Abstract
Dioxin is an extremely potent carcinogen. In highly exposed people, the most commonly observed toxicity is chloracne, a pathological response of the skin. Most of the effects of dioxin are attributed to its activation of the aryl hydrocarbon receptor (AHR), a transcription factor that binds to the Ah receptor nuclear translocator (ARNT) to regulate the transcription of numerous genes, including CYP1A1 and CYP1B1. In cultures of normal human epidermal keratinocytes dioxin accelerates cell differentiation, as measured by the formation of cornified envelopes. We show that this acceleration is mediated by the AHR; also, that dioxin increases the expression of several genes known to be regulated by ARNT, which have critical roles in the cornification and epidermal barrier function of the skin. Importantly, we demonstrate that all of these responses are opposed by ligand-activation of the EGF receptor (R), an important regulator of keratinocyte cell fate. In the CYP1A1 enhancer, EGFR activation prevents recruitment of the p300 coactivator, although not affecting the binding of the AHR or ARNT. The total cellular level of p300 protein does not decrease, and overexpression of p300 relieves EGFR-mediated repression of transcription, indicating that p300 is a critical target for the repression of the AHR complex by EGFR signaling. These results provide a mechanism by which 2,3,7,8-tetrachlorodibenzo-p-dioxin is able to disrupt epidermal homeostasis and identify EGFR signaling as a regulator of the AHR. This signaling may modulate the incidence and severity of chloracne and be of therapeutic relevance to human poisonings by dioxin.
Keywords: dioxin, ARNT, psoriasis, cornification
In the United States, >300 million people are exposed to dioxin (2,3,7,8-tetrachlorodibenzo-p-dioxin, TCDD) and dioxin-like compounds, primarily through food consumption. TCDD is among the most toxic pollutants known, and is classified as carcinogenic to humans (1). In addition to this direct human health risk of dioxin exposure, the risk characterization of dioxin broadly affects decisions concerning important public health policies, ranging from the safety of breast feeding to the need to dredge contaminated rivers, such as the Hudson (2, 3).
Animal studies have shown that most of the biological effects of TCDD are mediated through its binding to the aryl hydrocarbon receptor (AHR). The AHR is a cytoplasmic basic helix–loop–helix transcription factor. On ligand binding, the AHR moves to the nucleus and binds with the AHR nuclear translocation protein (ARNT), the best understood nuclear protein dimerization partner of the AHR. The AHR complex activates transcription through binding to specific DNA recognition sequences that have been identified upstream of the mRNA initiation site of genes, including the CYP1 family of cytochrome P450, CYP1A1, CYP1A2, and CYP1B1 (4).
In humans, chloracne is the most visible and consistent response to dioxin exposure. This chronic condition is characterized by metaplasia and hyperkeratinization of the ducts of the sebaceous gland, resulting in comedone formation, along with hyperproliferation and differentiation of the interfollicular squamous epithelium (5). As for other endpoints of dioxin toxicity, the mechanism of chloracne is not known. Also, the severity and persistence of chloracne does not correlate with the half-life or concentration of the chloracnegen, indicating that additional factors likely modulate individual susceptibility (5).
Submerged cultures of normal human epidermal keratinocytes (NHEKs) retain many of the regulatory, structural, and biochemical components that constitute normal differentiation of the epidermis (6–8). NHEKs respond to treatment with TCDD through changes in gene expression, cell proliferation, and differentiation (9–12). Previous studies showing that TCDD alters the expression of several secreted growth-regulatory proteins (10, 11) led us to consider the effects of the integrated signals of TCDD, cell density, and growth factors on NHEKs. Here, we identify EGF receptor (R) signaling as a repressor of the AHR, preventing the ability of TCDD to enhance gene transcription and accelerate terminal differentiation of NHEKs. Also, we show that TCDD enhances the expression of genes involved in late, but not early, differentiation, and that this enhanced expression is inhibited by EGF.
Results
Inhibition of CYP1 Induction by EGFR Signaling.
