Abstract
Preliminary findings indicate that PprI is a regulatory protein that stimulates transcription and translation of recA and other DNA repair genes in response to DNA damage in the extremely radioresistant bacterium Deinococcus radiodurans. To define the repertoire of proteins regulated by PprI and investigate the in vivo regulatory mechanism of PprI in response to γ radiation, we performed comparative proteomics analyses on wild type (R1) and a pprI knock-out strain (YR1) under conditions of ionizing irradiation. Results of two-dimensional electrophoresis and MALDI-TOF MS or MALDI-TOF/TOF MS indicated that in response to low dose γ ray exposure 31 proteins were significantly up-regulated in the presence of PprI. Among them, RecA and PprA are well known for their roles in DNA replication and repair. Others are involved in six different pathways, including stress response, energy metabolism, transcriptional regulation, signal transduction, protein turnover, and chaperoning. The last group consists of many proteins with uncharacterized functions. Expression of an additional four proteins, most of which act in metabolic pathways, was down-regulated in irradiated R1. Additionally phosphorylation of two proteins was under the control of PprI in response to irradiation. The different functional roles of representative PprI-regulated genes in extreme radioresistance were validated by gene knock-out analysis. These results suggest a role, either directly or indirectly, for PprI as a general switch to efficiently enhance the DNA repair capability and extreme radioresistance of D. radiodurans via regulation of a series of pathways.
The Gram-positive nonpathogenic bacterium Deinococcus radiodurans is characterized by extreme resistance to ionizing radiation, UV irradiation, desiccation, and a variety of DNA-damaging agents without resulting in lethality or mutagenesis (1, 2). This dramatic capability is ascribed to its outstanding efficiency in reconstructing a functional genome with high fidelity from hundreds of double strand breaks (DSBs)1 generated by DNA-damaging agents (2, 3), whereas few other organisms can tolerate DSBs (4). Exponentially growing D. radiodurans is able to withstand 50–100 times more ionizing radiation than Escherichia coli and can survive a 15-kGy acute ionizing radiation dose with no loss of viability. Its ability of continuous growth without any delay when exposed to a maximum of 60 Gy/h γ ray (5) has made it one of the most distinguished candidates for bioremediation of radioactive wastes and contaminants (6, 7). More than 50 years of research has provided many lines of evidence supporting the benefits of the extreme radioresistance of D. radiodurans from its highly efficient DNA damage repair system and its remarkable antioxidation system (1, 8–15). However, the mechanism underlying its radioresistance is still not completely understood (4, 9). Intriguingly this bacterium not only possesses most of the DNA repair genes found in other organisms but also contains many proteins of yet to be determined functions that have been revealed by genome sequencing and comparative genomics (16, 17). These function-unknown proteins may play crucial roles in radioresistance (9, 18) as implied by several studies in this bacterium (19–23).
Several years ago, our group and that of John Battista (19, 22) identified and validated a novel protein, PprI (also named IrrE), essential in D. radiodurans radioresistance. It strongly enhanced catalase activities and promoted the expression of RecA and PprA (19). Expression of the novel gene, driven by the promoter of D. radiodurans groEL (DR0607), also significantly enhanced the resistance of E. coli to γ irradiation. We found that the expression of PprI in E. coli also increased the expression of RecA and improved catalase activity (24). Both microarray and Western blotting analysis demonstrated that ionizing radiation did not increase PprI transcription or translation levels (19, 25–27). To our knowledge, no homologous protein has been identified through database searches in other organisms except in the closely related Deinococcus geothermalis, which is also an exceptionally radioresistant bacterium. Sequence analysis revealed that PprI contained a functional domain (DUF955) consisting of neutral zinc metallopeptidases and a lacI-type helix-turn-helix that led us, in our previous study, to propose it as a transcriptional regulator (19). However, the precise molecular mechanism by which PprI contributes to radioresistance remains unclear. Identification of the PprI regulatory network should provide insight into the role of PprI in the defense of γ radiation insults. In the current study, we conducted a systematic proteome-wide analysis to identify proteins and pathways regulated by PprI in bacterial cells recovering from radiation damage.
Beyond the previous focus on proteomic changes of D. radiodurans in response to irradiation (28–32), we systematically examined the changes at the proteome level in the wild type strain (R1), compared with the pprI-deleted mutant strain (YR1), in response to 1 kGy of ionizing radiation using two-dimensional electrophoresis (2-DE) and MALDI-TOF MS or MALDI-TOF/TOF MS to identify components of the pprI-mediated responsive pathways for bacterial cell survival. The majority of proteins identified as being regulated by PprI were involved in transcription, translation, DNA replication and repair, signal transduction, protein turnover and chaperoning, energy production and conversion, and metabolism. We found that upon radiation PprI turned on various pathways to funnel the cellular efforts into efficient repair and recovery of its intact genome for cellular survival.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Culture Media—
The D. radiodurans R1 strain and E. coli strain JM109 were available in our laboratory. D. radiodurans cultures were grown at 32 °C in TGY broth (0.5% Bacto tryptone, 0.1% glucose, 0.3% Bacto yeast extract) with aeration or on TGY plates supplemented with 1.5% agar, whereas E. coli was grown at 37 °C in LB broth (1.0% Bacto tryptone, 0.5% Bacto yeast extract, 1.0% NaCl) or on LB plates solidified with 1.5% agar. D. radiodurans cells were transformed using the modified CaCl2 technique as described previously (33).
Construction of Function-deficient Mutants of D. Radiodurans and Survival Curves under γ Radiation—
The construction of the PprI function-deficient mutant strain YR1 was described previously (34). Briefly a DNA fragment from the plasmid pRADK (34) containing the kanamycin resistance gene driven by the groEL (DR0607) promoter was reversely inserted into the pprI (DR0167) gene in the D. radiodurans genome.
To validate the functional involvement of the identified gene products in radioresistance, we deleted individual genes and tested the radiation resistance capacity of the mutants. Mutants of DR1473, DR2317, DRA0018, and DRA0283 were constructed using the same protocol (34) and designated M1473, M2317, MA0018, and MA0283, respectively (Table I). Primers used for the construction of these mutants are listed in supplemental Table S1. Survival curves under γ irradiation of four mutants, including M1473, M2317, MA0018, and MA0283, were carried out using a published protocol (34).
Table I.
Strains and plasmids
| Strains and plasmids | Characteristics | Source |
|---|---|---|
| Plasmids | ||
| pRADK | Shuttle plasmids between E. coli and D. radiodurans, AmpR, KanR, ChlR | Gao et al. (34) |
| pRADA0283myc | pRADK containing c-Myc-tagged dra0283 | This study |
| pRAD1343myc | pRADK containing c-Myc-tagged dr1343 | This study |
| Strains | ||
| E. coli | ||
| DH5α | SupE44, ΔlacU169, hsdR17, recA1 endA1, gryA96, thi-1, relA1 | Lab stock |
| D. radiodurans | ||
| R1 | Wild type strain (ATCC 13939) | Lab stock |
| YR1 | As R1 but pprI(DR0167)-deleted | Gao et al. (34) |
| MA0018 | As R1 but DRA0018-deleted | This study |
| MA0283 | As R1 but DRA0283-deleted | This study |
| M1473 | As R1 but DR1473-deleted | This study |
| M2317 | As R1 but DR2317-deleted | This study |
| R1pRADK | R1 containing pRADK | This study |
| DRA0283myc | R1 containing pRADA0283myc | This study |
| DR1343myc | R1 containing pRAD1343myc | This study |
Postirradiation Growth and Pulse Field Gel Electrophoresis—
To find an appropriate time point to obtain bacterial samples for proteomics analysis, we investigated cell growth and genome restitution during postirradiation recovery. Pulse field gel electrophoresis of D. radiodurans R1 and YR1 strains was conducted as described previously (35). Strains were grown to A600 = 0.3 and harvested. Cells were washed once with 0.9% NaCl, resuspended in MgSO4 (10 mm), acutely irradiated (1 kGy) at room temperature, and incubated in fresh media, and A600 values were measured at several time points. Concomitantly samples (5 ml) were prepared as DNA-agarose plugs and sequentially treated with lysozyme, proteinase K, and restriction enzyme NotI, and plugs were then sent for pulsed field gel electrophoresis (22 h at 14 °C) using the CHEF-MAPPER electrophoresis system (Bio-Rad). The main electrophoresis parameters were set as 6 V/cm, 40-s linear pulse, and a switching angle of 120° (−60° to +60°).
Cell Growth and Irradiation Treatment for Proteomics Analysis—
The R1 and YR1 strains were transferred to fresh TGY medium and grown (32 °C) with continuous shaking until early stationary phase (A600 = 0.8). Cells were harvested by centrifugation, washed twice with PBS (pH 7.4), and resuspended in MgSO4 (10 mm). Suspensions were halved: one-half was acutely irradiated on ice with 1 kGy 60Co γ rays; the other was used as the non-irradiated control. After treatment, cells were collected by centrifugation, resuspended in fresh TGY medium, incubated (32 °C for 60 min) with continuous shaking, then washed twice with PBS, and pelleted. The cell pellet was snap frozen in liquid nitrogen and stored (−80 °C).
Sample Preparation for 2-D PAGE—
Deep frozen cells were resuspended in lysis buffer (9 m urea, 4% (w/v) CHAPS, 65 mm DTT, 2% IPG buffer pH 3–10 linear, 1 mm PMSF, 40 mm Tris base) and lysed with a Biospec Minibeadbeater (Bartlesville, OK). Lysates were immediately placed on ice to inhibit proteolysis. Cell debris were removed by centrifugation (30,000 × g for 30 min), and the clear supernatant was stored (−80 °C) in aliquots until analysis. Protein concentrations were measured using the Bio-Rad protein assay reagent.
