Abstract
It is generally agreed that reactive oxygen species (ROS) contribute to skin aging, skin disorders, and skin diseases. Skin possesses an extremely efficient antioxidant system. This antioxidant activity is conferred by two systems: antioxidant enzymes and small molecules that can scavenge ROS by donating electrons. No gene-encoded secreted ROS scavengers have been reported. Amphibian skin is a multifunctional organ acting in defense, respiration, and water regulation, although it seems susceptible. Amphibian skins are easily harmed by biological or non-biological attacks such as microorganism infection or radiation injury. Among vertebrates, skins of amphibian are exposed to more dangers of radiation injury than others. Radiation toxicity occurs by directly attacking the genetic material and/or by generating ROS. In addition, amphibian skin respiration and inflammatory response also induce ROS generation. It is rational to hypothesize that amphibian skins should have potent free radical scavenging and radioprotective ability for their survival. Rana pleuraden is distributed in Southwest of China; it lives in the subtropical plateau (altitude around 2300 m) where there is strong ultraviolet radiation and long duration of sunshine. By peptidomics and genomics approaches, a large amount of antioxidant peptides belonging to 11 different groups with variable structures were isolated from the skin secretions of R. pleuraden. Their free radical scavenging and anti-inflammatory abilities were studied. All of these peptide share highly homologous preproregions, although mature antioxidant peptides have very divergent primary structures, suggesting the possibility of a common ancestor. Some peptides were also found to have multifunctional properties, such as combined antioxidant, anti-inflammatory, and antimicrobial activities. According to our knowledge, no gene-encoded specific antioxidant peptides have been reported except metallothionein. Our work possibly reveals a new skin antioxidant system. The current work also provides a large amount of peptide candidates with medical-pharmaceutical significance.
Skin is exposed to oxidative stress from both endogenous and exogenous sources, and very few tissues in the body are subjected to similar degrees of exposure as is skin (1, 2). Skin has developed various mechanisms to cope with the increasing oxidative stress (3–5). The skin antioxidant defense system can be classified into two major groups: the enzyme group and the low molecular weight antioxidant (LMWA)1 group. The first group includes superoxide dismutase, catalase, peroxidase, and some supporting enzymes such as glucose-6-phosphate dehydrogenase and glutathione reductase (3–10). The second group includes a large number of compounds capable of preventing oxidative damage by scavenging ROS (2–5, 8, 10). They share a similar chemical trait that allows them to donate an electron to ROS. LMWAs are from cell synthesis (e.g. GSH, NADH, and carnosine), waste products (e.g. uric acid), and dietary resources (e.g. carotene, polyphenols, and lipoic acid) (2–5, 8, 10). No gene-encoded LMWA has been reported.
Among vertebrates, amphibians may have the most susceptible skins because they are directly exposed to various living environments. Amphibians, being the first group of organisms forming a connecting link between land and water, are forced to adopt and survive in a variety of conditions laden with pathogens and predators (11). Amphibian skins play key roles in the everyday survival of amphibians and their ability to exploit a wide range of habitats and ecological conditions, although their naked skins seem to be susceptible to biological or non-biological injuries, such as microorganism infection, parasitization, predation, and physical harm including radiation and aseptic wounds (11). Therefore, they are endowed with an excellent chemical defense system composed of pharmacological and antimicrobial gene-encoded peptides (11–13). Amphibian skins are attractive subjects for chemical prospecting to reveal the detailed molecular defensive mechanism and high level of biochemical diversity. Some amphibian skin peptides, such as antimicrobial peptides, tachykinins, dermorphins, cholecystokinin, and bradykinins, have been studied extensively (14–25). They have pharmacological effects including cardiotoxic, neurotoxic, and antimicrobial activities (11). All the properties clearly and adversely affect potential predators or pathogens. In contrast to defensive peptides against biological injuries, few peptides against non-biological injuries have been reported. Radiation injury may be one of the most important non-biological stresses for amphibian skins. Amphibians living in the plateau where there is strong ultraviolet light and a long duration of sunshine may have more risks to encounter radiation injury.
Rana pleuraden with a body length up to 50 cm lives in the Yunnan-Guizhou subtropical plateau (most of its life environments are in an altitude around 2300 m and have a long duration of sunshine (2400 h/year). We predicted that R. pleuraden skin has strong free radical scavenging and radioprotective ability. In this work, antioxidant peptides from skin secretions of R. pleuraden were investigated by coupling proteomics analysis with cDNA trapping. Fourteen peptide groups (13 novel) containing 36 members were identified. Eleven of the peptide groups could exert antioxidant activities. This is the first report of gene-encoded antioxidant peptides secreted from skins.
EXPERIMENTAL PROCEDURES
Collection of Frog Skin Secretions
Adult specimens of R. pleuraden of both sexes (n = 6; weight range, 30–40 g) were collected in Yunnan Province of China. Skin secretions were collected as follows. Frogs were put into a cylinder container containing a piece of absorbent cotton saturated with anhydrous ether. Upon exposure to anhydrous ether for 1–2 min, the frog skin surface was seen to exude copious secretions. Skin secretions were collected by washing the dorsal region of each frog with 0.1 m NaCl solution (containing protease inhibitor mixture, Sigma). The collected solutions (total volume, 100 ml) were quickly centrifuged, and the supernatants were lyophilized. All the experiments were approved by Kunming Institute of Zoology, Chinese Academy of Sciences.
Peptide Fractionation
Lyophilized skin secretion samples (50 mg) were dissolved in 2 ml of 0.1 m phosphate buffer, pH 6.0, containing protease inhibitor mixture and filtered through a 10-kDa-cutoff Centriprep filter (Millipore, Bedford, CA). Concentrated filtrate was applied to a C18 reversed-phase (RP) HPLC (Hypersil BDS C18, 30 × 0.46 cm) column. The absorbance of the eluate was monitored at 280 and 220 nm. Each eluted peak was collected for antioxidant and antimicrobial activity assays.
Structural Analysis
Complete peptide sequencing was undertaken by Edman degradation on an Applied Biosystems pulsed liquid-phase sequencer, model 491. The fractions with antioxidant activities from RP-HPLC were placed in a MALDI plate (Kratos Analytical). Mass fingerprints were obtained using a MALDI-TOF mass spectrometer (AXIMA CFR, Kratos Analytical) in positive ion and linear mode. The specific parameters were as follows: the ion acceleration voltage was 20 kV, the accumulating time of single scanning was 50 s, and polypeptide mass standard (Kratos Analytical) served as external standard. The accuracy of mass determinations was within 0.1%.
Free Radical Scavenging Activity
2,2-Diphenyl-1-picrylhydrazyl (DPPH) Scavenging—
Free radical scavenging activity was determined by using a stable DPPH radical (Sigma) following a previously reported method of von Gadow et al. (27) with a slight modification. The assay mixture contained 1.9 ml of 5 × 10−5 m DPPH radical dissolved in ethanol and 0.1 ml of sample solution (concentration range, 0.5–10 mg/ml). After a 30-min incubation period at room temperature, the absorbance was read against a blank at 517 nm. Inhibition of free radical by DPPH in percent (I%) was calculated according to Equation 1,
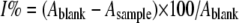 |
(Eq. 1) |
where Ablank is the absorbance of the control reaction (containing all reagents except the test compound) and Asample is the absorbance of the test compound. Butylated hydroxytoluene (BHT; Sigma) was used as positive control.
2,2′-Azinobis(3-ethylbenzothiazoline-6-sulfonic Acid) (ABTS+) Scavenging—
Free radical scavenging activity was also determined by measuring reduction of radical ABTS+ according to the manufacturer's instructions with the kit GMS10114.4 (Genmed Scientifics Inc., Shanghai, China). The total formation of products (i.e. the reduced form of ABTS and the purple antioxidin-RP1 modification) and the total consumption of ABTS radical were determined by linear regression analysis. The concentrations of ABTS and ABTS free radical were calculated by using ɛ340 = 4.8 × 104 m−1 cm−1 and ɛ415 = 3.6 × 104 m−1 cm−1, respectively (18, 19). The purple antioxidin-RP1 modification was monitored by using A550 (18).
NO Scavenging—
Scavenging of NO was determined by incubating 5 mm sodium nitroprusside dihydrate (SNP; Sigma) in PBS with different concentrations of samples at 25 °C. After 120 min, 0.5 ml of incubation solution was withdrawn and mixed with 0.5 ml of Griess reagent (Promega). The absorbance was measured at 550 nm. The amount of nitrite was calculated from a standard curve constructed using sodium nitrite. All absorbances were monitored using an Ultrospec 2100 pro UV/visible spectrophotometer (Amersham Biosciences).