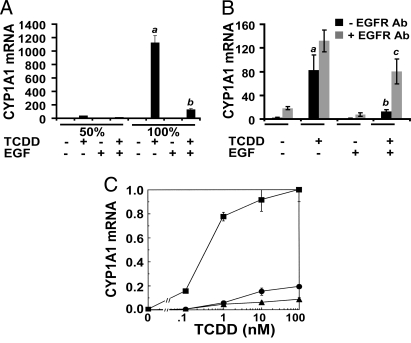
In response to treatment of keratinocytes with TCDD, the greatest increases in levels of CYP1A1 and CYP1B1 mRNA were observed in confluent cultures where the medium supplements, bovine pituitary extract and EGF, were excluded. Analysis of each supplement showed that the addition of EGF caused the diminished response to TCDD within these cultures (Fig. 1A). Although not visible in this figure because of the scale of the y axis, the basal level of CYP1A1 mRNA (Fig. 1A, −TCDD −EGF) is elevated 9-fold by an increase in cell density from 50% to 100%. In parallel experiments, we determined 7-ethoxycoumarin O-deethylase (ECOD) activity, a measure of TCDD-inducible CYP1A1 protein (13). In the absence of TCDD, basal ECOD was not detectable (<0.1 pmol of product per mg of protein per h) in cultures with or without supplemental EGF. In treated cells (10 nM TCDD, 48 h), ECOD activity (n = 3, mean ± SD) was 319.4 ± 62.5 pmol·mg−1·h−1 and 14.4 ± 2.5 pmol·mg−1·h−1 in the absence and presence of supplemental EGF (10 ng/mL), respectively (P < 0.01). To determine whether this repression by EGF occurred in more than one individual, we analyzed NHEKs from a second individual, as well as a pooled sample from 3 other individuals. EGF caused a similar repression of AHR-mediated CYP1 mRNA expression in both the second individual (99% repression) and the pooled sample (89% repression) (Fig. S1A). Also, pretreatment in a concentration-dependent manner with either EGF or TGFα diminished the TCDD-mediated increases in CYP1A1 and CYP1B1 mRNA (Fig. S1 B and C). For both CYP1A1 and CYP1B1 mRNA, the 50% effective concentration (EC50) for the response to EGF or TGFα was ≈0.7 and 0.8 nM, respectively, consistent with EGFR-mediated effects (14). Also, a neutralizing EGFR Ab was able to inhibit the EGF-mediated repression of CYP1A1 mRNA (Fig. 1B).
Fig. 1.
The effects of EGF on TCDD-inducible gene expression in cultures of NHEKs are mediated through the EGFR. (A) NHEKs were grown to a density of either 50% or 100% before basal medium, or medium with EGF (10 ng/mL), was added 24 h before treatment; mRNA was isolated after treatment with either 0.1% DMSO or TCDD (10 nM) for 24 h. Real-time PCR was used to determine the level of CYP1A1 mRNA. Levels of mRNA [mean (n = 3) ± SD] are expressed in units relative to the minimum, given a value of 1. (B) Cells were grown to confluence, and basal medium was added for 24 h. The EGFR-neutralizing Ab LA1 (10 μg/mL) was added 2 h before EGF (1 ng/mL). EGF was added for 1 h, followed by TCDD (10 nM) for 2 h. Levels of mRNA [mean (n = 3) ± SD] are expressed in units relative to the minimum, given a value of 1. In A and B, the a indicates that the value from treatment with TCDD is significantly different from the DMSO control; the b indicates that cotreatment with TCDD and EGF is significantly different from with TCDD alone; and the c indicates that the addition of EGFR Ab significantly changed the mRNA level in TCDD plus EGF treatment group. In all cases, P < 0.02 by Student's t test. (C) Dose–response of CYP1A1 mRNA induction. Cells were treated with either 0.1% DMSO or TCDD (at the indicated concentrations; x axis) for 24 h. Levels of mRNA (y axis) are expressed in units relative to the maximum, given a value of 1. Filled squares, basal medium; filled circles, with EGF (10 ng/mL); filled triangles, with TGFα (50 ng/mL). Each point indicates the mean value; bars, SD; n = 5 per group.