2-DE—
2-DE was performed according to the manufacturer's instructions (Amersham Biosciences). Briefly each protein sample in the lysis buffer was diluted to 500 μl with rehydration solution (9 m urea, 2% CHAPS, 30 mm DTT, 0.5% IPG buffer pH 4–10 linear, 0.002% bromphenol blue). Immobiline DryStrip gels (pH 4–7, 24 cm; Amersham Biosciences) were rehydrated with 500 μl of mixture solution in 24-cm strip holders and electrofocused with an Ettan IPGphor Isoelectric Focusing System (Amersham Biosciences). The focusing protocol was performed as follows: 50 μA/strip at 20 °C, 30 V for 12 h, 500 V for 1 h, 1000 V for 1 h, and 8000 V for 7 h. After isoelectric focusing, strips were equilibrated (30 min) with gentle shaking in SDS equilibration buffer (6 m urea, 30% glycerol, 2% SDS, 1% DTT, 2.5% iodoacetamide, 50 mm Tris-HCl buffer, pH 8.8, 0.002% bromphenol blue) and then separated by SDS-PAGE (12.5%). The second dimension SDS electrophoresis was performed using a Hoefer SE 600 unit (Amersham Biosciences). Four gels were run simultaneously: non-irradiated R1 and YR1 and radiation-treated R1 and YR1 (n = 4/sample).
Silver Staining and Data Analysis—
All resolved protein spots in 2-D gels were visualized by silver staining using a Silver Staining kit (Amersham Biosciences). Other standard chemicals required were purchased from Sigma. Stained gels were scanned on an ImageScanner (Amersham Biosciences), and images were analyzed with ImageMaster 2D Elite software supplied by the manufacturer. A D. radiodurans wild type R1 spot file served as the experiment reference pattern. On average, over 1000 spots were detected on each 2-D gel image. Among these, ∼950 of the most reproducible spots were included in the data analysis. Protein spots separated on 2-D gels were quantitated in terms of their relative volume (spot volume/total spot volume). For comparison, the value of each spot was divided by that of each corresponding spot volume from the untreated R1 strain. Statistical significance of the differences in expression profiles of D. radiodurans with or without the γ irradiation was evaluated by t test with significance set at p < 0.05; all statistical calculations utilized Microsoft Excel software. Only those spots with a 2-fold or greater change in expression levels were considered significant and selected for spot picking, trypsin digestion, and mass spectrometry analysis to identify their protein content.
In-gel Trypsin Digestion—
Approximately 200 μg of protein from each sample (non-irradiated R1 and YR1 and irradiated R1 and YR1) was separated by isoelectric focusing. After separation by a second SDS-PAGE, proteins were detected by Coomassie Blue R-250 staining (15 h). Protein spots demonstrating different expression patterns compared with controls (induction rate ≥2) were excised from gels and processed for mass spectrometric analysis. Excised spots were reduced at room temperate with tris(2-carboxyethyl)phosphine (Pierce), alkylated with iodoacetamide (Sigma), and digested (20 h) in situ with trypsin (Sigma). Peptides were extracted by addition of a 50% acetonitrile, 5% TFA solution, and extracted solutions were concentrated to 4 μl in a lyophilizer (VirTis, Gardiner, NY). Peptides were treated with ZipTips (Millipore, Bedford, MA) before application to the sample plate in cases where the signal to noise ratio on MALDI-TOF spectra was not ideal. A protein-free gel piece was similarly processed and used as the control to identify autoproteolysis products derived from trypsin.
Mass Spectrometry Analysis and Database Searching—
For most of the 2-D gel protein samples (supplemental Table S3), we determined the identity of protein spots in the Research Center for Proteome Analysis, Chinese Academy of Sciences, Shanghai, China. The digested sample was mixed with an equal volume of cyano-4-hydroxycinnamic acid (10 mg/ml; Sigma) saturated with 50% acetonitrile in 0.05% TFA and analyzed by MALDI-TOF MS using an AutoFlex TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). The working mode was set with positive ion reflection mode, an accelerating voltage of 20 kV, and 150-ns delayed extraction time. The spectrum masses ranging from 700 to 3500 Da were acquired with laser shots at 200/spectrum. A Peptide Mixture-1 kit (Bruker Daltonics) was used for external calibration. The matrix and autolytic peaks of trypsin were used for internal calibration. Monoisotopic mass was analyzed with FlexAnalysis 2.0 (Bruker Daltonics) and automatically collected with a signal to noise ratio >4 and a peak quality index >30. The known contaminant ions (human keratin and tryptic autodigest peptides) were excluded. For interpretation of the mass spectra, monoisotopic peptide masses were input into Mascot 2.0 (Matrix Science) for analysis with BioTools 2.1 (Bruker Daltonics).
The rest of the protein samples (supplemental Table S3) was analyzed with a Voyager DE STR MALDI-TOF mass spectrometer (Applied Biosystems, Foster City, CA) in the Zhejiang University Mass Spectrometry Facility (Hangzhou, China) using a procedure as described previously (30). Briefly the digested samples were spotted onto a MADLI target with an equal volume of cyano-4-hydroxycinnamic acid (10 mg/ml; Sigma) saturated with 50% acetonitrile in 0.05% TFA. The data were acquired on a Voyager DE STR MALDI-TOF mass spectrometer. The instrument settings were reflector mode with 160-ns delay extraction time, positive polarity, and 20-kV accelerating voltage. The spectrum masses were usually acquired with laser shots at 200/spectrum and ranged from 1000 to 4000 Da. External calibration was performed using a Peptide Mass Standard kit (Perspective Biosystems, Framingham, MA). The matrix and the autolytic peaks of trypsin were used as internal standards for mass calibration. The acquired data were processed with base-line correction, noise removal (5%), and peak deisotoping using Data Explorer 4.0 (Applied Biosystems) following exclusion of known contaminant ions (human keratin and tryptic autodigest peptides). The processed data were input into Mascot 2.0 (Matrix Science) for protein identity searching.
Many of processed spectra from the AutoFlex MALDI-TOF/TOF mass spectrometer were searched against the NCBI nonredundant protein database (updated on May 26, 2005), which contained 2,471,633 sequences. The search was restricted to “Bacteria (Eubacteria)” as taxonomy, which contained 944,772 sequences. The remaining spectra were searched against the NCBI nonredundant protein database (updated on August 5, 2005), which contained 2,739,666 sequences. The search was restricted to “Other Bacteria” as taxonomy, which contained 171,641 sequences. The spectra from the Voyager DE STR mass spectrometer were searched against the NCBI nonredundant protein database (updated on August 29, 2005), which contained 2,794,673 sequences. The search was restricted to “Other Bacteria” as taxonomy, which contained 180,331 sequences. The other main search parameters for all PMF searches were as follows: type of search, peptide mass fingerprint; enzyme, trypsin; fixed modifications, carbamidomethylcysteine; variable modifications, methionine oxidation; mass values, monoisotopic; protein mass, unrestricted; peptide mass tolerance, 100 ppm (the PMF search of seven proteins allowed a 150-ppm mass tolerance; see supplemental Table S2); fragment mass tolerance, ±1Da; peptide charge state, 1+; and maximum missed cleavages, 1. Proteins whose scores were greater than 65 were considered significant (p < 0.05), and only D. radiodurans proteins with the best score in each Mascot search were accepted as successful identifications.
Unidentified samples by MALDI-TOF MS were submitted to an AutoFlex MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) in the “LIFT” mode of the instrument. Tryptic digests were prepared using an AnchorChip sample plate (Bruker Daltonics) according to the manufacturer's protocol. Both MS and MS/MS data were acquired with an N2 laser at 25-Hz sampling rate. PMF data and MS/MS data were processed with FlexAnalysis 2.0. The combined data of PMF and MS/MS data were submitted by BioTools 2.1 to Mascot 2.0 for protein identification against the NCBI nonredundant protein database (updated on August 5, 2005), which contained 2,739,666 sequences. The search was again restricted to “Other Bacteria” as taxonomy, which contained 171,641 sequences. The main search parameters were: type of search, MS/MS ion search; enzyme, trypsin; fixed modifications, carbamidomethylcysteine; variable modifications, methionine oxidation; mass values, monoisotopic; protein mass, unrestricted; peptide mass tolerance, 100 ppm; fragment mass tolerance, ±0.5 Da; maximum missed cleavages, 1; and instrument type, MALDI-TOF/TOF. Individual ion scores greater than 36 indicate identity or extensive homology (p < 0.05) as described by Mascot. Only D. radiodurans proteins with the best score in each Mascot search were accepted as successful identifications.
Immunoprecipitation and Western Blotting—
To validate the nature of phosphorylation of affected proteins identified based on migration difference on 2D gel, two genes, DR1343 and DRA0283, were amplified and tagged with a c-Myc DNA fragment by PCR (primers are listed in supplemental Table S1). The NdeI- and BamHI-digested PCR products were ligated to the plasmid pRADK (34), and the resulting recombinant plasmids and pRADK were transformed into the R1 strain; the resulting strains were designated DR1343myc, DRA0283myc, and R1pRADK. Cells were grown to early stationary phase, harvested, and washed twice with TBS (50 mm Tris-HCl, pH 7.6, 150 mm NaCl), dispersed in a lysis buffer (TBS, Roche Applied Science protease inhibitor mixture, Sigma phosphatase inhibitor mixtures 1 and 2), lysed by sonicator, and centrifuged (15,000 × g for 30 min). The cell lysate was incubated with polyclonal anti-c-Myc antibodies (Sigma) in the presence of protein A-Sepharose beads (Amersham Biosciences) overnight at 4 °C. After washing with TBST (TBS with the addition of 0.1% Tween 20), precipitated proteins were used for Western blotting; mouse monoclonal anti-phosphoserine antibody and anti-phosphotyrosine antibodies (Sigma) were used to detect DRA0283 and DR1343, respectively. The identity of phosphorylated proteins was confirmed using an anti-c-Myc antibody to detect the c-Myc-tagged proteins (DRA0283 and DR1343) immunoprecipitated by mouse monoclonal anti-phosphoserine antibody or anti-phosphotyrosine antibodies.