Reducing Power
The reducing power was evaluated according to Oyaizu (28) and Kaur et al. (29). Different amounts of tested samples were suspended in distilled water and mixed with 2.5 ml of 0.2 m phosphate buffer, pH 6.6, and 2.5 ml of 1% K3Fe(CN)6. The mixture was incubated at 50 °C for 20 min, 2.5 ml of 10% TCA was added to the mixture, and the mixture was centrifuged at 3000 rpm for 10 min. The upper layer of the solution (2.5 ml) was mixed with distilled water (2.5 ml) and FeCl3 (0.5 ml; 0.1%), and the absorbance was measured at 700 nm. An increase in absorbance of the reaction mixture indicated reducing power. Butylated hydroxyanisole (BHA) was used as a positive control.
Anti-inflammatory Activity Assay
Cytokines assays were detected by antibody sandwich ELISAs using the kits from Adlitteram Diagnostic Laboratories, Inc. according to the manufacturer's instructions.
Antimicrobial Activity Testing
Microbe-killing testing was performed according to our previous methods (16, 17). Standard bacterial and fungal strains used in antimicrobial activity assays, Gram-positive bacterium Staphylococcus aureus (ATCC2592), Gram-negative bacteria Escherichia coli (ATCC25922) and Bacillus dysenteriae, and fungus Candida albicans (ATCC2002) were obtained from Kunming Medical College. Bacteria were first grown in Luria-Bertani (LB) broth to an A600 of 0.8. A 10-μl aliquot of the bacteria was then taken and added to 8 ml of fresh LB broth with 0.7% agar and poured over a 90-mm Petri dish containing 25 ml of 1.5% agar in LB broth. After the top agar hardened, a 20-μl aliquot of the test sample filtered on a 0.22-μm Millipore filter was dropped onto the surface of the top agar and completely dried before being incubated overnight at 37 °C. In the case that an examined sample contained antimicrobial activity, a clear zone formed on the surface of the top agar representing inhibition of bacterial growth. The minimal inhibitory concentration (MIC) was determined in liquid LB medium as previously reported (16, 17). The MIC at which no visible growth occurred was recorded.
SMART cDNA Synthesis
Total RNA was extracted using TRIzol (Invitrogen) from the skin of a single sample of amphibian. cDNA was synthesized by SMART™ techniques by using a SMART PCR cDNA synthesis kit (Clontech). The first strand was synthesized by using cDNA 3′ SMART CDS Primer II A, 5′-AAGCAGTGGTATCAACGCAGAGTACT30N−1N-3′ (N = A, C, G, or T; N − 1 = A, G, or C), and SMART II An oligonucleotide, 5′-AAGCAGTGGTATCAACGCAGAGTACGCGGG-3′. The second strand was amplified using Advantage polymerase by 5′ PCR primer II A, 5′-AAGCAGTGGTATCAACGCAGAGT-3′.
cDNA Cloning
The cDNA synthesized by SMART techniques was used as template for PCR to screen the cDNAs encoding antioxidant peptides. The oligonucleotide primers (as listed in supplemental Table S1) in the antisense direction were designed according to the amino acid sequences determined by Edman degradation. Primer II A as mentioned under “SMART cDNA Synthesis” was used as the sense direction primer. The DNA polymerase was Advantage polymerase from Clontech. The PCR conditions were 2 min at 94 °C followed by 30 cycles of 10 s at 92 °C, 30 s at 50 °C, and 40 s at 72 °C. Finally the PCR products were cloned into pGEM®-T Easy vector (Promega, Madison, WI). DNA sequencing was performed on an Applied Biosystems DNA sequencer, model ABI PRISM 377. Diversity screening of cDNA encoding possible antioxidant peptides was performed by using primers based on the predicted signal peptide sequences of precursors encoding purified antioxidant peptides that have been cloned by specific primers.
Purification of Purple Antioxidin-RP1 Modification and MALDI-TOF MS Analysis
The purple product of antioxidin-RP1 with ABTS free radical was purified by a Sephadex G-25 Fine column (volume, 30 × 1.6 cm) eluted with 2 mm NH4HCO3, pH 8.5, at a flow rate of 0.3 ml/min, and 0.3-ml fractions were collected manually (supplemental Fig. S2). The absorbance at 280, 340, 415, and 550 nm was monitored, respectively. The fraction containing purple antioxidin-RP1 modification was purified further by RP-HPLC using a C18 column. The purified purple antioxidin-RP1 modification was subjected to MALDI-TOF MS analysis.
Nuclear Magnetic Resonance (NMR)
The NMR sample of free peptide was prepared by dissolving 1 mg of antioxidin-RP1 powder in 550 μl of PBS buffer (90% H2O, 10% D2O, pH 5.2). The purple complex was prepared by dissolving 0.5 mg of antioxidin-RP1 powder in 550 μl of PBS buffer including 1 mm ABTS+, pH 5.2. All NMR experiments were performed on a Varian Unity INOVA 600-MHz spectrometer equipped with four radiofrequency channels, pulse shaping, and z axis pulsed field gradient capabilities. Two-dimensional phase-sensitive NMR spectra, including total correlation spectroscopy (TOCSY; mixing time, 75 ms) and nuclear Overhauser enhancement spectroscopy (mixing time, 300 ms), were recorded at 298 K. The Watergate approach was used for suppression of the solvent peak. Quadrature detection in the F1 dimension was achieved using the state time-proportional phase incrementation approach. Data were collected with 256 and 4096 complex data points in the t1 and t2 dimensions, respectively, and signals were averaged over 32 transients. Data were processed using the program NMRPipe/NMRDraw (30) and analyzed by Sparky (31). Linear prediction in the t1 dimension was used before the Fourier transformation. The average chemical shift changes of backbone protons were calculated according to the formula δave = [rad]ΔδHN2 + ΔδHA2[/rad].
Alkylation of Cysteine
Thiol groups were alkylated according to a previous method (32). Peptides (0.18 mm) were incubated with 22 mm iodoacetamide (IAA) in PBS (10 mm sodium phosphate, pH 7.4, 120 mm NaCl, 3 mm KCl) for 1 h at room temperature in the dark and then desalted by Sephadex G-25 Fine (column volume, 30 × 1.6 cm) eluted with 2 mm NH4HCO3, pH 8.5.
Synthetic Peptides
All of the peptides (supplemental Table S2) used in this work were synthesized by AC Scientific Inc. (Xi An, China) with a confirmed purity higher than 95% by HPLC and MALDI-TOF mass spectrometry.
RESULTS
Purification and Primary Structure Determination of Antioxidant Peptides—
More than 10 peaks were eluted from the supernatant of the R. pleuraden skin secretions by C18 RP HPLC as illustrated in supplemental Fig. S1A. The peaks are indicated as 1–14 in supplemental Fig. S1A. Peaks 1, 2, 4–8, and 11–13 were found to exert significant antioxidant activities. These peaks were collected to be subjected to purity analysis by MALDI-TOF mass spectrometry. As illustrated in supplemental Fig. S1, C–P, most of the peaks are high in purity except peak 2 and peak 13. Most of the mass spectrometry showed (M + H)+, (M + Na)+, or (M + K) + signals. Peak 2 and peak 13 were purified further by C18 RP HPLC (data not shown). All the amino acid sequences of the purified peptides with antioxidant activities were analyzed by Edman degradation.
Ten purified antioxidant peptides were named pleurain-A1, pleurain-D4, pleurain-E1, pleurain-J1, pleurain-K1, pleurain-M1, pleurain-P1, and antioxidin-RP2, respectively. An antimicrobial peptide named pleurain-C1 was also purified. Their complete amino acid sequences were determined by Edman degradation as listed in Table I. The calculated masses of these peptides based on the amino acid sequences determined by Edman degradation matched well with the observed masses analyzed by MALDI-TOF mass spectrometry as illustrated in supplemental Fig. S1, C, D, F–J, and M–O. Most of the mass differences are less than 0.5 Da, and the largest mass difference is −0.9 Da in the case of pleurain-M1. All these antioxidant peptides are composed of 14–36 amino acid residues with a molecular mass of 1–4 kDa. Pleurain-A1 has been reported as an antimicrobial peptide in our previous work (26). Other peptide families are novel.
Table I.