Experiments using actinomycin D showed that treatment with EGF did not decrease the half-life of the CYP1A1 mRNA. The rate of decay was similar in each set of samples, with an apparent half-life of 24 h (Fig. S1D). Also, EGF did not change the level of cellular AHR protein (Fig. S1E). We studied the TCDD-concentration response for CYP1A1 mRNA induction in cells treated in basal medium or medium supplemented with EGF or TGFα (Fig. 1C). Treatment with EGF did not simply displace the EC50 dose–response curve for TCDD to the right by a factor of 10 or even 100. Rather, <20% of the fractional response was observed in cells treated with EGF or TGFα at concentrations of TCDD 100-fold greater than the EC50. Also, there was no indication that maximal response could be achieved at the highest concentrations of solubility of TCDD in the medium, ≈1,000 nM. When coupled with data demonstrating that AHR protein levels were not decreased by EGF, these data indicate that EGF and TGFα affect the ability of the AHR to transduce its signal in response to treatment with TCDD.
Inhibition of AHR-Regulated Transcription by EGF.
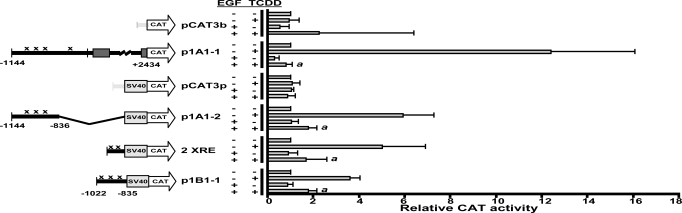
The results from 2 independent nuclear run-on experiments showed that EGF decreased TCDD-mediated increases in the rates of transcription of CYP1A1 and CYP1B1 to nearly basal levels (Fig. S2). ARNT, which is not transcriptionally regulated by the AHR, was not affected by EGF treatment. To locate the upstream regulatory sequences of CYP1 critical for transducing EGF-mediated repression of transcription, we tested CAT reporter constructs (Fig. 2). The 2 control reporter vectors, pCAT3b, which does not contain a SV40 promoter, and pCAT3p, which does, did not respond significantly to treatments with TCDD or EGF. The construct p1A1-1, which contains the complete CYP1A1 sequence from −1144 to +2434, including 4 XREs and the endogenous CYP1A1 promoter, responded strongly to TCDD, and this response was repressed to basal levels by treatment with EGF. The response to TCDD was still significant when the CYP1A1 sequence was decreased to −1144 to −836 in p1A1-2, which contains 3 XREs, but not the CYP1A1 promoter, as was the suppression by treatment with EGF. This result indicates that the CYP1A1 enhancer is necessary for repression by treatment with EGF. Similarly, the construct p1B1-1, which contains the CYP1B1 sequence from −1022 to −835 including 3 XREs, was sufficient for repression by EGF treatment. Also, analysis of the construct 2 XRE, consisting of the minimal synthetic 2 XRE sequence, showed that this sequence alone was sufficient for repression of TCDD-mediated transcription by EGF.
Fig. 2.
EGF inhibits the TCDD-mediated increase in transcription. Levels of CAT activity (mean ± SD, n = 4–5) are expressed in units relative to the control treatment (without EGF and TCDD), given a value of 1; pCAT3 basic, pCAT3b; pCAT promoter, pCATp. Xenobiotic-responsive elements (XREs) in the constructs are indicted by the letter x. The a indicates that the value from treatment with TCDD and EGF is significantly different from value with TCDD and without EGF; P < 0.05 by Student's t test.
Inhibition of Recruitment of the p300 Coactivator to the CYP1A1 Enhancer, but Not AHR/ARNT DNA Binding, by EGFR Ligands.