RESULTS
Sampling Time Point for Proteomics Analysis—
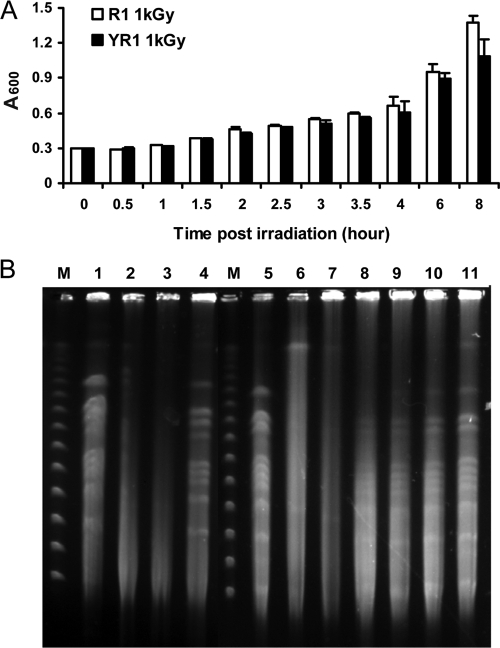
After irradiation, the growth of wild type D. radiodurans was halted, whereas the high molecular genomic DNA in both wild type (R1) and the pprI-deleted (YR1) strains was detectably degraded for a period of 1 h immediately after irradiation and before growth was reinitiated (Fig. 1), indicating active DNA repair as described previously (18). At 90 min, both cell types started to grow slowly (Fig. 1A). At that time, the wild type bacteria almost completely recovered its genome, whereas the mutant had just started to recover (Fig. 1B). Samples were taken at 60 min, which was considered a suitable time point to perform proteomics analysis to detect components of the PprI-mediated responsive pathways for the bacterial cell survival.
Fig. 1.
Cell growth and genome recovery after γ radiation. A, growth of D. radiodurans cultures for strains R1 (□) and YR1 (▪) after 1 kGy of γ radiation. Each data point represents an average of triplicate experiments and error bars represent standard deviations. B, genome recovery of wild type R1 and the pprI knock-out strain YR1 after exposure to irradiation. M, pulse field gel electrophoresis markers (New England Biolabs); lane 1, unirradiated R1, lanes 2–4, R1 postirradiation time 0, 45, and 90 min, respectively; lane 5, unirradiated YR1; lanes 6–11, R1 postirradiation time 0, 45, 90, 180, 270, and 360 min, respectively.
Thirty-one Proteins Are Induced Only in the Presence of PprI during Recovery of D. radiodurans from Irradiation Damage—
To study responses to a low dose of ionizing irradiation, R1 and YR1 strains in the early stationary phase were irradiated (1 kGy) with γ rays. Protein extracts were subjected to 2-D gel electrophoresis. Over 950 protein spots were resolved on each gel, accounting for ∼30% of all hypothetical ORFs in D. radiodurans (supplemental Fig. S1). To identify proteins in the PprI regulon in response to radiation damage, we focused on proteins that displayed a >2-fold increase or decrease in the irradiated R1 strain but not in the irradiated YR1 strain compared with untreated R1 and YR1 strains, respectively. We found that the expression levels of 31 proteins increased significantly in R1 but not in YR1 in response to γ radiation. Additionally the expression levels of five proteins decreased remarkably in R1 but increased or remained unchanged in YR1 (supplemental Table S2). These altered protein expression profiles were grouped into various functional categories based on NCBI's Clusters of Orthologous Groups (COGs) of proteins, including groups for information storage and processing, signal transduction and cellular processing, metabolism, and poorly characterized proteins.
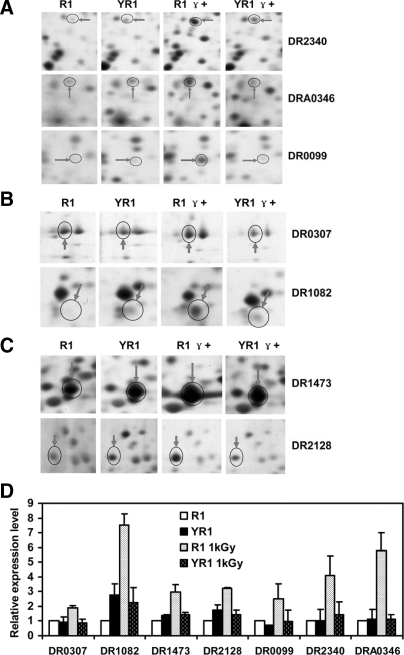
Seven proteins were classified under the functional group for information storage and processing, including DR0307, DR1082, DR1473, DR2128, DR0099, DR2340, and DRA0346 (Fig. 2). A DNA damage repair system is critical to the extreme radioresistance of D. radiodurans. Here we observed that the expression of three proteins involved in this system was affected by PprI in response to γ radiation: DR0099, DR2340, and DRA0346 (Fig. 2, A and D). These results were similar to previous studies in which the wild type strain was up-regulated following irradiation (18, 25, 27, 30). Expression of DR2340 (RecA protein) and DRA0346 (PprA) was significantly enhanced in the irradiated wild type D. radiodurans but not in the absence of PprI. During the radiation recovery process, there was a 3-fold increase in the expression of DR0099 (single strand DNA-binding protein (SsB)) in the wild type strain following irradiation, but no change was noted in the absence of PprI. DR0307 and DR1082 were included in the subgroup of translation, ribosomal structure, and biogenesis. DR0307 (elongation factor EF-G) and DR1082 (light-repressed protein A) were observably up-regulated in the presence of PprI following irradiation. However, stimulation of their expression was suppressed in YR1 (Fig. 2, B and D). Analogically DR1473 (phage shock protein A) and DR2128 (DNA-directed RNA polymerase α unit), categorized in the transcription subgroup, also exhibited a similar phenotype (Fig. 2, C and D).
Fig. 2.
Enhanced expression of proteins involved in information storage and processing pathways in wild type D. radiodurans in response to γ radiation (1 kGy). R1, wild type strain without irradiation; YR1, pprI knock-out strain without irradiation; R1 γ+, wild type strain with irradiation; YR1 γ+, pprI knock-out strain with irradiation. Protein spots in 2-D gels are shown for DR0099, DR2340, and DRA0346 (SsB, RecA, and PprA, respectively) (A); DR0307 and DR1082 (elongation factor G and light-repressed protein A, respectively) (B); and DR1473 and DR2128 (phage shock protein A and DNA-directed RNA polymerase α subunit, respectively) (C). D, relative protein expression levels (see “Experimental Procedures” for calculations). Each data point represents an average of quadruplicate experiments, and error bars represent standard deviations.
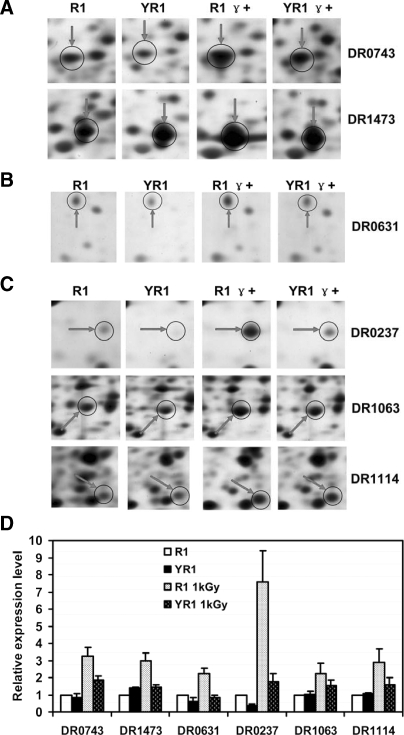
In the category of cellular processes and signaling, the expression levels of six proteins (DR0743, DR1473, DR0631, DR0237, DR1063, and DR1114) increased in response to irradiation stress in the presence of PprI (Fig. 3). DR0743 and DR1473, members of the signal transduction mechanism subgroup, responded strongly to irradiation in R1 (Fig. 3A). DR0743, a response regulator, showed an over 3-fold induction following irradiation in the presence of PprI in the R1 strain, but this induction was significantly lower in the pprI knock-out strain. The phage shock protein A (DR1473) not only functions in the transcription group but also operates in signal transduction. Although the cell division protein FtsZ (DR0631) was up-regulated significantly in the R1 strain following irradiation, its expression did not increase in the mutant YR1 strain (Fig. 3, B and D). Both DR0237 and DR1063 are peptidyl-prolyl cis-trans isomerases, and DR1114 is a member of the HSP20 heat shock protein family; all three proteins participate in protein turnover. The results indicated that the response of these three proteins to γ radiation was under the control of PprI for their expression increased remarkably after radiation in the PprI-containing wild type strain but did not reach similar levels in the pprI knock-out strain (Fig. 3, C and D).
Fig. 3.