Antioxidant and antimicrobial peptides from skin of R. pleuraden
AM, antimicrobial; AO, antioxidant; —, no assay.
| Name | Sequence | Function |
|---|---|---|
| Pleurain-A1 | SIITMTKEAKLPQLWKQIACRLYNTC | AM, AO |
| Pleurain-A2 | SIITMTKEAKLPQSWKQIACRLYNTC | |
| Pleurain-A3 | SIITMTREAKLPQLWKQIACRLYNTC | |
| Pleurain-A4 | SIITTTKEAKLPQLWKQIACRLYNTC | |
| Pleurain-B1 | FLGGLLASLLGKI | AM |
| Pleurain-B2 | FLGGLLSGIFKHL | |
| Pleurain-B3 | FLGGLLFGIFKHL | |
| Pleurain-B4 | FLGGLLSGLFKHL | |
| Pleurain-B5 | FLGGLLSSIFGHL | |
| Pleurain-C1 | YPELQQDLIARLL | AM |
| Pleurain-C2 | FPELQQDLIARLL | |
| Pleurain-D1 | FLSGILKLAFKIPSVLCAVLKNC | AM, AO |
| Pleurain-D2 | FLSGILKLASKIPSVLCAVLKNC | |
| Pleurain-D3 | FFSGILKLVFKIPSVLCAVLKNC | |
| Pleurain-D4 | FRSGILKLASKIPSVLCAVLKNC | |
| Pleurain-E1 | AKAWGIPPHVIPQIVPVRIRPLCGNV | AM, AO |
| Pleurain-G1 | GFWDSVKEGLKNAAVTILNKIKCKISECPPA | AM, AO |
| Pleurain-G2 | GLWDSVKEGLKNAAVTILNKIKCKIFECPPA | |
| Pleurain-G3 | GLWDSVKEGLKNAAVTILNKIKCKISECPPA | |
| Pleurain-G4 | GLWDSVKEGFKNAAVTLLNKIKCKISACPPA | |
| Pleurain-G5 | GIFDSIKEGFKNAAVTLLDKIKCKISACPPA | |
| Pleurain-G6 | GIFDSIKEGFKNAAVTLLNKIKCKISDCPPA | |
| Pleurain-G7 | GIFDSIKEGFKNAAVTLLNKIKCKISECPPA | |
| Pleurain-G8 | GIFDSIKEGFKNAAVTLLNKIK | |
| Pleurain-G9 | GIFDSIKEGFKNAAVTLLNKIKCKNF | |
| Pleurain-J1 | FIPGLRRLFATVVPTVVCAINKLPPG | AM, AO |
| Pleurain-J2 | FIPGLRRLSATVVPTVVCAINKLPPG | |
| Pleurain-K1 | DDPDKGMLKWKNDFFQEF | AO |
| Pleurain-M1 | GLLDSVKEGLKKVAGQLLDTLKCKISGCTPA | AM, AO |
| Pleurain-M2 | GLLDSVKEGLKKAAGQLLDTLKCEISGCTPA | |
| Pleurain-N1 | GFFDRIKALTKNVTLELLNTITCKLPVTPP | AO |
| Pleurain-P1 | SFGAKNAVKNGLQKLRNQCQANNYQGPFCDIFKKNP | AO |
| Pleurain-Q1 | SFLCDLKILATNAAKNAGQCVVTTLSCKLCGTC | — |
| Pleurain-R1 | CVHWMTNTARTACIAP | AM, AO |
| Antioxidin-RP1 | AMRLTYNKPCLYGT | AO |
| Antioxidin-RP2 | SMRLTYNKPCLYGT |
Antioxidant Peptide Diversity—
The cDNAs encoding the precursors of these antioxidant peptides isolated from skin secretions of R. pleuraden were cloned as illustrated in Fig. 1. All the precursors share similar overall structures including highly conserved signal peptide regions and variable mature peptide regions. According to the conserved signal peptides, common primers (5′-CCAAA(G/C)ATGTTCACC(T/A)TGAAGAAA(T/C)-3′) (sense) and primer IIA (antisense) mentioned under “SMART cDNA Synthesis“) were used to screen possible antioxidant peptide diversity. 115 cDNAs encoding 36 different peptides belonging to 14 groups were isolated (GenBank™ accession numbers EU138029–EU138143 and EU780583). The precursors encoding nine of these 10 isolated peptides from the frog skin secretions have been cloned. Only pleurain-R1 precursor has not been cloned. The deduced mature peptides from the cDNA sequences have amino acid sequences identical to those of these purified antioxidant peptides. The four members of pleurain-A have been reported in our previous work (26). Pleurain-D has four members. Pleurain-J, pleurain-M, and antioxidin-RP have two members. Only one member has been found in other groups. In addition, four novel groups of peptides (pleurain-B containing five members, pleurain-G containing nine members, pleurain-N containing one members, and pleurain-Q containing one member) were cloned from the skin cDNA library. Pleurain-B and pleurain-N has been proved to exert only antimicrobial activity and antioxidant activity, respectively, and pleurain-G contains both antimicrobial and antioxidant activities as described later. The function of pleurain-Q has not been investigated. In total, 11 groups (pleurain-A, -D, -E, -G, -J, -K, -M, -N, -P, -R, and antioxidin-RP) of peptides containing antioxidant activity have been identified from the skin of R. pleuraden, suggesting the diversity of antioxidant peptides in amphibian skin. Interestingly all the groups of antioxidant peptides contain proline residues, suggesting that proline may have an important role for their antioxidant capability.
Fig. 1.
The alignment of precursors encoding amphibian antioxidant and antimicrobial peptides. The region of signal peptide is indicated by a bar. Mature peptides are underlined. The amino acid residues identical to the first line are shaded. The precursor sequences of amphibian antimicrobial peptides are from Refs. 14, 15, and 17.
Free Radical Scavenging Activity—
Because of their relative stability, easy measurement, and good reproducibility, DPPH and ABTS+ radicals are commonly used to evaluate antioxidant capacity (33). Both assays are based on decolorization by monitoring absorbance decreases at the characteristic wavelength of 520 nm for the DPPH system and 734 nm for the ABTS system. The ABTS system is usually correlated with the DPPH system because these two radicals have a similar chemical property of hydrogen/electron donation (33). As listed in Table II, all 11 antioxidant peptides could scavenge the free radical of DPPH and/or ABTS+. The antioxidant peptides react with DPPH and ABTS+ and convert these colored stable free radicals into colorless compounds. The amount of reduced DPPH and ABTS+ can be quantified by measuring a decrease in absorbance at 517 and 734 nm, respectively. At a concentration of 80 μg/ml, pleurain-N1, -R1, and antioxidin-RP1 showed strong DPPH free radical scavenging ability, whereas pleurain-N1, -P1, and antioxidin-RP1 showed strong ABTS+ free radical scavenging ability. Most of these antioxidant peptides also quenched NO released by an NO donor of SNP. Incubation of SNP solution in PBS at 25 °C for 120 min resulted in the release of NO. Among them, antioxidin-RP1 had the strongest ability to quench NO release as well as to reduce DPPH and ABTS+ free radicals.
Table II.
Antioxidant activities of peptides (80 μg/ml)
Values are means ± S.D. of three replicate analyses. NA, no activity.