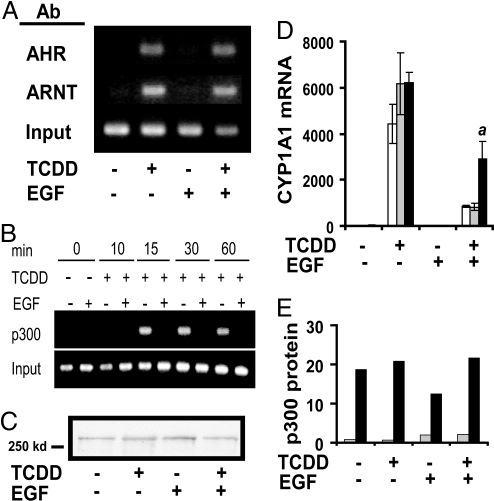
By EMSA, TCDD-mediated AHR–DNA interactions were not affected by EGF or TGFα (Fig. S3). Also, ChIP assays showed that binding of AHR and ARNT to the endogenous CYP1A1 enhancer was not affected by treatment with EGF (Fig. 3A). Because EGF was able to repress transcription driven by the minimal synthetic 2 XRE, but did not decrease the binding of AHR or ARNT to CYP1A1 XREs, we investigated the effect of EGF on TCDD-mediated coactivator recruitment. Transcription by the AHR complex has been shown to be regulated by several coactivators, including the histone acetyltransferase p300 (15). ChIP assays revealed that TCDD increased the association of p300 with the CYP1A1 enhancer region after 15 min of treatment, and that the presence of p300 was sustained throughout. Importantly, the recruitment of p300 to the enhancer was prevented by treatment with EGF (Fig. 3B). Immunoblots of nuclear extracts from NHEKs demonstrated that p300 protein levels were not altered (Fig. 3C). Overexpression of p300 using a recombinant adenovirus significantly relieved the EGF-mediated repression of CYP1A1 mRNA expression (Fig. 3D). Immunoblots indicated that expression of recombinant p300 protein was increased >10-fold over the endogenous levels (Fig. 3E).
Fig. 3.
EGF decreases binding of p300, but not AHR or ARNT, to the CYP1A1 XRE. Basal medium, or medium with EGF (10 ng/mL) was added 24 h before treatment. (A and B) ChIP assay of the CYP1A1 enhancer under the indicated conditions. (A) Cells were treated with either 0.1% DMSO or TCDD (10 nM) for 0.5 h. The Abs used during immunoprecipitation are indicated at left. Levels of input DNA are shown in the lowest inset. (B) Cells were treated with either 0.1% DMSO or TCDD (10 nM) for the indicated amount of time. The samples were subjected to ChIP analysis by using p300 Ab. Levels of input DNA are shown in the lowest row. (C) Immunoblot of nuclear extracts (10 μg per lane) incubated with p300 Ab. Cells were treated with either 0.1% DMSO or TCDD (10 nM) for 30 min. (D) Messenger RNA from uninfected cells (white bars), null vector-infected cell (gray bars), or p300-infected cells (black bars) was isolated after 24 h of treatment with 0.1% DMSO or TCDD (10 nM). Real-time PCR was used to determine the level of CYP1A1 mRNA, and the lowest value for each set of experiments was given a value of 1. The values are a mean of triplicate samples ± SD. The a indicates that p300 infection (black bars) produced significantly more CYP1A1 mRNA than the null vector infection (gray bars); P < 0.03 by Student's t test. (E) Quantification of immunoblots of p300 in whole cell extracts from null vector-infected cells (gray bars) or p300-infected cells (black bars). Extracts were isolated after 24 h of treatment with 0.1% DMSO or TCDD (10 nM).
Inhibition of AHR-Mediated Keratinocyte Differentiation by EGF.