Enhanced expression of proteins cataloged in cellular processes and signaling pathway in wild type D. radiodurans in response to 1 kGy of γ radiation. R1, wild type strain without irradiation; YR1, pprI knock-out strain without irradiation; R1 γ+, wild type strain with irradiation; YR1 γ+, pprI knock-out strain with irradiation. Protein spots in 2-D gels are shown for DR0743 and DR1473 (response regulator and phage shock protein A, respectively) (A); DR0631 (cell division protein FtsZ) (B); and DR0237, DR1063, and DR1114 (peptidyl-prolyl cis-trans isomerase, peptidyl-prolyl cis-trans isomerase C, and heat shock protein, HSP20 family, respectively) (C). D, relative protein expression levels (see “Experimental Procedures” for calculations). Each data point represents an average of quadruplicate experiments, and error bars represent standard deviations.
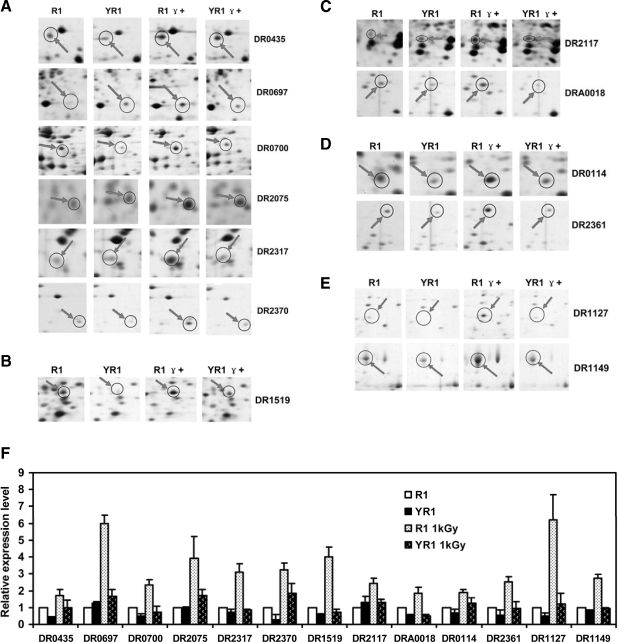
Metabolic pathways play an irreplaceable role in the recovery of D. radiodurans after irradiation. Proteins important for energy production and conversion, amino acid metabolism, nucleotide metabolism, lipid metabolism, and the ion transport system were identified as role players in response to radiation exposure under the control of PprI (Fig. 4). Of particular significance, in the category of energy production and conversion, was the 2–6-fold up-regulation of the expression level of six proteins in R1, but not in YR1, after exposure to irradiation: DR0435 (putative cytochrome complex iron-sulfur subunit), DR0697 (V-type ATP synthase, E subunit), DR0700 (V-type ATP synthase, A subunit), DR2075 (ferredoxin), DR2317 (potassium channel, β subunit), and DR2370 (pyruvate dehydrogenase complex) (Fig. 4, A and D). Similar phenotypes were observed for five additional proteins, including DR1519 (ketol-acid reductoisomerase) that functions in amino acid coenzyme metabolism, DR2117 (adenylate kinase) and DRA0018 (5`-nucleotidase) that are important in nucleotide metabolism, DR0114 (enoyl-CoA hydratase) and DR2361 (acyl-CoA dehydrogenase) that are involved in lipid metabolism, and DR1127 (hypothetical protein) and DR1149 (Na+/H+ antiporter) with suggested functions in inorganic ion metabolism (Fig. 4, B–F). These findings suggested that PprI plays an important role in the regulation of protein expression in multiple metabolic pathways.
Fig. 4.
Enhanced expression of proteins cataloged in the D. radiodurans metabolic pathways in response to γ radiation (1 kGy). R1, wild type strain without irradiation; YR1, pprI knock-out strain without irradiation; R1 γ+, wild type strain with irradiation; YR1 γ+, pprI knock-out strain with irradiation. A, protein spots in 2-D gels for DR0435 (cytochrome complex iron-sulfur), DR0697 (V-type ATP synthase, E subunit), DR0700 (V-type ATP synthase, A subunit), DR2075 (ferredoxin), DR2317 (potassium channel β subunit), and DR2370 (pyruvate dehydrogenase complex). The six proteins are cataloged in the subgroup of energy production and conversion. B, DR1519 (ketol-acid reductoisomerase) in 2-D gels. C, DR2117 and DRA0018 (adenylate kinase and 5`-nucleotidase, respectively). D, DR0114 and DR2361 (enoyl-CoA hydratase and acyl-CoA dehydrogenase, respectively). E, DR1127 and DR1149 (hypothetical protein and Na+/H+ antiporter, respectively). F, relative expression levels of proteins (see “Experimental Procedures” for calculations). Each data point represents an average of quadruplicate experiments, and error bars represent standard deviations.
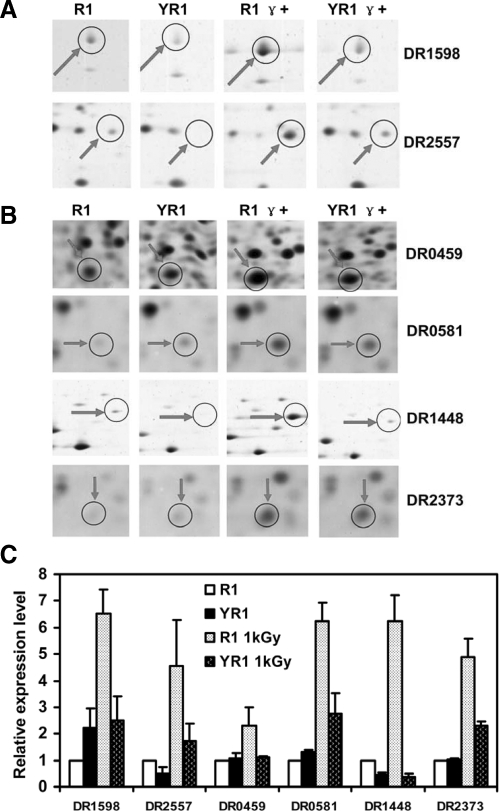
The last group consisted of proteins cataloged as function-poorly understood proteins. Cellular activity characteristics of proteins in this category were unclear. Two proteins in this category, DR1598 (protease) and DR2557 (hypothetical protein), exhibited increases in the levels of expression in response to irradiation and in the presence of PprI. Previous genome sequencing revealed many function-unknown proteins in D. radiodurans (16) that are proposed to play important roles in radioresistance of this bacterium (9, 12, 18). Many of these proteins are not cataloged in NCBI's COGs and are considered hypothetical proteins. Our results suggested that four hypothetical proteins were induced by γ radiation in R1 compared with YR1 (Fig. 5). These included DR0459, DR0581, DR1448, and DR2373.
Fig. 5.
Enhanced expression of proteins involved in the poorly characterized group in wild type D. radiodurans in response to γ radiation (1 kGy). R1, wild type strain without irradiation; YR1, pprI knock-out strain without irradiation; R1 γ+, wild type strain with irradiation; YR1 γ+, pprI knock-out strain with irradiation. A, protein spots in 2-D gels for DR1598 and DR2557 (protease and hypothetical protein, respectively). B, spots of four hypothetical proteins in 2-D gels, DR0459, DR0581, DR1448, and DR2373, that are not cataloged by COGs. C, relative expression levels of proteins (see “Experimental Procedures” for calculations). Each data point represents an average of quadruplicate experiments, and error bars represent standard deviations.
Four Proteins Were Repressed Only in the Presence of PprI in Response to Irradiation Damage—
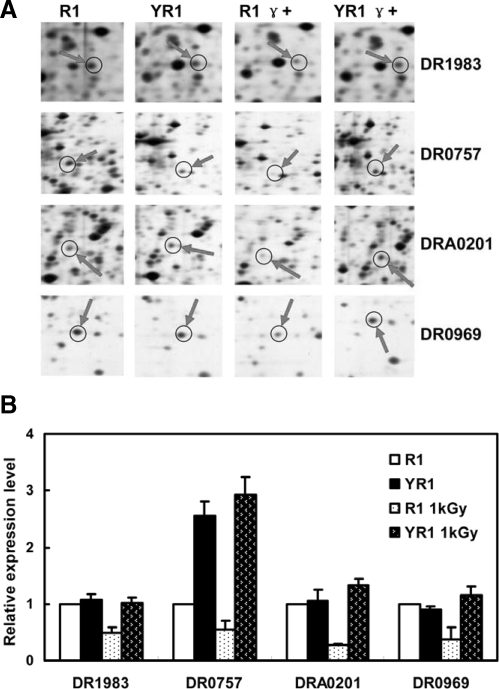
After irradiation, the expression of a number of proteins involved in different pathways is repressed in D. radiodurans as a defense mechanism (25, 27). Here the expression of four proteins remained unchanged in the absence of PprI following irradiation; however, those proteins were significantly repressed by irradiation in R1 compared with the unirradiated R1 strain, including DR1983 (ribosomal protein S1), DR0757 (citrate synthase), DRA0201 (NH3-dependent NAD+ synthase), and DR0969 (hypothetical protein) (Fig. 6).
Fig. 6.
Remarkable reduction in protein expression in D. radiodurans in response to γ radiation (1 kGy). R1, wild type strain without irradiation; YR1, pprI knock-out strain without irradiation; R1 γ+, wild type strain with irradiation; YR1 γ+, pprI knock-out strain with irradiation. A, protein spots in 2-D gels for DR1983 (ribosomal protein S1), DR0757 (citrate synthase), DRA0201 (NH3-dependent NAD+ synthase), DR0969 (hypothetical protein), and DR2337 (hypothetical protein). B, relative expression levels of proteins (see “Experimental Procedures” for calculations). Each data point represents an average of quadruplicate experiments and error bars represent standard deviations.