| Samples | Free radical scavenging capacity
|
Fe3+ reducing power (absorbance at 700 nm) | ||
|---|---|---|---|---|
| ABTS+ | DPPH | NO | ||
| % | ||||
| H2O | 0 ± 0.0 | 0 ± 6.22 | NA | NA |
| BHA | 70.1 ± 6.5 | 90.4 ± 13.9 | 46.5 ± 7.4 | 0.27 ± 0.06 |
| NFAT inhibitor | 15.2 ± 1.7 | 9.5 ± 5.4 | NA | 0.06 ± 0.02 |
| Galanin (porcine) fragment 1–16 | 8.9 ± 3.3 | 8.2 ± 5.5 | NA | 0.06 ± 0.01 |
| Somatostatin fragment 9–23 | 17.6 ± 3.6 | 11.3 ± 2.2 | 2.9 ± 0.3 | 0.08 ± 0.04 |
| Pleurain-A1 | 74.8 ± 2.2 | NA | 21.1 ± 5.2 | 0.15 ± 0.04 |
| M5G-pleurain-A1 | 49.6 ± 4.3 | NA | 10.7 ± 3.6 | 0.09 ± 0.02 |
| P12G-pleurain-A1 | 60.1 ± 4.8 | NA | 3.1 ± 0.3 | 0.13 ± 0.04 |
| W15G-pleurain-A1 | 70.7 ± 2.0 | NA | 11.3 ± 2.7 | 0.12 ± 0.07 |
| Y23G-pleurain-A1 | 37.1 ± 3.4 | NA | 5.5 ± 1.9 | 0.07 ± 0.01 |
| M5G/P12G/W15G/Y23G-pleurain-A1 | No antioxidant activity | |||
| Pleurain-D4 | NA | 30.3 ± 12.7 | 35.7 ± 3.6 | 0.21 ± 0.02 |
| P13G-pleurain-D4 | NA | 5.1 ± 1.3 | 2.7 ± 0.3 | 0.03 ± 0.01 |
| Pleurain-E1 | 6.3 ± 8.5 | 69.1 ± 9.6 | 11.5 ± 2.9 | 0.29 ± 0.02 |
| W4G-pleurain-E1 | 5.7 ± 2.4 | 52.3 ± 3.2 | 10.7 ± 4.5 | 0.24 ± 0.05 |
| P7G/P8G/P12G/P16G/P24G-pleurain-E1 | 3.3 ± 0.5 | 27.6 ± 5.7 | 5.1 ± 3.2 | 0.11 ± 0.04 |
| C23G-pleurain-E1 | 2.5 ± 0.7 | 20.4 ± 2.9 | 2.9 ± 1.3 | 0.09 ± 0.01 |
| W4G/P7G/P8G/P12G/P16G/P24G/C23G-pleurain-E1 | No antioxidant activity | |||
| Pleurain-G1 | 66.3 ± 1.8 | 58.5 ± 3.3 | 41.3 ± 7.7 | 0.19 ± 0.01 |
| W3G-pleurain-G1 | 41.7 ± 8.2 | 30.7 ± 4.5 | 27.1 ± 2.2 | 0.12 ± 0.03 |
| P29G/P30G-pleurain-G1 | 30.1 ± 3.9 | 21.5 ± 2.6 | 19.2 ± 5.3 | 0.08 ± 0.05 |
| W3G/P29G/P30G-pleurain-G1 | NA | 5.6 ± 1.4 | NA | 0.02 ± 0.01 |
| Pleurain-J1 | 60.1 ± 0.0 | 76.4 ± 9.6 | NA | 0.37 ± 0.04 |
| P3G/P14G/P24G/P25G-pleurain-J1 | 23.6 ± 2.5 | 25.9 ± 4.4 | NA | 0.15 ± 0.03 |
| C18G-pleurain-J1 | 20.2 ± 3.2 | 19.9 ± 6.7 | NA | 0.11 ± 0.05 |
| P3G/P14G/P24G/P25G/C18G-pleurain-J1 | No antioxidant activity | |||
| Pleurain-K1 | 18.6 ± 2.2 | 13.6 ± 3.9 | NA | 0.24 ± 0.11 |
| P3G-pleurain-K1 | 11.3 ± 2.9 | 9.2 ± 1.8 | NA | 0.17 ± 0.06 |
| M7G-pleurain-K1 | 6.5 ± 1.3 | NA | NA | 0.09 ± 0.01 |
| W10G-pleurain-K1 | 15.1 ± 2.6 | 10.9 ± 3.3 | NA | 0.21 ± 0.05 |
| P3G/M7G/W10G-pleurain-K1 | No antioxidant activity | |||
| Pleurain-M1 | 29.8 ± 0.0 | 31.1 ± 17.3 | NA | 0.13 ± 0.00 |
| P30G-pleurain-M1 | 3.1 ± 1.1 | NA | NA | 0.01 ± 0.00 |
| Pleurain-N1 | 87.8 ± 1.5 | 91.4 ± 3.3 | 52.4 ± 11.2 | 0.35 ± 0.04 |
| C23G-pleurain-N1 | 40.3 ± 3.7 | 42.1 ± 2.2 | 30.6 ± 1.0 | 0.16 ± 0.02 |
| P26G/P29G/P30G-pleurain-N1 | 30.5 ± 5.9 | 27.7 ± 4.5 | 25.4 ± 3.8 | 0.13 ± 0.05 |
| C23G/P26G/P29G/P30G-pleurain-N1 | No antioxidant activity | |||
| Pleurain-P1 | 41.9 ± 2.0 | 83.9 ± 3.8 | 33.1 ± 6.2 | 0.23 ± 0.06 |
| Y24G-pleurain-P1 | 17.3 ± 3.5 | 37.6 ± 7.2 | 17.1 ± 2.9 | 0.10 ± 0.02 |
| P27G/P36G-pleurain-P1 | 20.5 ± 6.7 | 45.3 ± 3.2 | 22.2 ± 4.7 | 0.15 ± 0.01 |
| Y24G/P27G/P36G-pleurain-P1 | 10.1 ± 0.5 | 13.8 ± 4.1 | NA | 0.06 ± 0.01 |
| Pleurain-R1 | 92.4 ± 1.5 | 21.4 ± 3.0 | 17.5 ± 3.9 | 0.34 ± 0.09 |
| W4G-pleurain-R1 | 79.3 ± 9.9 | 20.6 ± 5.1 | 15.6 ± 2.1 | 0.31 ± 0.03 |
| M5G-pleurain-R1 | 38.7 ± 6.2 | 13.3 ± 4.9 | 8.2 ± 1.6 | 0.16 ± 0.05 |
| P16G-pleurain-R1 | 45.6 ± 9.5 | 11.3 ± 5.8 | 9.6 ± 3.4 | 0.18 ± 0.01 |
| W4G/M5G/P16G-pleurain-R1 | No antioxidant activity | |||
| Antioxidin-RP1 | 95.9 ± 4.7 | 96.6 ± 5.1 | 66.4 ± 9.8 | 0.41 ± 0.07 |
| M2G-antioxidin-RP1 | 73.5 ± 8.2 | 72.7 ± 10.5 | 50.1 ± 6.7 | 0.36 ± 0.02 |
| Y6G-antioxidin-RP1 | 55.5 ± 2.1 | 46.8 ± 3.5 | 32.4 ± 3.3 | 0.23 ± 0.05 |
| Y12G-antioxidin-RP1 | 85.7 ± 7.1 | 79.6 ± 9.5 | 52.6 ± 6.2 | 0.37 ± 0.07 |
| P9G-antioxidin-RP1 | 82.1 ± 4.9 | 70.5 ± 2.8 | 45.0 ± 2.9 | 0.37 ± 0.01 |
| C10G-antioxidin-RP1 | 50.1 ± 2.2 | 42.3 ± 5.6 | 27.4 ± 5.5 | 0.22 ± 0.00 |
| M2G/Y6G/Y12G/P9G/C10G-antioxidin-RP1 | No antioxidant activity | |||
Reducing Power—
Ferric ions (Fe3+) reducing antioxidant capacity were determined by the Fe3+–Fe2+ transformation method. As listed in Table II, most of these antioxidant peptides have significant reducing power when compared with the standard (BHA). The results demonstrate the electron donor properties of these antioxidant peptides for neutralizing free radicals by forming stable products. In vivo, the reducing reaction terminates the radical chain reactions that may otherwise be very damaging (34). Among them, antioxidin-RP1 is the strongest in reducing power as well as in free radical scavenging abilities.
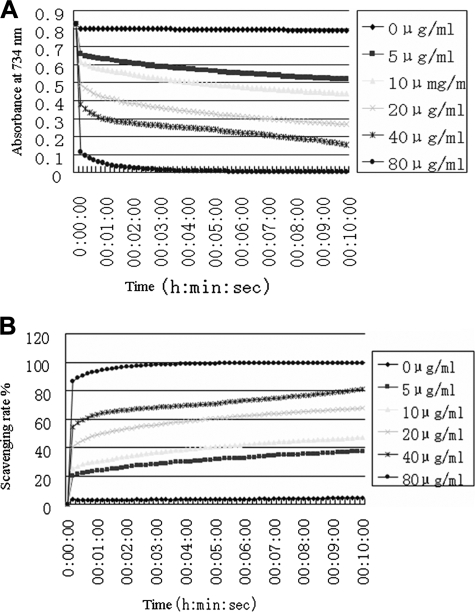
DPPH Free Radical Scavenging Kinetics—
As the results reported above show, antioxidin-RP1 has the strongest antioxidant ability. Its free radical scavenging kinetics was investigated in our experiments. As illustrated in Fig. 2, antioxidin-RP1 could rapidly scavenge DPPH in a dose-dependent manner. It could get rid of DPPH immediately when it makes contact with DPPH. At the concentration of 80 μg/ml, antioxidin-RP1 could scavenge 85% of DPPH within 5 s and scavenge nearly 100% of DPPH within 4 min. 20 μg/ml antioxidin-RP1 could scavenge 50% of DPPH within 2 min. Even at a concentration down to 5 μg/ml, 40% of DPPH was scavenged within 10 min by antioxidin-RP1. All these results indicated that antioxidin-RP1 is a strong and highly active antioxidant peptide.
Fig. 2.
DPPH free radical scavenging kinetics of antioxidin-RP1. A, DPPH was scavenged by antioxidin-RP1 in a dose- and time-dependent manner. B, percent rate of DPPH scavenged by antioxidin-RP1 is dose- and time-dependent.