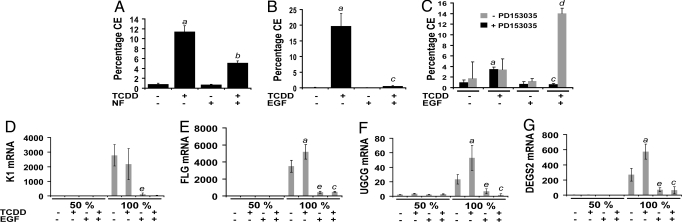
As previously reported (9), treatment of keratinocytes with TCDD significantly increased the percentage of cornified envelopes (CEs), and accelerated the rate of terminal differentiation (Fig. 4A). The formation of CEs-a highly cross-linked structure of loricrin, involucrin, and additional proteins is a quantitative marker of keratinocyte differentiation (16). Inhibition of the TCDD-mediated increase in terminal differentiation by α-naphthoflavone (1 μM), an AHR antagonist at this concentration, indicated that the increase in terminal differentiation by TCDD was mediated by the AHR (Fig. 4A). This increase was completely blocked by EGF (Fig. 4B). Also, when cells were treated with EGF, cotreatment of the cells with TCDD and the EGFR tyrosine kinase inhibitor PD153035 resulted in an increased efficacy to promote cell differentiation compared with TCDD alone (Fig. 4C).
Fig. 4.
Opposing effects of TCDD and EGF on differentiation of NHEKs. (A) NHEKs were grown to confluence, and basal medium, or medium with α-naphthoflavone (NF, 1 μM) was added 24 h before treatment. CEs were isolated after treatment with either 0.1% DMSO or TCDD (10 nM) for 5 days. (B) NHEKs were grown to confluence, and basal medium with or without EGF (10 ng/mL) was added 24 h before treatment. CEs were isolated after treatment with either 0.1% DMSO or TCDD (10 nM) for 5 days. (C) NHEKs were grown to confluence, and basal medium, or medium with EGF (10 ng/mL), was added 24 h before treatment. CEs were isolated after treatment with either 0.1% DMSO or TCDD (10 nM) in the presence or absence of PD153035 (300 nM) for 5 days. (A–C) The values for CEs are a mean of triplicate samples ± SD. (D–G) NHEKs were grown to a cell density of either 50% or 100% confluence before basal medium, or medium with EGF (10 ng/mL), was added for 24 h before treatment. Total mRNA was isolated after treatment with either control vehicle (0.1% DMSO) or TCDD (10 nM) for 24 h. Real-time PCR was used to determine the relative expression of each indicated gene (y axis). Levels of mRNA [mean (n = 3) ± SD] are expressed in units relative to the minimum, given a value of 1. (A–G) The a indicates that the value from treatment with TCDD is significantly different from the DMSO control; the b indicates that the value from cotreatment with TCDD and NF is significantly different from TCDD alone; the c indicates that the value from cotreatment with TCDD and EGF is significantly different from with TCDD alone; the d indicates that the value from treatment with TCDD, EGF, and PD153035 is significantly different from treatment with TCDD and EGF; and the e indicates that treatment with EGF is significantly different from DMSO control treatment. For each of these comparisons, P ≤ 0.01 by Student's t test. Comparisons made in D–G are within the group grown to a cell density of 100% confluence.
Increases in the Expression of ARNT-Regulated Genes Involved in Cornification by TCDD.
The expression of keratin (K)1, one the earliest markers of keratinocyte differentiation, is known to be regulated by cell density (17). Although treatment with EGF inhibited the density-dependent expression of K1, TCDD had no effect on the expression of this gene, whose product is involved in early differentiation (Fig. 4D). However, the levels of mRNA of 3 genes were significantly elevated by treatment with TCDD for 24 h (Fig. 4 E–G). These genes included filaggrin (FLG), ceramide glucosyltransferase (UGCG), and sphingolipid C4-hydroxylase/delta 4-desaturase (DEGS2), all known to be regulated by ARNT during cornification (late differentiation) and to contribute to epidermal barrier function (18, 19). Also, treatment with EGF markedly inhibited both the cell density-dependent and TCDD-mediated expression of each of these 3 genes (Fig. 4 E–G).
Discussion
This study shows that EGF or TGFα opposed AHR-mediated biological effects in human keratinocytes, including gene transcription and cell differentiation. Given that the activation of the AHR in NHEKs has previously been shown to increase expression of TGFα (11), and that our work shows that EGFR ligands block AHR signaling, we conclude that EGFR activation likely modifies AHR signaling in an autocrine/paracrine manner.