PprI Affects Post-translational Modification of Two Proteins—
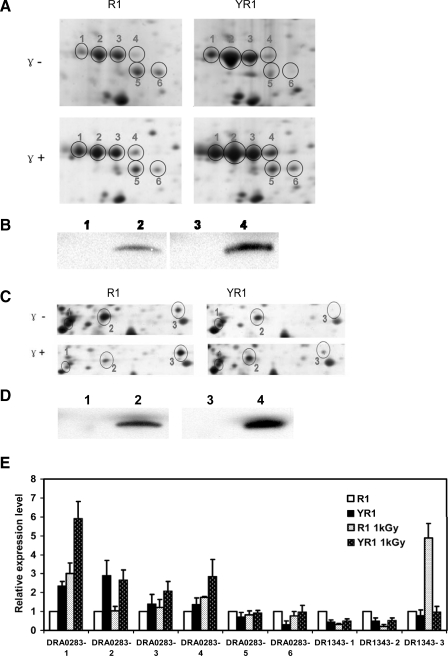
Interestingly six protein spots in different positions were identified by MS as isoforms of a single protein: DRA0283, which encodes a serine protease (Fig. 7A). The differing migration patterns suggested post-translational modifications. Deletion of pprI in the YR1 mutant strain led to an increase in levels of isoforms 1, 2, 3, and 4 but a decrease in isoforms 5 and 6. In response to irradiation, isoforms 1 and 4 increased significantly. Irradiation dramatically up-regulated isoforms 1 and 2 in YR1 but yielded a much smaller increase in the R1 strain. The isoforms (except isoforms 5 and 6) shared the same molecular weight but differed in isoelectric points, suggesting that these proteins might be phosphorylated. To confirm this hypothesis, the c-Myc-tagged DRA0283 was expressed in D. radiodurans and immunoprecipitated with polyclonal anti-c-Myc antibody. Western blotting confirmed that the serine protease was indeed phosphorylated in vivo (Fig. 7B). To verify this result, immunoprecipitation was used to pull down the protein utilizing a monoclonal anti-phosphoserine antibody, and the protein was subsequently detected with anti-c-Myc antibody (Fig. 7B). Similarly DR1343 (glyceraldehyde-3-phosphate dehydrogenase) also yielded multiple spots in the same gel (Fig. 7, C and E). Three spots corresponded to DR1343; two of these were remarkably reduced in response to irradiation in R1 but remained almost unchanged in YR1. The third spot, DR1343, exhibited an increase after irradiation in the wild type strain but not in the mutant lacking the pprI gene. DR1343 phosphorylation was validated using the method described above (Fig. 7D). These findings suggested that PprI plays a role in the regulation of specifically expressed isoforms of these proteins in response to γ radiation via phosphorylation.
Fig. 7.
Post-translational modifications in D. radiodurans in response to γ radiation (1 kGy). A, DRA0283 (serine protease) isoforms in R1 and YR1 with or without irradiation. B, determination of phosphorylation of DRA0283. The control strain R1pRADK (lane 1) and the c-Myc-tagged DRA0283 strain DRA0283myc (lane 2) were immunoprecipitated by the anti-c-Myc antibody and detected with monoclonal anti-phosphoserine antibody. The control strain R1pRADK (lane 3) and the c-Myc-tagged DRA0283 strain DRA0283myc (lane 4) were immunoprecipitated by monoclonal anti-phosphoserine antibody and detected with the anti-c-Myc antibody. C, DR1343 (glyceraldehyde-3-phosphate dehydrogenase) changes in R1 and YR1 with or without irradiation. D, determination of phosphorylation of DR1343. The control strain R1pRADK (lane 1) and c-Myc-tagged DR1343 strain DR1343myc (lane 2) were immunoprecipitated by the anti-c-Myc antibody and detected with monoclonal anti-phosphoserine antibody. The control strain R1pRADK (lane 3) and the c-Myc-tagged DR1343 strain DR1343myc (lane 4) were immunoprecipitated by monoclonal anti-phosphoserine antibody and detected with anti-c-Myc antibody. E, relative expression of post-translationally modified DRA0283 and DR1343 in R1 and YR1 with or without irradiation (see “Experimental Procedures” for calculations). Each data point represents an average of quadruplicate experiments, and error bars represent standard deviations.
Deletion of Response Genes Causes Significant Radiation Sensitivity—
Our data indicated that disruption of pprI resulted in the lack of up-regulation of dozens of its downstream proteins in response to radiation. These proteins were categorized into different functional pathways. To investigate whether proteins downstream of PprI contributed to radioresistance in response to γ radiation, four genes (DRA0018, DRA0283, DR1473, and DR2317), representative of four functional pathways, were eliminated in individual knock-out mutant strains. Survival curves of the resulting knock-outs were measured in response to irradiation (Fig. 8). Results demonstrated that the four newly constructed mutants displayed greater sensitivity to irradiation compared with the wild type strain. DR1473 (phage shock protein A), cataloged in transcription and signal transduction functional groups, is a stress response protein (17) and only demonstrated ∼2.5% of the radioresistance displayed by the wild type strain at an 8-kGy dose. Knock-out strains MA0018, MA0283, and M2317, corresponding to genes DRA0018 (5`-nucleotidase), DRA0283 (serine protease), and DR2317 (potassium channel β subunit), also displayed a defect in radioresistance with an approximate average 6-fold decrease compared with the R1 strain. The serine protease functions in post-translational modification, the 5`-nucleotidase participates in nucleotide metabolism, and the potassium channel β subunit was cataloged under energy metabolism and ion transport. These results indicated that proteins involved in various pathways play a role in radioresistance of D. radiodurans.
Fig. 8.
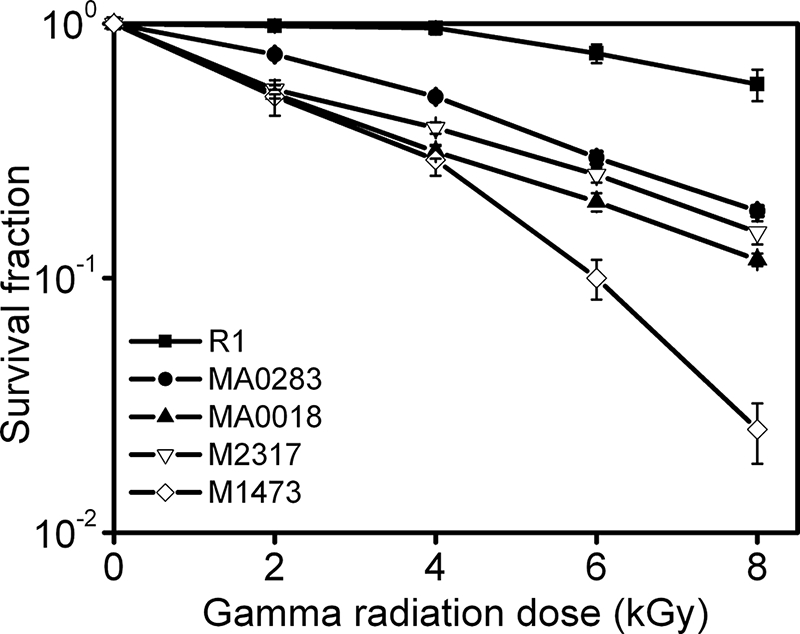
Representative survival curves of D. radiodurans strains R1, MA0283, MA0018, M2317, and M1473 following exposure to γ radiation. R1 (▪), wild type strain; MA0283 (•), deletion mutant of DRA0283; MA0018 (▴), deletion mutant of DRA0018; M2317 (▿), deletion mutant of DR2317; M1473 (⋄), deletion mutant of DR1473. Each data point represents an average of triplicate experiments, and error bars represent standard deviations.
DISCUSSION
We used 2-DE with IPG and mass spectra to investigate the proteomic changes in wild type and pprI knock-out strains following γ ray irradiation. By observing the genomic DNA recovery and cellular survival processes, we found that the whole genome repair process was going on at 1-h postirradiation period in the wild type and pprI-deleted mutant strains. Therefore this time point was used to prepare protein samples to determine which proteins were involved in cellular responses to acute irradiation damage. In this study, over 950 protein spots were visualized in each silver-strained gel; 40 proteins showed at least a 2-fold up-regulation in the R1 in response to 1 kGy of irradiation, including two isoforms of DR1343 and DRA0283. Although there are multiple studies specifically describing the proteomic changes in D. radiodurans wild type strain following γ radiation (30–32), in the current study, differences in the number of proteins identified were observed. One study reported that nine proteins were induced and at least 13 were diminished following 6 kGy of irradiation (32). However, only one protein was successfully identified as elongation factor EF-Tu because of limited genomic sequence information at that time. Poor resolution is not uncommon in 2-DE using carrier ampholyte-generated pH gradients (36) and can affect protein identification. More recently, 17 proteins were identified in another 2-DE-based proteomics study focusing on protein recycling in D. radiodurans in the postirradiation recovery (31), including several major molecular chaperones, tricarboxylic acid cycle enzymes, stress response proteins, and hypothetical proteins. In this case, the authors treated stationary cells (A600 = 3.0) with 6 kGy of irradiation, and diluted cells (A600 = 0.5) were observed for dynamics of protein expression during the cellular recovery process. Differences in cell status, irradiation dose, incubation time, quantity of sampling, IEF pH ranges, and staining methods all affect the number of proteins identified. We previously identified induced expression of 26 proteins in D. radiodurans wild type strain KD8301 cultured with 2 μg/ml streptomycin (30); 21 were identified by mass spectrometer. Nine identified proteins were also up-regulated in the current study, including DNA repair proteins SsB and PprA, stress response proteins, and metabolic enzymes. Global transcriptome analysis of the bacterium recovering from ionizing radiation has also provided useful information regarding cellular genomic expression profiling in response to the severe damage following irradiation (18, 25, 27). Previous research showed that 832 genes were induced and 451 genes were repressed over 2-fold in wild type strain recovering from 15 kGy of irradiation (25). The coverage of genes by microarray (94% (25)) is usually much higher than that by 2-DE (30% in this study). Levels of induced transcripts in microorganisms do not always correlate to protein levels in vivo (37, 38). However, compared with previous microarray studies, ∼60% of radiation-responding proteins in our study were consistent with induced transcripts identified in early and mid-phases after irradiation damage (18, 25, 27).