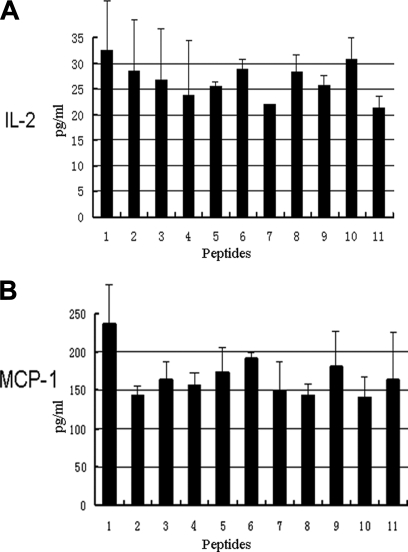
Anti-inflammatory Activities of Antioxidant Peptides—
Various proofs have indicated the cross-talking between inflammation and oxidants (35). Considering the antioxidant activities of these peptides isolated from the skin of R. pleuraden, we hypothesized that these antioxidant peptides have anti-inflammatory functions. As illustrated in Fig. 3, they could exert anti-inflammatory effects. At a concentration of 5 μg/ml, they could significantly inhibit secretions of IL-2 and MCP-1 induced by lipopolysaccharides from E. coli. IL-2 and MCP-1 are known proinflammatory cytokines (36). These antioxidant peptides from amphibian skins possibly scavenge oxidants to decrease cytokine secretion (IL-2 and MCP-1) and to inhibit inflammatory responses.
Fig. 3.
Antioxidant peptides (2 μg/ml) inhibit secretions of IL-2 (A) and MCP-1 (B) induced by lipopolysaccharides (2 μg/ml). 1, control; 2, pleurain-D4; 3, pleurain-E1; 4, pleurain-G1; 5, pleurain-J1; 6, pleurain-K1; 7, pleurain-M1; 8, pleurain-N1; 9, pleurain-N1; 10, pleurain-P1; 11, antioxidin-RP1. Error bars represent the standard error of the mean in three separate experiments.
Antimicrobial Activities—
The precursors of the current antioxidant peptides have been found to share conserved signal peptides with antimicrobial peptide precursors from ranid amphibians, suggesting that these antioxidant peptides may have antimicrobial activities. The 11 groups of antioxidant peptides and another two groups of peptides (pleurain-B1 and -C1) identified by cDNA screening were assayed for their antimicrobial abilities. The antimicrobial activities of pleurain-A have been reported in our previous work. As listed in Table III, there are nine groups of peptides (Pleurain-B, -C, -D, -E, -G, -J, -M, -N, and -R) that exert antimicrobial functions. All nine groups of peptides with antimicrobial activities are of novel peptide families; no similar peptides could be found by BLAST (basic local alignment search tool) search. The current results added nine more families of novel antimicrobial peptides. Seven of them have both antioxidant and antimicrobial activities, and they are bifunctional peptides. Pleurain-B and -C have only antimicrobial activities. Antioxidin-RP, pleurain-N, and pleurain-P have no antimicrobial activities, and they seem to be specific antioxidant peptides.
Table III.
Antimicrobial activities of peptides from R. pleuraden skins and their mutants
NA, no activity.
| Antimicrobial activity (MIC)
|
||||
|---|---|---|---|---|
| E. coli | S. aureus | Bacillus subtilis | C. albicans | |
| μg/ml | ||||
| Antioxidin-RP1 | NA | NA | NA | NA |
| M2G-antioxidin-RP1 | NA | NA | NA | NA |
| Y6G-antioxidin-RP1 | NA | NA | NA | NA |
| Y12G-antioxidin-RP1 | NA | NA | NA | NA |
| P9G-antioxidin-RP1 | NA | NA | NA | NA |
| C10G-antioxidin-RP1 | NA | NA | NA | NA |
| M2G/Y6G/Y12G/P9G/C10G-antioxidin-RP1 | NA | NA | NA | NA |
| Pleurain-A1 | 60 | 15 | 120 | 30 |
| M5G-pleurain-A1 | 60 | 15 | 120 | 30 |
| P12G-pleurain-A1 | 120 | 15 | 120 | 60 |
| W15G-pleurain-A1 | 120 | 30 | 120 | 60 |
| Y23G-pleurain-A1 | 30 | 15 | 60 | 15 |
| M5G/P12G/W15G/Y23G-pleurain-A1 | 120 | 15 | 120 | 30 |
| Pleurain B1 | 25 | 3.1 | 3.1 | 1.6 |
| Pleurain C1 | 50 | 6.3 | 3.1 | 25 |
| Pleurain D1 | 25 | 12.5 | 50 | 25 |
| Pleurain-D4 | 50 | 12.5 | 50 | 25 |
| P13G-pleurain-D4 | 50 | 25 | 50 | 25 |
| Pleurain E1 | 25 | 6.3 | NA | 25 |
| W4G-pleurain-E1 | 50 | 12.6 | NA | 25 |
| P7G/P8G/P12G/P16G/P24G-pleurain-E1 | 25 | 12.6 | NA | 50 |
| C23G-pleurain-E1 | 25 | 3.2 | NA | 25 |
| W4G/P7G/P8G/P12G/P16G/P24G/C23G-pleurain-E1 | 50 | 6.3 | NA | 50 |
| Pleurain G1 | 50 | 50 | 100 | 12.5 |
| W3G-pleurain-G1 | 100 | 50 | 100 | 12.5 |
| P29G/P30G-pleurain-G1 | 50 | NA | 100 | 25 |
| W3G/P29G/P30G-pleurain-G1 | 100 | NA | 100 | 25 |
| Pleurain J1 | 12.5 | 12.5 | 50 | 6.3 |
| P3G/P14G/P24G/P25G-pleurain-J1 | 12.5 | 12.5 | 100 | 12.5 |
| C18G-pleurain-J1 | 6.3 | 6.3 | 50 | 6.3 |
| P3G/P14G/P24G/P25G/C18G-pleurain-J1 | 12.5 | 12.5 | 100 | 6.3 |
| Pleurain K1 | NA | NA | NA | NA |
| P3G-pleurain-K1 | NA | NA | NA | NA |
| M7G-pleurain-K1 | NA | NA | NA | NA |
| W10G-pleurain-K1 | NA | NA | NA | NA |
| P3G/M7G/W10G-pleurain-K1 | NA | NA | NA | NA |
| Pleurain M1 | 6.3 | 6.3 | 25 | 12.5 |
| P30G-pleurain M1 | 6.3 | 6.3 | 25 | 12.5 |
| Pleurain N1 | 50 | 12.5 | NA | 12.5 |
| C23G-pleurain-N1 | 25 | 12.5 | NA | 6.3 |
| P26G/P29G/P30G-pleurain-N1 | 100 | 25 | NA | 12.5 |
| C23G/P26G/P29G/P30G-pleurain-N1 | 50 | 25 | NA | 25 |
| Pleurain P1 | NA | NA | NA | NA |
| Y24G-pleurain-P1 | NA | NA | NA | NA |
| P27G/P36G-pleurain-P1 | NA | NA | NA | NA |
| Y24G/P27G/P36G-pleurain-P1 | NA | NA | NA | NA |
| Pleurain R1 | 100 | 25 | 100 | 50 |
| W4G-pleurain-R1 | 100 | 12.5 | 100 | 50 |
| M5G-pleurain-R1 | 100 | 12.5 | 100 | 50 |
| P16G-pleurain-R1 | 200 | 25 | 200 | 50 |
| W4G/M5G/P16G-pleurain-R1 | 200 | 25 | 100 | 50 |
All the Antioxidant Peptide Precursors Share Similar N-terminal Preproregions and Common Defensive Signal Peptides with Antimicrobial Peptides from Ranidae Amphibians—
Sequence comparison of antioxidant peptides of R. pleuraden with antimicrobial peptide precursors of Ranidae amphibians is illustrated in Fig. 1. All the antioxidant peptides have similar precursor structures. Furthermore their precursors are similar to antimicrobial peptide precursors of ranid amphibians. They share a similar overall structure that is composed of an N-terminal peptide followed by an acidic spacer peptide and a C-terminal mature peptide. Their signal peptides (composed of 22 amino acid residues) are highly conserved with 70–100% identity (Fig. 1). In all of the precursors, there is a dibasic enzymatic processing site insertion (-Lys-Arg- or -Arg-Arg-) between the spacer peptide and mature peptide. It is the same with other defensive peptide precursors of amphibians such as antimicrobial peptides (16–19), suggesting that these antioxidant peptide precursors have a biosynthesis pathway similar to that of antimicrobial peptides.