Inhibitor studies demonstrated that gene transcription and cell differentiation effects of EGF were mediated through the EGFR. Mechanistic studies revealed that EGF inhibited transcription; however, assembly of the AHR-ARNT complex at the enhancer of CYP1A1 was not altered. Further studies demonstrated that p300 recruitment was decreased by EGFR signaling. A role for p300 in AHR-regulated transcription was first indicated by studies in which E1A repressed CYP1A1 induction (20). Since then, p300 was shown to bind to ARNT in the AHR–ARNT complex (20, 21). Also, a recent study showed that TCDD-mediated elevations of CYP1A1 and CYP1B1 mRNAs were inhibited when p300 was decreased by mRNA interference (22), indicating that this coactivator has a role in AHR-mediated transcription. Here, as previously reported (23), the recruitment of p300 to the CYP1A1 XREs was detected within 15 min of TCDD treatment. Although EGF prevented recruitment of p300 to the CYP1A1 enhancer, we demonstrated that this effect was not accompanied by a decrease in the total level of nuclear p300 protein. However, by overexpressing p300, we were able to significantly relieve the EGFR-mediated repression of CYP1A1, indicating that some form of p300 was limited in its availability for recruitment to the AHR complex. A possible mechanism for the loss of p300 recruitment to the AHR complex is one of limiting cofactors, where transcription factors compete for cofactor binding (24). This mechanism is supported by a study showing that multiple signaling pathways and >10 transcription factors (25), some of which are known to use p300 (26), are activated by EGF. Also, studies in M12 cells showed that a single transcription factor downstream of EGFR, Egr1, bound to the promoters of 288 genes (27). An alternate possibility is that EGFR signaling may cause a posttranslational modification of p300, preventing it from binding to the AHR complex. Evidence supporting this mechanism comes from studies of K16 gene expression in keratinocytes, where Erk2-mediated phosphorylation of C-terminal serine residues was shown to control p300 recruitment to the K16 promoter (28). The observed relief from EGF-mediated repression by p300 overexpression may represent the fraction of the pool of expressed p300 that escapes posttranslational modifications. For the AHR, LPS-stimulated activation of nuclear factor (NF)-κΒ has been shown to repress AHR-mediated transcription and overexpression of p300 or SCR-1 relieved this effect (29). Because EGF is known to activate NF-κΒ (30), we considered this mechanism. However, neither inhibition of NF-κΒ with ammonium pyrrolidinedithiocarbamate, nor the cell-permeant inhibitory peptide SN50 relieved the repression by EGF (Fig. S4). These results indicate that NF-κΒ is not responsible for the effects observed here in NHEKs. In summary, our data indicate that p300 is a key component of the AHR transcriptional complex and is a critical target for the repression of this complex by EGFR signaling.
In addition to the regulation of CYPs, the AHR mediates numerous toxic effects of TCDD. Because chloracne is the hallmark of dioxin exposure in people, and increased interfollicular differentiation is a consistent endpoint of this effect (5), we studied the effect of TCDD on keratinocyte differentiation. Our results indicate that the AHR mediates this effect of TCDD, which was inhibited to near-basal levels by treatment with EGF. Although EGF is known to inhibit keratinocyte differentiation (16, 31), we know of no other study showing that EGF inhibits the action of a differentiation-promoting chemical. EGFR tyrosine kinase inhibitors are known to promote keratinocyte differentiation (32), and such chemicals are under development for the treatment of psoriasis. Here, our results showing the enhanced efficacy of an EGFR tyrosine kinase inhibitor and TCDD to promote cell differentiation in the presence of EGF indicate that combined drug therapy may provide an improved benefit over single-agent therapy for psoriasis.
To evaluate the stage of differentiation affected by dioxin, we determined the effects of cell density, TCDD, and EGF on the expression of genes that are either early or late markers of this process. The expression of both K1 (early) and FLG (late) was increased by cell density and inhibited by EGF, supporting the concept that EGF acts to inhibit multiple prodifferentiation signals in keratinocytes. The EGF effect on density-dependent gene expression has been reported in the literature (31). For K1, a recent study shows that EGF suppresses K1 by repressing the expression of Notch 1, an important regulator of cell-fate decisions in the epidermis (33). Although the expression of K1 was not altered by TCDD treatment, the expression of FLG was significantly increased over density alone by TCDD. The effect of TCDD on FLG, an important protein of the cornified layer, but not on K1, indicates that the action of TCDD may be restricted to cornification.