Previous studies indicate that PprI plays a crucial role in the extreme radioresistance of D. radiodurans as a general switch (19, 34). We proposed that this protein regulated the expression of many proteins in response to γ radiation. To test such a hypothesis, we compared the proteomic changes of the R1 strain with YR1 after radiation exposure. Gene expression induced by irradiation has been suggested to function in response to irradiation to help D. radiodurans recover from lethal DNA damage (25, 30). To determine the pathways regulated by PprI in response to irradiation, we selected out those genes with a statistically significant changed expression profile in R1 following irradiation in comparison with YR1; 37 proteins met this criterion. The variety of functions found among these proteins spanned across most groups of the NCBI's COGs, including subgroups of translation, transcription, replication and DNA repair, signal transduction mechanism, post-translational modification and protein turnover, cell cycle control, energy production and conversion, carbohydrate metabolism, nucleotide metabolism, amino acid metabolism, lipid metabolism, coenzyme metabolism, ion channel and transport, and genes of unknown function.
Although γ radiation can induce double and single strand breaks as well as base damage, often times leading to unrepairable damage in many organisms, D. radiodurans can repair the genome perfectly through the use of a highly resourceful and multipurpose repair system (1, 4, 9, 39). Three proteins in the functional groups of replication and DNA repair were identified as highly expressed in irradiated R1 compared with non-irradiated R1 and irradiated YR1, including well known DNA repair proteins RecA, SsB, and PprA. Mutants of RecA and PprA have been previously proven to be very sensitive to ionizing irradiation (20, 40). As the key protein in homologous recombination, the RecA-dependent DSB DNA repair pathway was highlighted in D. radiodurans radioresistance (1, 9). Unlike the induction of the E. coli RecA protein, neither of two LexA homologues (DRA0344 and DRA0074) is involved in the induction of RecA in D. radiodurans (28, 41). Our previous studies showed that PprI and RecX participated in the regulation of RecA as positive and negative regulators, respectively, in response to γ radiation (19, 42). Interestingly a novel protein named Deinococcus radiodurans Response Regulator A (DrRRA) (DR2418) has also been identified for its positive role in the control of RecA expression under stress-free growth conditions, although its expression was constant under radiation stresses regardless of the presence of DrRRA (23). This may suggest that there is a novel regulatory mechanism of RecA in this bacterium. Our comparative proteomics study indicated a 5-fold increase of RecA expression in the presence of PprI in the irradiated R1 strain, whereas it was unchanged in YR1 following γ ray treatment. Overexpression of RecA cannot restore the radioresistance of a pprI mutant strain (19, 43), indicating that other proteins involved in processing of damaged DNA ends from irradiation are important in the regulation of PprI. Limited content of RecA does not affect the survival of D. radiodurans after γ irradiation but delays the DNA repair process (43), leading us to propose that this repair delay may kill the bacterium. Well known for its multiple roles in replication, recombination, and repair (44), the SsB was also newly identified here as being under the control of PprI in response to γ radiation and is proposed to be important for chromosome repair. Content of SsB in D. radiodurans is higher and works more actively compared with that in E. coli (9). Failure of SsB induction after irradiation may affect the efficiency of DNA repair in the pprI knock-out strain. The novel protein PprA has been hypothesized to play a critical role in the not quite clearly understood non-homologous end-joining pathway in the bacterium due to failure of efficient DNA ligation (20). However, we also found that overexpression of RecA or PprA in a pprI-deleted mutant only complemented partial radioresistance (19), indicating that PprI regulates genes, in addition to homologous recombination repair and non-homologous end-joining pathways, to resist potentially lethal damage as a result of irradiation.
D. radiodurans possesses many stress response proteins to manage various stresses (17); several of these are under the control of PprI in the recovery from radiation, including DR0743 (response regulator), DR1082 (light-repressed protein A), DR1473 (phage shock protein A), DR1114 (HSP20), and DR1127 (hypothetical protein). DR1473 was predicted as a phage shock protein controlling membranes integrity (17), and its null mutant in this study was found to be significantly sensitive to irradiation, indicating that normal action of the protein was necessary in the recovery of D. radiodurans from irradiation. The hypothetical protein DR1127 was predicted as a toxic anion resistance protein (17). We recently found that the disruption of DR1127 caused D. radiodurans to be significantly more sensitive to γ radiation and H2O2 oxidative stress. Additionally it was shown that the DR1127 protein could bind to double-stranded DNA in vitro, protecting it from oxidative damage (45) and suggesting a role for this novel protein in the PprI-mediated pathways in response to irradiation.
Protection of proteins from irradiation-originated oxidation is facilitated by a high concentration cellular Mn2+ ions, which is considered essential for the radioresistance of the bacterium (46). Nevertheless severe ionizing radiation may still damage proteins, and the damaged cellular components need more enzymes to participate in repair. Therefore, protein recycling is another important activity in rapid recovery of D. radiodurans from radiation damage, including protein degradation and resynthesis (31, 32, 47). Here we identified several proteins involved in the functional groups of protein recycling that were under the control of PprI in response to irradiation, including transcription (DR1473, DR1970, and DR2128), translation (DR0307 and DR1082), posttranslational modifications, and protein turnover (DR0237, DR1063, DR1114, and DRA0283). With the exception of DR1082, all were predicted to be constitutively and highly expressed and were proposed to attribute themselves to the extreme radioresistance (47). DR2128, α subunit of DNA-directed RNA polymerase, plays a role in gene expression. Previous studies showed that DNA-directed RNA polymerases have a RecA-like expression pattern after γ ray exposure (25, 27, 29), indicating that the protein may participate in the recovery of the bacterium from severe radiation damage. The elongation factor G from E. coli, functioning in the elongation phase of protein synthesis, was also found to play a role as a chaperone in protein folding and renaturation after stress (48). In the current study, DR0307 (elongation factor G) in D. radiodurans was induced by radiation only in the presence of PprI, implicating its downstream role in the severe loss of radioresistance caused by pprI deletion. DR0237 and DR1063 are peptidyl-prolyl cis-trans isomerases, and their induction in response to γ radiation also suggests a relationship with PprI in radioresistance. Similarly DR1114 (heat shock protein, HSP20 family) is also part of the PprI response to irradiation damage. All three proteins work on protein folding: peptidyl-prolyl cis-trans isomerases catalyze slow protein folding reactions (49), and HSP20 belongs to small heat shock proteins, which can prevent irreversible protein denaturation of heat-damaged proteins (50). From the category of protein resynthesis, turnover, and protection, a serine protease, DRA0283, downstream of the PprI regulatory pathways was proposed to participate in the degradation of damaged proteins in D. radiodurans following γ radiation (47). Furthermore we validated that this serine protease contributed to D. radiodurans radioresistance by comparing the survival rates between the DRA0283 knock-out strain and the wild type strain under γ radiation stress. Another protease, DR1598, was strongly induced in radiation recovery and might be a member of the protease team involved in the degradation of damaged proteins in the defense against radiation. We suggest that proteins in this group are strong contributors to the radioresistance of D. radiodurans and are under the tight regulation of PprI via posttranslational modification in response to ionizing radiation.
Metabolic configuration is thought to contribute to the radioresistance of D. radiodurans (11). Thus, proteins identified here belonging to metabolism, including energy acquisition, nucleotide metabolism, amino acid metabolism, lipid metabolism, coenzyme metabolism, and ion transport, make up a majority of the proteins under the control of PprI in response to γ radiation. Energy is required in various cellular activities, including DNA damage repair. Considering that induced V-type synthases DR0697 and DR0700 provide enough energy for recovery (25) in the presence of PprI, V-type synthase in D. radiodurans might also be regulated by PprI. Similarly four other proteins function to provide energy including DR0435 (cytochrome complex iron-sulfur), DR2075 (ferredoxin), DR2370 (pyruvate dehydrogenase complex), and DR2317. The DR2317 gene is the putative β subunit of the potassium channel, and our data suggested that disruption of this gene incurred a decrease in D. radiodurans radioresistance. Consistent with what has been described previously (25), the citrate synthase DR0757, involved in the tricarboxylic acid cycle, was repressed in the presence of PprI to avoid production of additional free radicals. In addition, a 5`-nucleotidase was noted for its enhanced expression, and its knock-out strain was validated here to be more sensitive to γ radiation, suggesting that nucleotide metabolism joins in the recovery from radiation damage.
Our data indicate that PprI may play a critical role in the post-translational modification of at least two proteins: DRA0283 and DR1343. The serine protease DRA0283, involved in protein recycling in D. radiodurans when recovering from irradiation (31), was here shown to contribute to radioresistance. This study confirmed that the protein was phosphorylated in vivo. However, there are still two other lower molecular weight isoforms (isoforms 5 and 6), indicative of a post-translational modification other than phosphorylation, such as a partial deletion of the N or C terminus. DR1343 (glyceraldehyde-3-phosphate dehydrogenase) is a housekeeping protein functioning in basic catabolic processes, including translational regulation, DNA replication, and DNA repair (51). Glyceraldehyde-3-phosphate dehydrogenase possesses phosphotransferase/kinase activity and can phosphorylate itself as well as other proteins (52). We found that the protein phosphorylated in D. radiodurans and the levels of its phosphorylated isoform were related to the presence of PprI in response to γ radiation.