Amino Acid Residue Replacement to Investigate the Molecular Basis of Antioxidant Peptides—
It has been mentioned above that all the groups of antioxidant peptides contain proline residue in their sequences. Furthermore sulfur-containing amino acids (methionine and cysteine) (32, 37), a phenol-containing amino acid (tyrosine) (32), tryptophan (37), and proline (38) have been suggested to contribute antioxidant functions. All these amino acid residues (proline, methionine, free cysteine, tyrosine, and tryptophan) in the 11 groups of antioxidant peptides were replaced by glycine (supplemental Table S2), respectively, and the antioxidant and antimicrobial activities of these mutants were assayed. In the peptide families of pleurain-A, -D, -G, -M, and -R, we did not replace cysteines that form intramolecular disulfides. Similar disulfides have been confirmed to have important roles in the antimicrobial activity of amphibian antimicrobial peptides (12, 14, 15, 17). As listed in Tables II and III, the replacement of proline, methionine, free cysteine, or tyrosine obviously decreased their antioxidant capability, whereas tryptophan replacement just had a slight effect on the antioxidant capability. Replacing all these amino acid residues eliminated their antioxidant function. For the peptides containing both antioxidant and antimicrobial activities (pleurain-A, -D, -E, -G, -J, -M, -N, and -R), the amino acid replacements had no obvious effects on their antimicrobial capability, although their antioxidant functions had been intensively destroyed or even completely eliminated. Some interesting details were found to confirm that some of the important structural features are sequence/peptide-specific among the different peptides. For example, pleurain-M1 has a single proline, and its replacement by glycine reduced its free radical scavenging activity (via ABTS assay) by 10-fold (Table I), yet it had no effect on antimicrobial activity (Table III). In contrast, pleurain-K1 has a methionine, proline, and tryptophan. The mutation data about pleurain-K1 is a bit more complex, showing little change in antioxidant activity with the elimination of tryptophan and a more nuanced decrease in activity for substitution of proline (less than 2-fold reduction in antioxidant activity) and methionine (∼3-fold reduction) but nothing like the drop in activity with pleurain-M1, which has only one proline. These results suggest that the proline in different positions among these antioxidant peptides plays different roles. Furthermore the control peptides containing proline, methionine, free cysteine, tyrosine, and tryptophan (nuclear factor of activated T cells (NFAT) inhibitor, secretin human, and somatostatin fragment 9–23) showed weak antioxidant capability (Table II and supplemental Table S2). All the results suggest that these antioxidant peptides are sequence-specific.
The Investigation of Antioxidant Mechanism—
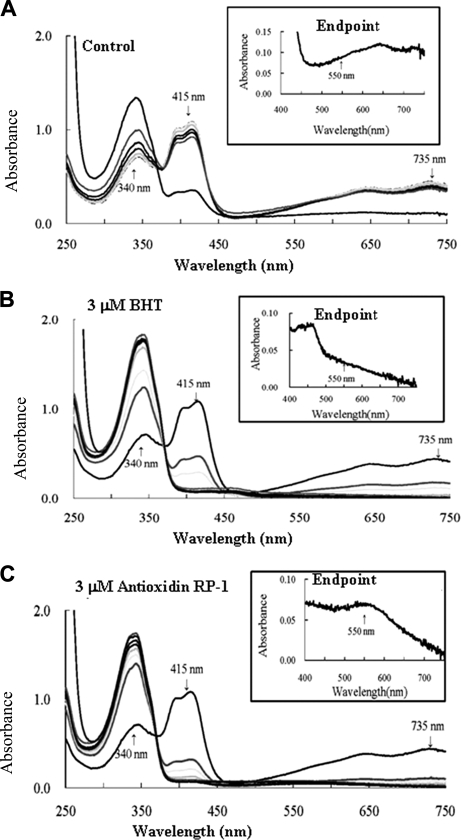
Antioxidin-RP1 has been found to be the most potent antioxidant peptide and to contain most of the amino residues responsible for antioxidant function (methionine, free cysteine, tyrosine, and proline). This peptide was selected to react with ABTS radical to study the possible antioxidant mechanism.
As illustrated in Fig. 4, the ABTS radical was readily reduced by antioxidin-RP1. Overlaid scans of the solution showed a decrease of the ABTS radical-specific 415 and 735 nm peaks and a concomitant increase of the ABTS-specific peak at 340 nm. Although the ABTS radical displayed a slender autoscavenging activity (Fig. 4A), antioxidin-RP1 could significantly catalyze this process (Fig. 4C); even its catalysis rate is much faster than that of a commercial antioxidant factor (BHT) (Fig. 4B). The reduction process by antioxidin-RP1 is a two-phase reaction. An initial faster phase over several seconds was followed by a second slower phase that was still ongoing after 20 min (Figs. 2A and 4C). Most of the ABTS radical was reduced after 2 s by antioxidin-RP1. Remaining ABTS radicals were reduced by adding 60 mm NaN3. Two end products were found; they are reduced ABTS represented by the 340 nm peak and a novel peak at 550 nm (Fig. 4C, inset) that gave the solution a purple color (supplemental Fig. S2A). No product with the absorbance at 550 nm was found in the BHT-ABTS reaction system (Fig. 4B), indicating that BHT may have a different radical scavenging mechanism than antioxidin-RP1.
Fig. 4.
The investigation of antioxidant mechanism. Water (A), BHT (B), and antioxidin-RP1 (C), respectively, were added to standard ABTS+ solutions to a final concentration of 3 μm; the absorbance spectrum was read at 0 s, 2 s, 30 s, 2 min, 5 min, 10 min, and 20 min; and finally an end point scan was read immediately after the addition of sodium azide.
The purple compound was purified by Sephadex G-25 and RP HPLC and then was subjected to MALDI-TOF mass spectrometry analysis (supplemental Fig. S2, A–C). As illustrated in supplemental Fig. S2, A and B, the purple compound was first co-eluted with the antioxidin-RP1 (absorbance at 280), whereas the remaining antioxidin-RP1, ABTS radicals, and ABTS were eluted later. Because of the co-elution of the purple compound with antioxidin-RP1, we hypothesized that the purple compound can bind with antioxidin-RP1. The MALDI-TOF mass spectrometry analysis of the purple compound gave a mass (m/z) of 1898.91 (supplemental Fig. S2C). The mass of 1898.91 is larger than the mass (1629.80) of antioxidin-RP1 and the mass (515) of ABTS, but it is nearly equal to the total mass (1900.8) of antioxidin-RP1 plus ABTS free radical (271). This result indicated that the purple compound consists of possible adducts of antioxidin-RP1 with ABTS free radical.
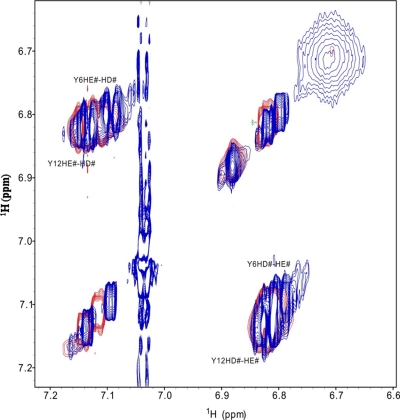
It has been proved that phenols can react with ABTS radicals to form purple compounds that show a broad absorbance around 550 nm (32, 39, 40). Only tyrosine contains phenol side chain. There are two tyrosine residues (Tyr6 and Tyr12) in antioxidin-RP1, and they possibly react with ABTS radicals to form the antioxidin-RP1-ABTS purple adduct. The total mass (1900.8) of antioxidin-RP1 plus ABTS free radical is 2 mass units larger than the observed mass of 1898.91 (supplemental Fig. S2C). The 2 mass units account for the oxidized tyrosine residue as confirmed by a previous report (32). As listed in Table II, Tyr6 replacement in antioxidin-RP1 extremely decreased the antioxidant capability, whereas Tyr12 replacement had only a slight effect on the antioxidant capability, suggesting that Tyr6 takes part in the reaction with ABTS radicals. Because ABTS radicals interact with aromatic regions of phenols (32, 39, 40), the superposition of the two side-chain aromatic regions (Tyr6 and Tyr12) of TOCSY spectra was recorded on free antioxidin-RP1 and in the complex with ABTS+ by two-dimensional NMR (Fig. 5). The NMR chemical shift also indicated that Tyr6 is responsible for the ABTS radical link. Fig. 5 shows that the residue Tyr6 had larger chemical shift changes than did Tyr12 in aromatic regions. Furthermore Tyr6 replacement in antioxidin-RP1 resulted in loss of purple compound formation, whereas Tyr12 replacement had no effect on the purple compound formation (data not shown).
Fig. 5.
Superposition of TOCSY spectra of side-chain aromatic regions of antioxidin-RP1 free (red) and in the complex with ABTS+ (blue). # means two aromatic protons have the same chemical shift, no difference. HE# represents HE1 and HE2; HD# represents HD1 and HD2.