Two independent studies have shown that targeted ablation of Arnt in the mouse epidermis results in early postnatal death associated with a failure of epidermal permeability barrier function (18, 19). In each study, histology showed small changes in the architecture of the cornified layers. Biochemical analyses revealed changes in lipid metabolism affecting permeability, specifically alterations of the 4-sphingenine scaffolds of ceramides (18), and ceramide metabolism, including down-regulation of the UGCG pathway in the Arnt null epidermis (19). In one study, microarray analysis revealed that the expression of FLG was regulated by ARNT (19). Because ARNT is the heterodimer partner of the AHR, and the expression of FLG was elevated by TCDD treatment of NHEKs, we examined the expression of 2 additional genes critical to sphingolipid and ceramide biosynthesis in skin and regulated by Arnt (19, 20). This work showed that the expression of UCGC and DEGS2 (DES2) was increased by TCDD treatment and repressed by EGF. Ceramides are the major lipid component of the epidermal permeability barrier. DEGS2 is expressed during keratinocyte differentiation and is important to the synthesis of ceramides on their extrusion from lamellar granules into the extracellular space of the upper layers of the epidermis (34). Because the phenotype of epidermal Arnt null mice is much more severe (early postnatal death; see refs. 18 and 19) than the reported phenotype of Ahr null mice (35, 36), the effects of Arnt ablation in the epidermis cannot be completely codependent on the AHR. However, it is possible that a subset of ARNT-regulated genes is regulated by the AHR-ARNT heterodimer and participates in epidermal differentiation. It is also possible that the effects of TCDD on epidermal differentiation are aberrant due to inappropriate activation of the AHR, formation of AHR–ARNT complexes, and disruption of ARNT signaling.
In conclusion, the results of our work demonstrate previously undescribed regulation of AHR-mediated effects by EGFR signaling. The effects of TCDD on the expression of ARNT-regulated genes important to epidermal barrier function provide new insights into the actions of TCDD as a chloracnegen. It remains to be studied whether EGFR signaling by the mechanism described here affects the incidence or severity of dioxin toxicity in vivo, and to elaborate the effects of TCDD in the skin.
Materials and Methods
Keratinocyte Cell Culture.
Neonatal foreskin NHEKs, purchased from Lonza, were grown in keratinocyte-SFM (Invitrogen). Unless otherwise indicated, confluent fifth-passage NHEKs were incubated in complete medium for 2 days before pretreatment with EGF for 24 h. Where multiple treatments are indicated, all subsequent treatments took place in the continued presence of the supplement.
Biochemical Analyses.
We isolated mRNA by using RNA Stat-60 (Tel-Test). Real-time PCR was carried out by using iQ SYBR Green Supermix (Bio-Rad) and the following primers (5′-3′). CYP1A1: CATCCCCCACAGCACAACAAGAGA and GCAGCAGGATAGCCAGGAAGAGAA; K1: TGTCTGGAGAATGTGCCCCGAACG and CCGCCGCCACCTCCAGAACCAT; FLG: GACACCCCGGATCCTCTCACC and AGCTGCCATGTCTCCAAACTAAAC; UGCG: CCTGCGGGAGCGTTGTC and TTGTTGAGGTGTAATCGGGTGTAG; and DEGS2: GCGGGTGTACAGGCTGGCAAAAGA and ACAAGGGCAGCAGTCCAGAGCACA. Cyclophilin was used as the reference for sample normalization using primers GCAGAGGGTTAAGGCGCAGACTAC and TAAGGTGGGCAGAGAAGGGGTTTT. After verifying that cyclophilin and target mRNA primers had similar amplifying efficiencies, the comparative method, using the formula 2−ΔΔCT, was used to quantify relative mRNA levels and carry out statistical analysis (37). All statistical comparisons were performed by using SigmaStat 3.5 (SPSS).