A cross-protection among different stresses was hypothesized because common proteins are induced by heat shock, γ irradiation, and desiccation in D. radiodurans (18, 53, 54). In the current study, several proteins induced by irradiation in the presence of PprI were also identified in previous studies to be involved in the heat shock response, including four stress response proteins (DR0743, DR1114, DR1082, and DR1473), two DNA repair proteins (DR2340 and DRA0346), and two metabolic proteins (DR1598 and DR1967). This indicates overlap among different responsive pathways. D. radiodurans becomes extremely sensitive to H2O2, mitomycin C, UV light, and γ rays in the absence of PprI (19, 22, 43, 55). On the other hand, both mitomycin C and γ radiation result in DSBs. Although participation of RecA is essential in DNA DSB repair, overexpression of RecA protein in pprI-deleted strain completely restores the resistance to mitomycin C but not to γ radiation (43), indicating that the PprI-mediated cellular response differs depending on the stress type.
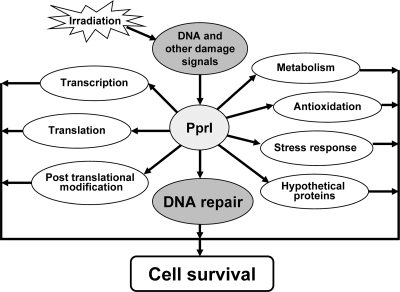
We propose that the extreme radioresistance of D. radiodurans stems from its efficient and systematic mobilization and operation. Our hypothetical functional response diagram of PprI in D. radiodurans in response to radiation (Fig. 9) is based on our current data and on the observation that PprI is indispensable for D. radiodurans radioresistance. Although the initial causes of cellular stresses (ionizing radiation) and end consequences (cellular survival) as well as the enhanced expression of protein components in different pathways are clear, the interrelationship among different functional pathways is missing and awaits further studies. As illustrated by the network diagram, irradiation causes many cellular damage events, including DNA damage (DSBs and single strand breaks), directly or indirectly by production of reactive oxygen species. PprI may directly sense the DNA damage and/or other cellular damage signals. Activated PprI, acting as a transcriptional factor, may switch on defense systems, including the DNA repair system as well as transcription, translation, and post-translational modification systems that function to increase the production of defense proteins and activate other defense pathways. The metabolism of energy is particularly important to provide energy to DNA repair and cellular recovery processes from radiation damage. Antioxidation and stress response pathways assist the reduction of damage from potential stresses by these processes (19). The current work demonstrates that PprI acts as a hub to dozens of upstream and downstream pathways to realize the extreme radioresistance and cellular survival capacity of D. radiodurans.
Fig. 9.
Proposed model for D. radiodurans survivorship in part due to activation of the PprI-mediated functional pathways in response to irradiation. Irradiation produces various cellular damage signals. PprI may directly sense those signals or be activated by other intermediate sensors. The activated PprI partly controls the stress response pathway and also switches on many defense pathways. The DNA repair system is one of the major pathways that is activated by PprI. It enhances the scavenging capacity for reactive oxygen species (19) by switching on catalase E. It up-regulates protein expression in several protein synthesis and recycling pathways. The activated transcription, translation, and post-translational systems subsequently work together to produce more defense proteins, proteases responsible for degrading damaged proteins, and proteins working in energy metabolism, all of which participate in cell survival from radiation damage directly or indirectly.
Acknowledgments
We thank Li Zheng and Valerie Chavez in the Shen laboratory and Chunchao Zhang at the University of Michigan for stimulating discussions and critical reading of the manuscript. We also thank the Research Center for Proteome Analysis of the Chinese Academy of Sciences for instructions in proteomics analysis.
Footnotes
Published, MCP Papers in Press, October 24, 2008, DOI 10.1074/mcp.M800123-MCP200
The abbreviations used are: DSB, double strand break; PprI, inducer of pleiotropic proteins promoting DNA repair; RecA, recombinase A; PprA, pleiotropic protein promoting DNA repair A; SsB, single strand DNA-binding protein; 2-DE, two-dimensional electrophoresis; COGs, clusters of orthologous groups; Gy, grays; kGy, kilogray(s); 2-D, two-dimensional; NCBI, National Center for Biotechnology Information; PMF, peptide mass fingerprint.
This work was supported by a grant from the National Basic Research Program of China (2004 CB 19604), a grant from the National Hi-Tech Development Program (2007AA021305), a key project from the National Natural Science Foundation of China (30830006), and the project “Application of Nuclear Techniques in Agriculture” from the Chinese Ministry of Agriculture (200803034) (to Y. J. H.); by a scholarship from China Scholarship Council (to H. M. L.); and by United States Congress-directed Department of Defense Grant 1435-04-06GT63257 (to B. H. S.).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Minton, K. W. ( 1994) DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol. Microbiol. 13, 9–15 [DOI] [PubMed] [Google Scholar]
- 2.Battista, J. R., Earl, A. M., and Park, M. J. ( 1999) Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol. 7, 362–365 [DOI] [PubMed] [Google Scholar]
- 3.Lin, J., Qi, R., Aston, C., Jing, J., Anantharaman, T. S., Mishra, B., White, O., Daly, M. J., Minton, K. W., Venter, J. C., and Schwartz, D. C. ( 1999) Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science 285, 1558–1562 [DOI] [PubMed] [Google Scholar]
- 4.Narumi, I. ( 2003) Unlocking radiation resistance mechanisms: still a long way to go. Trends Microbiol. 11, 422–425 [DOI] [PubMed] [Google Scholar]
- 5.Venkateswaran, A., McFarlan, S. C., Ghosal, D., Minton, K. W., Vasilenko, A., Makarova, K., Wackett, L. P., and Daly, M. J. ( 2000) Physiologic determinants of radiation resistance in Deinococcus radiodurans. Appl. Environ. Microbiol. 66, 2620–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brim, H., McFarlan, S. C., Fredrickson, J. K., Minton, K. W., Zhai, M., Wackett, L. P., and Daly, M. J. ( 2000) Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat. Biotechnol. 18, 85–90 [DOI] [PubMed] [Google Scholar]
- 7.Lange, C. C., Wackett, L. P., Minton, K. W., and Daly, M. J. ( 1998) Engineering a recombinant Deinococcus radiodurans for organopollutant degradation in radioactive mixed waste environments. Nat. Biotechnol. 16, 929–933 [DOI] [PubMed] [Google Scholar]
- 8.Battista, J. R. ( 1997) Against all odds: the survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 51, 203–224 [DOI] [PubMed] [Google Scholar]
- 9.Cox, M. M., and Battista, J. R. ( 2005) Deinococcus radiodurans—the consummate survivor. Nat. Rev. Microbiol. 3, 882–892 [DOI] [PubMed] [Google Scholar]
- 10.Daly, M. J., Gaidamakova, E. K., Matrosova, V. Y., Vasilenko, A., Zhai, M., Venkateswaran, A., Hess, M., Omelchenko, M. V., Kostandarithes, H. M., Makarova, K. S., Wackett, L. P., Fredrickson, J. K., and Ghosal, D. ( 2004) Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306, 1025–1028 [DOI] [PubMed] [Google Scholar]
- 11.Ghosal, D., Omelchenko, M. V., Gaidamakova, E. K., Matrosova, V. Y., Vasilenko, A., Venkateswaran, A., Zhai, M., Kostandarithes, H. M., Brim, H., Makarova, K. S., Wackett, L. P., Fredrickson, J. K., and Daly, M. J. ( 2005) How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol. Rev. 29, 361–375 [DOI] [PubMed] [Google Scholar]
- 12.Makarova, K. S., Omelchenko, M. V., Gaidamakova, E. K., Matrosova, V. Y., Vasilenko, A., Zhai, M., Lapidus, A., Copeland, A., Kim, E., Land, M., Mavrommatis, K., Pitluck, S., Richardson, P. M., Detter, C., Brettin, T., Saunders, E., Lai, B., Ravel, B., Kemner, K. M., Wolf, Y. I., Sorokin, A., Gerasimova, A. V., Gelfand, M. S., Fredrickson, J. K., Koonin, E. V., and Daly, M. J. ( 2007) Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLoS ONE 2, e955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahradka, K., Slade, D., Bailone, A., Sommer, S., Averbeck, D., Petranovic, M., Lindner, A. B., and Radman, M. ( 2006) Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443, 569–573 [DOI] [PubMed] [Google Scholar]
- 14.Daly, M. J. ( 2006) Modulating radiation resistance: insights based on defenses against reactive oxygen species in the radioresistant bacterium Deinococcus radiodurans. Clin. Lab. Med. 26, 491–504 [DOI] [PubMed] [Google Scholar]