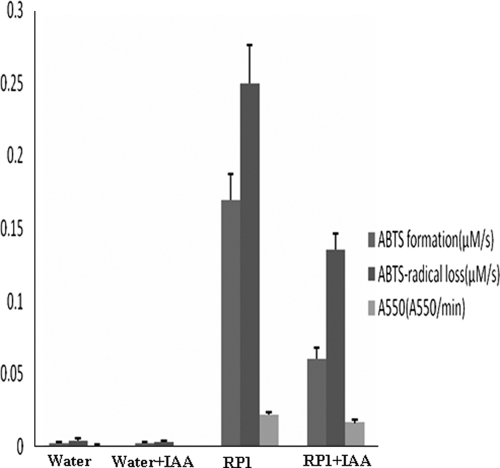
As listed in Table II, another significant effect on the antioxidant capability of antioxidin-RP1 is Cys10 replacement. As illustrated in Fig. 6, alkylation of antioxidin-RP1 (Cys10 alkylation) by IAA caused a significant, but incomplete, decrease of the reduction rate of ABTS radical. The formation of purple ABTS-antioxidin-RP1 was also significantly decreased. This suggests that the Cys10 thiol group is responsible for both the binding of ABTS to antioxidin-RP1 and the reduction of ABTS radical. Furthermore Fig. 6 also illustrates that the antioxidant capability of antioxidin-RP1 is from both the reduction of ABTS radical and the formation of purple ABTS-antioxidin-RP1. All the results indicated that multiple amino residues in antioxidin-RP1 likely act in consort in antioxidant function.
Fig. 6.
The comparison of antioxidant capability between antioxidin-RP1 and alkylated antioxidin-RP1 by IAA. RP1, antioxidin-RP1.
DISCUSSION
As an organ directly exposed to ROS from the environment as well as endogenous sources, skin is thus a major candidate and target for oxidative stresses, so as a result it is under pressure to develop an efficient antioxidant defense system (1–5). Two antioxidant systems have been identified (6–10). One is the gene-encoded antioxidant enzyme system; another one is organic small molecules (LMWAs) that are not of gene-encoded origin. They can directly or indirectly scavenge ROS.
Amphibian skins are morphologically and pharmacologically rational organs with multiple defensive functions. Their skin glands have rich secretions containing pharmacological molecules with a large amount of diversity (11). For example, antimicrobial peptides with various structures and antimicrobial spectrums have been identified from amphibian skins. ROS may be one of the most important stresses for amphibian naked skins. R. pleuraden is a ranid amphibian living in the subtropical plateau with strong radiation and a long duration of sunshine that make it an ideal model to study the interaction between skins and ROS. By a peptidomics and genomics approach coupled with function testing, 36 peptides belonging to 14 groups (including 13 novel groups) were identified from skins of R. pleuraden. Eleven groups contain antioxidant functions (Tables I and II and Fig. 2). They could exert functions of free radical scavengers. They are composed of 14–36 amino acid residues. All of them are cationic small peptides except pleurain-K1. The current antioxidant agents are different from the two known groups of antioxidant agents, antioxidant enzymes and organic small molecules (LMWAs). Antioxidant enzymes are the gene-encoded proteins; LMWAs are organic small molecules that are not of gene-encoded origin. The current antioxidant peptides have gene-encoded origins as antioxidant enzymes do, but they have no enzyme activity; instead they act as direct free radical scavengers like LMWAs. These peptides may comprise a novel antioxidant array, and they were proposed as the third antioxidant system. A peptide group (metallothionein) has been found to contain antioxidant activity, but it is significantly different from the current antioxidant groups. Metallothionein is considered an intracellular protein because of a lack of secretion signal sequences (41), whereas the current antioxidant groups are secreted peptides; they contain obvious secretion signal sequences in their precursors as described below.
cDNA cloning indicated that all these antioxidant peptides are released from small precursor proteins containing 51–79 amino acid residues. A genomics approach revealed that they have similar precursors, although the mature antioxidant peptides have variable primary structures. All of the precursors are composed of a signal peptide, acidic spacer peptide, and mature antioxidant peptide. They share a highly conserved signal peptide and acidic spacer peptide (Fig. 1). The remarkable similarity of preproregions of precursors that give rise to very different mature peptides suggests that the corresponding genes form a multigene family originating from a common ancestor. The diversification of antioxidant peptide loci potentially might have evolved in response to selective pressure exerted by variable ROS. In addition, the antioxidant mechanism investigation by amino acid residue replacement or modification and the activity comparison with control peptides provided proof to support that these antioxidant peptides are sequence-specific (Table II and Figs. 4–6). All these results indicated these peptides are encoded by specific genes to exert antioxidant functions.
As important defensive peptides in amphibian skins, antimicrobial peptides have been studied intensively. It was found that the precursors of the current antioxidant peptides share a highly conserved signal peptide and spacer peptide with amphibian antimicrobial peptide precursors. Based on the similarity, we tested antimicrobial functions of these antioxidant peptides. Among the 11 groups of antioxidant peptides, seven groups have antimicrobial activities, and thus they are bifunctional peptides (Tables I and III). Only four groups are specific antioxidant peptides. The precursor similarity of amphibian antioxidant peptides and antimicrobial peptides and the presence of bifunctional peptides with both antioxidant and antimicrobial activities suggest that these defensive peptides might have a common evolutionary origin. They have a similar bibasic (-Lys-Arg- or -Arg-Arg-) processing site for releasing mature antioxidant or antimicrobial peptides that also suggests their evolutionary relationship. In addition to their possible evolutionary connectivity, antioxidant and antimicrobial peptides share another two similarities. One similarity is the extreme diversity. Amphibian antimicrobial peptide diversity has been studied intensively. The current work also implied the diversity of antioxidant peptides in amphibian skin, although only one amphibian species was investigated in this work. In a single species, 11 groups of antioxidant peptide groups including 28 members were found. Another similarity is that both antioxidant and antimicrobial peptides can rapidly exert their biological functions. Amphibian antimicrobial peptides can kill microbes in a short time (within a few minutes) (42). As reported in the current work, amphibian antioxidant peptides can scavenge free radicals within several seconds. The rapidly acting kinetics likely makes amphibians try their utmost to protect skins from infection or oxidant stress. Different from antimicrobial peptides acting only as a defense against biological injury, antioxidant peptides can take part in both biological and non-biological injuries. These defensive peptides most likely act in consort in oxidant and microbial defenses.
Footnotes
Published, MCP Papers in Press, November 20, 2008, DOI 10.1074/mcp.M800297-MCP200
The abbreviations used are: LMWA, low molecular weight antioxidant; ROS, reactive oxygen species; RP, reversed-phase; DPPH, 2,2-diphenyl-1-picrylhydrazyl; BHT, butylated hydroxytoluene; ABTS+, 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid); SNP, sodium nitroprusside dihydrate; BHA, butylated hydroxyanisole; LB, Luria-Bertani; MIC, minimal inhibitory concentration; NMR, nuclear magnetic resonance; TOCSY, total correlation spectroscopy; IAA, iodoacetamide; NFAT, nuclear factor of activated T cells.
