W. L. ( 2006) Acetylation of estrogen receptor α by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol. Endocrinol. 20, 1479–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marino, M., Ascenzi, P., and Acconcia, F. ( 2006) S-Palmitoylation modulates estrogen receptor α localization and functions. Steroids 71, 298–303 [DOI] [PubMed] [Google Scholar]
- 39.Jiang, M. S., and Hart, G. W. ( 1997) A subpopulation of estrogen receptors are modified by O-linked N-acetylglucosamine. J. Biol. Chem. 272, 2421–2428 [DOI] [PubMed] [Google Scholar]
- 40.Tateishi, Y., Kawabe, Y., Chiba, T., Murata, S., Ichikawa, K., Murayama, A., Tanaka, K., Baba, T., Kato, S., and Yanagisawa, J. ( 2004) Ligand-dependent switching of ubiquitin-proteasome pathways for estrogen receptor. EMBO J. 23, 4813–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lonard, D. M., Nawaz, Z., Smith, C. L., and O'Malley, B. W. ( 2000) The 26S proteasome is required for estrogen receptor-α and coactivator turnover and for efficient estrogen receptor-α transactivation. Mol. Cell 5, 939–948 [DOI] [PubMed] [Google Scholar]