- 15.Chen, H., Xu, G., Zhao, Y., Tian, B., Lu, H., Yu, X., Xu, Z., Ying, N., Hu, S., and Hua, Y. ( 2008) A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS ONE 3, e1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White, O., Eisen, J. A., Heidelberg, J. F., Hickey, E. K., Peterson, J. D., Dodson, R. J., Haft, D. H., Gwinn, M. L., Nelson, W. C., Richardson, D. L., Moffat, K. S., Qin, H., Jiang, L., Pamphile, W., Crosby, M., Shen, M., Vamathevan, J. J., Lam, P., McDonald, L., Utterback, T., Zalewski, C., Makarova, K. S., Aravind, L., Daly, M. J., Minton, K. W., Fleischmann, R. D., Ketchum, K. A., Nelson, K. E., Salzberg, S., Smith, H. O., Venter, J. C., and Fraser, C. M. ( 1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286, 1571–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makarova, K. S., Aravind, L., Wolf, Y. I., Tatusov, R. L., Minton, K. W., Koonin, E. V., and Daly, M. J. ( 2001) Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65, 44–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka, M., Earl, A. M., Howell, H. A., Park, M. J., Eisen, J. A., Peterson, S. N., and Battista, J. R. ( 2004) Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 168, 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua, Y., Narumi, I., Gao, G., Tian, B., Satoh, K., Kitayama, S., and Shen, B. ( 2003) PprI: a general switch responsible for extreme radioresistance of Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 306, 354–360 [DOI] [PubMed] [Google Scholar]
- 20.Narumi, I., Satoh, K., Cui, S., Funayama, T., Kitayama, S., and Watanabe, H. ( 2004) PprA: a novel protein from Deinococcus radiodurans that stimulates DNA ligation. Mol. Microbiol. 54, 278–285 [DOI] [PubMed] [Google Scholar]
- 21.Harris, D. R., Tanaka, M., Saveliev, S. V., Jolivet, E., Earl, A. M., Cox, M. M., and Battista, J. R. ( 2004) Preserving genome integrity: the DdrA protein of Deinococcus radiodurans R1. PLoS Biol. 2, e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earl, A. M., Mohundro, M. M., Mian, I. S., and Battista, J. R. ( 2002) The IrrE protein of Deinococcus radiodurans R1 is a novel regulator of recA expression. J. Bacteriol. 184, 6216–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, L., Xu, G., Chen, H., Zhao, Y., Xu, N., Tian, B., and Hua, Y. ( 2008) DrRRA: a novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Mol. Microbiol. 67, 1211–1222 [DOI] [PubMed] [Google Scholar]
- 24.Gao, G., Tian, B., Liu, L., Sheng, D., Shen, B., and Hua, Y. ( 2003) Expression of Deinococcus radiodurans PprI enhances the radioresistance of Escherichia coli. DNA Repair (Amst.) 2, 1419–1427 [DOI] [PubMed] [Google Scholar]
- 25.Liu, Y., Zhou, J., Omelchenko, M. V., Beliaev, A. S., Venkateswaran, A., Stair, J., Wu, L., Thompson, D. K., Xu, D., Rogozin, I. B., Gaidamakova, E. K., Zhai, M., Makarova, K. S., Koonin, E. V., and Daly, M. J. ( 2003) Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 100, 4191–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao, G., Le, D., Huang, L., Lu, H., Narumi, I., and Hua, Y. ( 2006) Internal promoter characterization and expression of the Deinococcus radiodurans pprI-folP gene cluster. FEMS Microbiol. Lett. 257, 195–201 [DOI] [PubMed] [Google Scholar]
- 27.Chen, H., Xu, Z. J., Tian, B., Chen, W. W., Hu, S. N., and Hua, Y. J. ( 2007) Transcriptional profile in response to ionizing radiation at low dose in Deinococcus radiodurans. Prog. Nat. Sci. 17, 529–536 [Google Scholar]
- 28.Sheng, D., Zheng, Z., Tian, B., Shen, B., and Hua, Y. ( 2004) LexA analog (dra0074) is a regulatory protein that is irrelevant to recA induction. J. Biochem. 136, 787–793 [DOI] [PubMed] [Google Scholar]
- 29.Lipton, M. S., Pasa-Tolic, L., Anderson, G. A., Anderson, D. J., Auberry, D. L., Battista, J. R., Daly, M. J., Fredrickson, J., Hixson, K. K., Kostandarithes, H., Masselon, C., Markillie, L. M., Moore, R. J., Romine, M. F., Shen, Y., Stritmatter, E., Tolic, N., Udseth, H. R., Venkateswaran, A., Wong, K. K., Zhao, R., and Smith, R. D. ( 2002) Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. U. S. A. 99, 11049–11054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, C., Wei, J., Zheng, Z., Ying, N., Sheng, D., and Hua, Y. ( 2005) Proteomic analysis of Deinococcus radiodurans recovering from gamma-irradiation. Proteomics 5, 138–143 [DOI] [PubMed] [Google Scholar]
- 31.Joshi, B., Schmid, R., Altendorf, K., and Apte, S. K. ( 2004) Protein recycling is a major component of post-irradiation recovery in Deinococcus radiodurans strain R1. Biochem. Biophys. Res. Commun. 320, 1112–1117 [DOI] [PubMed] [Google Scholar]
- 32.Tanaka, A., Hirano, H., Kikuchi, M., Kitayama, S., and Watanabe, H. ( 1996) Changes in cellular proteins of Deinococcus radiodurans following gamma-irradiation. Radiat. Environ. Biophys. 35, 95–99 [DOI] [PubMed] [Google Scholar]
- 33.Meima, R., Rothfuss, H. M., Gewin, L., and Lidstrom, M. E. ( 2001) Promoter cloning in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 183, 3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao, G., Lu, H., Huang, L., and Hua, Y. ( 2005) Construction of DNA damage response gene pprI function-deficient and function-complementary mutants in Deinococcus radiodurans. Chin. Sci. Bull. 50, 311–316 [Google Scholar]
- 35.Mattimore, V., and Battista, J. R. ( 1996) Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178, 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Görg, A., Obermaier, C., Boguth, G., Harder, A., Scheibe, B., Wildgruber, R., and Weiss, W. ( 2000) The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21, 1037–1053 [DOI] [PubMed] [Google Scholar]
- 37.Gygi, S. P., Rochon, Y., Franza, B. R., and Aebersold, R. ( 1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao, H., Bausch, C., Richmond, C., Blattner, F. R., and Conway, T. ( 1999) Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181, 6425–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minton, K. W. ( 1996) Repair of ionizing-radiation damage in the radiation resistant bacterium Deinococcus radiodurans. Mut. Res. 363, 1–7 [DOI] [PubMed] [Google Scholar]
- 40.Gutman, P. D., Carroll, J. D., Masters, C. I., and Minton, K. W. ( 1994) Sequencing, targeted mutagenesis and expression of a recA gene required for the extreme radioresistance of Deinococcus radiodurans. Gene (Amst.) 141, 31–37 [DOI] [PubMed] [Google Scholar]
- 41.Narumi, I., Satoh, K., Kikuchi, M., Funayama, T., Yanagisawa, T., Kobayashi, Y., Watanabe, H., and Yamamoto, K. ( 2001) The LexA protein from Deinococcus radiodurans is not involved in RecA induction following gamma irradiation. J. Bacteriol. 183, 6951–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheng, D., Liu, R., Xu, Z., Singh, P., Shen, B., and Hua, Y. ( 2005) Dual negative regulatory mechanisms of RecX on RecA functions in radiation resistance, DNA recombination and consequent genome instability in Deinococcus radiodurans. DNA Repair (Amst.) 4, 671–678 [DOI] [PubMed] [Google Scholar]
- 43.Jolivet, E., Lecointe, F., Coste, G., Satoh, K., Narumi, I., Bailone, A., and Sommer, S. ( 2006) Limited concentration of RecA delays DNA double-strand break repair in Deinococcus radiodurans R1. Mol. Microbiol. 59, 338–349 [DOI] [PubMed] [Google Scholar]
- 44.Meyer, R. R., and Laine, P. S. ( 1990) The single-stranded DNA-binding protein of. Escherichia coli. Microbiol. Rev. 54, 342–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, Y., Tian, B., and Hua, Y. ( 2007) dr1127: a novel gene of Deinococcus radiodurans responsible for oxidative stress. Chin. Sci. Bull 52, 2081–2087 [Google Scholar]
- 46.Daly, M. J., Gaidamakova, E. K., Matrosova, V. Y., Vasilenko, A., Zhai, M., Leapman, R. D., Lai, B., Ravel, B., Li, S. M., Kemner, K. M., and Fredrickson, J. K. ( 2007) Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 5, e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlin, S., and Mrazek, J. ( 2001) Predicted highly expressed and putative alien genes of Deinococcus radiodurans and implications for resistance to ionizing radiation damage. Proc. Natl. Acad. Sci. U. S. A. 98, 5240–5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caldas, T., Laalami, S., and Richarme, G. ( 2000) Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J. Biol. Chem. 275, 855–860 [DOI] [PubMed] [Google Scholar]
- 49.Schmid, F. X. ( 1993) Prolyl isomerase: enzymatic catalysis of slow protein-folding reactions. Annu. Rev. Biophys. Biomol. Struct. 22, 123–142 [DOI] [PubMed] [Google Scholar]
- 50.Narberhaus, F. ( 2002) α-Crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol. Mol. Biol. Rev. 66, 64–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sirover, M. A. ( 1997) Role of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell function and in cell pathology. J. Cell. Biochem. 66, 133–140 [PubMed] [Google Scholar]
- 52.Sirover, M. A. ( 1999) New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432, 159–184 [DOI] [PubMed] [Google Scholar]
- 53.Schmid, A. K., Howell, H. A., Battista, J. R., Peterson, S. N., and Lidstrom, M. E. ( 2005) Global transcriptional and proteomic analysis of the Sig1 heat shock regulon of Deinococcus radiodurans. J. Bacteriol. 187, 3339–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmid, A. K., Lipton, M. S., Mottaz, H., Monroe, M. E., Smith, R. D., and Lidstrom, M. E. ( 2005) Global whole-cell FTICR mass spectrometric proteomics analysis of the heat shock response in the radioresistant bacterium Deinococcus radiodurans. J. Proteome Res. 4, 709–718 [DOI] [PubMed] [Google Scholar]
- 55.Tian, B., Zhang, S. W., Xu, Z. J., Sheng, D. H., and Hua, Y. J. ( 2006) Effects of PprI and RecX on antioxidant activity of. Deinococcus radiodurans. Wei Sheng Wu Xue Bao 46, 238–242 [PubMed] [Google Scholar]