This work was supported by Chinese National Natural Science Foundation Grant 30830021, Chinese Academy of Sciences Grants KSCXZ-YW-R-088 and KSCX2-YW-G-024, and Ministry of Science Grants 2007AA02Z192 and 2007AA100602.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Rieger, M. M., and Pains, M. ( 1993) Oxidative reactions in and on the skin: mechanism and prevention. Cosmet. Toiletries 108, 43–56 [Google Scholar]
- 2.Kohen, R., and Gati, I. ( 2000) Skin low molecular weight antioxidants and their role in aging and in oxidative stress. Toxicology 148, 149–157 [DOI] [PubMed] [Google Scholar]
- 3.Shindo, Y., Witt, E., and Packer, L. ( 1993) Antioxidant defense mechanisms in murine epidermis and dermis and their response to ultraviolet light. J. Investig. Dermatol. 100, 260–265 [DOI] [PubMed] [Google Scholar]
- 4.Schalbreuter, K. U., and Wood, J. M. ( 1989) Free radical reduction in the human epidermis. Free Radic. Biol. Med. 6, 519–532 [DOI] [PubMed] [Google Scholar]
- 5.Halliwell, B., and Gutteridge, J. M. C. ( 1989) in Free Radicals in Biology and Medicine, pp. 160–165, Oxford University Press, Oxford
- 6.Kohen, R., Chevion, S., Schwartz, R., and Berry, E. M. ( 1996) Evaluation of the total low molecular weight antioxidant activity of plasma in health and diseases: a new approach. Cell. Pharmacol. 3, 355–359 [Google Scholar]
- 7.Kohen, R., Fanberstein, D., and Tirosh, O. ( 1997) Reducing equivalents in the aging process. Arch. Gerontol. Geriatr. 24, 103–123 [DOI] [PubMed] [Google Scholar]
- 8.Kohen, R. ( 1999) Skin antioxidants: their role in aging and in oxidative stress—new approaches for their evaluation. Biomed. Pharmacother. 53, 181–192 [DOI] [PubMed] [Google Scholar]
- 9.Kohen, R., Beit-Yannai, E., Berry, E. M., and Tirosh, O. ( 1999) Evaluation of the overall low molecular weight antioxidant activity of biological fluids and tissues by cyclic voltammeter, in Methods in Enzymology (Packer, L., ed) Vol. 300, pp. 285–290, Academic Press, San Diego, CA [DOI] [PubMed]
- 10.Chevion, S., Berry, E. M., Kitrossky, N., and Kohen, R. ( 1997) Evaluation of plasma low molecular weight antioxidant capacity by cyclic voltammetry. Free Radic. Biol. Med. 22, 411–421 [DOI] [PubMed] [Google Scholar]
- 11.Clarke, B. T. ( 1997) The natural history of amphibian skin secretions, their normal functioning and potential medical applications. Biol. Rev. Camb. Philos. Soc. 72, 365–379 [DOI] [PubMed] [Google Scholar]
- 12.Zasloff, M. ( 2002) Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 13.Borgden, K. A. ( 2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250 [DOI] [PubMed] [Google Scholar]
- 14.Duda, T. F., Jr., Vanhoye, D., and Nicolas, P. ( 2002) Roles of diversifying selection and coordinated evolution in the evolution of amphibian antimicrobial peptides. Mol. Biol. Evol. 19, 858–864 [DOI] [PubMed] [Google Scholar]
- 15.Conlon, J. M., Kolodziejek, J., and Nowotny, N. ( 2004) Antimicrobial peptides from ranid frogs: taxonomic and phylogenetic markers and a potential source of new therapeutic agents. Biochim. Biophys. Acta 1696, 1–14 [DOI] [PubMed] [Google Scholar]
- 16.Lai, R., Zheng, Y. T., Shen, J. H., Liu, G. J., Liu, H., Lee, W. H., Tang, S. Z., and Zhang, Y. ( 2002) Antimicrobial peptides from skin secretions of Chinese red belly toad Bombina maxima. Peptides 23, 427–435 [DOI] [PubMed] [Google Scholar]
- 17.Li, J., Xu, X., Xu, C., Zhou, W., Zhang, K., Yu, H., Zhang, Y., Zheng, Y., Rees, H. H., Lai, R., Yang, D., and Wu, J. ( 2007) Anti-infection peptidomics of amphibian skin. Mol. Cell. Proteomics 6, 882–894 [DOI] [PubMed] [Google Scholar]
- 18.Simmaco, M., De Biase, D., Severini, C., Aita, M., Erspamer, G. F., Barra, D., and Bossa, F. ( 1990) Purification and characterization of bioactive peptides from skin extracts of Rana esculenta. Biochim. Biophys. Acta 1033, 318–32312 [DOI] [PubMed] [Google Scholar]
- 19.Lu, Y., Li, J., Yu, H., Xu, X., Liang, J., Tian, Y., Ma, D., Lin, G., Huang, G., and Lai, R. ( 2006) Two families of antimicrobial peptides with multiple functions from skin of rufous-spotted torrent frog, Amolops loloensis. Peptides 27, 3085–3091 [DOI] [PubMed] [Google Scholar]
- 20.Zhou, M., Chen, T., Walker, B., and Shaw, C. ( 2006) Lividins: novel antimicrobial peptide homologs from the skin secretion of the Chinese Large Odorous frog, Rana (Odorrana) livida. Identification by “shotgun” cDNA cloning and sequence analysis. Peptides 27, 2118–2123 [DOI] [PubMed] [Google Scholar]
- 21.Lee, W. H., Li, Y., Lai, R., Li, S., Zhang, Y., and Wang, W. ( 2005) Variety of antimicrobial peptides in the Bombina maxima toad and evidence of their rapid diversification. Eur. J. Immunol. 35, 1220–1229 [DOI] [PubMed] [Google Scholar]
- 22.Li, J., Liu, T., Xu, X., Wang, X., Wu, M., Yang, H., and Lai, R. ( 2006) Amphibian tachykinin precursor. Biochem. Biophys. Res. Commun. 350, 983–986 [DOI] [PubMed] [Google Scholar]
- 23.Liu, X., Wang, Y., Cheng, L., Song, Y., and Lai, R. ( 2007) Isolation and cDNA cloning of cholecystokinin from the skin of Rana nigrovittata. Peptides 28, 1540–1544 [DOI] [PubMed] [Google Scholar]
- 24.Conlon, J. M., and Aronsson, U. ( 1997) Multiple bradykinin-related peptides from the skin of the frog, Rana temporaria. Peptides 18, 361–365 [DOI] [PubMed] [Google Scholar]
- 25.Lai, R., Liu, H., Lee, W. H., and Zhang, Y. ( 2001) A novel bradykinin-related peptide from skin secretions of toad Bombina maxima and its precursor containing six identical copies of the final product. Biochem. Biophys. Res. Commun. 286, 259–263 [DOI] [PubMed] [Google Scholar]
- 26.Wang, X., Song, Y., Li, J., Liu, H., Xu, X., Lai, R., and Zhang K. ( 2007) A new family of antimicrobial peptides from skin secretions of Rana pleuraden. Peptides 28, 2069–2074 [DOI] [PubMed] [Google Scholar]
- 27.von Gadow, A., Joubert, E., and Hansmann, C. F. ( 1997) Effect of extraction time and additional heating on the antioxidant activity of Rooibos tea (Aspalathus lineraris) extracts. J. Agric. Food Chem. 45, 1370–1374 [Google Scholar]
- 28.Oyaizu, M. ( 1986) Studies on products of browning reaction prepared from glucosamine. Jpn. J. Nutr. 44, 307–315 [Google Scholar]
- 29.Kaur, G., Alam, M. S., Jabbar, Z., Javed, K., and Athar, M. ( 2006) Evaluation of antioxidant activity of Cassia siamea flowers. J. Ethnopharmacol. 108, 340–348 [DOI] [PubMed] [Google Scholar]
- 30.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. ( 1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 31.Goddard, T. D., and Kneller, D. G. ( 1999) SPARKY 3, University of California, San Francisco
- 32.Akerström, B., Maghzal, G. J., Winterbourn, C. C., and Kettle, A. J. ( 2007) The lipocalin α1-microglobulin has radical scavenging activity. J. Biol. Chem. 282, 31493–31503 [DOI] [PubMed] [Google Scholar]
- 33.Jung, H. A., Jeong, D. M., Chung, H. Y., Lim, H. A., Kim, J. Y., Yoon, N. Y., and Choi, J. S. ( 2008) Re-evaluation of the antioxidant prenylated flavonoids from the roots of Sophora flavescens. Biol. Pharm. Bull. 31, 908–915 [DOI] [PubMed] [Google Scholar]
- 34.Ak, T., and Gülçin, I. ( 2008) Antioxidant and radical scavenging properties of curcumin. Chem.-Biol. Interact. 174, 27–37 [DOI] [PubMed] [Google Scholar]
- 35.Portugal, M., Barak, V., Ginsburg, I., and Kohen, R. ( 2007) Interplay among oxidants, antioxidants, and cytokines in skin disorders: present status and future considerations. Biomed. Pharmacother. 61, 412–422 [DOI] [PubMed] [Google Scholar]
- 36.Xu, X., Yang, H., Ma, D., Wu, J., Wang, Y., Song, Y., Wang, X., Lu, Y., Yang, J., and Lai, R. ( 2008) Toward an understanding of the molecular mechanism for successful blood feeding by coupling proteomics analysis with pharmacological testing of horsefly salivary glands. Mol. Cell. Proteomics 7, 582–590 [DOI] [PubMed] [Google Scholar]
- 37.Bourdon, E., and Blache, D. ( 2001) The importance of proteins in defense against oxidation. Antioxid. Redox Signal. 3, 293–311 [DOI] [PubMed] [Google Scholar]
- 38.Krishnan, N., Dickman, M. B., and Becker, D. F. ( 2008) Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic. Biol. Med. 44, 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsudaira, P. ( 1987) Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262, 10035–10038 [PubMed] [Google Scholar]
- 40.Re, R., Pellegrini, N., Proteggente, A., Pannala, A., Yang, M., and Rice-Evans, C. ( 1999) Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 26, 1231–1237 [DOI] [PubMed] [Google Scholar]
- 41.Chung, R. S., Fung, S. J., Leung, Y. K., Walker, A. K., McCormack, G. H., Chuah, M. I., Vickers, J. C., and West, A. K. ( 2007) Metallothionein expression by NG2 glial cells following CNS. CMLS Cell. Mol. Life Sci. 64, 2716–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, J., Zhang, C., Xu, X., Wang, J., Yu, H., Lai, R., and Gong, W. ( 2007) Trypsin inhibitory loop is an excellent lead structure to design serine protease inhibitors and antimicrobial peptides. FASEB J. 21, 2466–2473 [DOI] [PubMed] [Google Scholar]