Protein Assays.
ECOD assays were performed as described (13). For immunoblot detection of AHR, nuclear extracts were prepared by using NE-PER kit (Pierce). Proteins (20 μg per lane) were separated by SDS/PAGE through a 7.5% gel and transferred to PVDF (Millipore) (38). The AHR protein was detected by using the Ab H211 (Santa Cruz Biotechnology). For detection of p300, whole cell extracts were prepared by incubating cells in RIPA (0.1% SDS/1% Nonidet P-40/5 mM EDTA/0.5% sodium deoxycholate/150 mM NaCl/50 mM Tris·Cl, pH 7.2/Halt protease inhibitor mixture; Pierce). Nuclear extracts were prepared by using the NE-PER kit (Pierce). Samples were separated on a 5% gel, transferred to PVDF (Millipore), and incubated with a p300 antibody (1 μg/mL) (Upstate). Lamin B1 Ab (Santa Cruz Biotechnology) was used to control for sample loading.
EMSA and ChIP Assays.
EMSAs were carried out according to a published method (39). ChIP assays were performed as described in ref. 42, except that DNA was purified by using Qiaquick DNA purification kit (Qiagen). The conditions for PCR included an initial 15-min 95 °C incubation, followed by cycling between 95 °C for 30 s, 61.4 °C for 30 s, and 72 °C for 30 s, and then a single incubation at 72 °C for 2 min. The PCR primers used were TAAGAGCCCCGCCCCGACTCCCT and TAGCTTGCGTGCGCCGGCGACAT, amplifying the sequence between −1088 and −886 (40), relative to the CYP1A1 transcription start site. PCR products were analyzed on 1.8% agarose gels. To analyze input DNA, DNA-protein cross-links were reversed by adding NaCl to 0.45 mM and heating to 95 °C for 15 min. RNase A was added, then samples were incubated at 37 °C for 30 min, and purified by using the QIAquick DNA purification kit (Qiagen). PCR was performed and analyzed by using the above primers and conditions.
Transcription Assays.
Nuclear run-on assays were performed as described in ref. 10. To create p1A1-1, KpnI-digested pRNH11C (kindly provided by R. N. Hines; see ref. 41) was cloned into KpnI-digested pCAT3 promoter (Promega) to create pJSR212; pJSR212 was digested with KpnI and inserted into KpnI-digested pCAT basic. To create p1A1-2, pJSR212 was digested with ApaI and SacI. The ends were filled in by using the Klenow fragment of DNA polymerase, and the insert was ligated back into pCAT3 promoter; 2 XRE was generated by annealing the following oligonucleotides, GCGAGCTCGGGATCCTTCTCACGCAACGCCTGGGCATCCTTCTCACGCAACGCCTGGGCACTCGAGCGG and CCGCTCGAGTGCCCAGGCGTTGCGTGAGAAGGATGCCCAGGCGTTGCGTGAGAAGGATCCCGAGCTCGC, which contain 2 consensus XREs, connected by a BamHI site, with SacI and XhoI sites on the ends. The SacI- and XhoI-digested double-stranded oligonucleotide was ligated into SacI- and XhoI-digested pCAT3 promoter; p1B1-1 was a kind gift from W. F. Greenlee (42).
Transcriptional Reporter Analysis.
Cells were cotransfected with a reporter construct and the β-gal vector, pCMV10 (kindly provided by C.-S. Hwang, The Johns Hopkins University). Confluent cells were incubated with DNA and Lipofectin (Invitrogen) for 12 h, then switched to complete medium for 18 h. Cells were put into basal medium for 6 h, followed by treatment medium for 24 h. Chloramphenicol acetyltransferase (CAT) activity was measured as described (39).
Adenovirus Infections.
Fourth-passage cells were grown to a density of ≈95%. Adenovirus constructs (Cell Biolabs) were added in complete medium at a multiplicity of infection of 1,000. After 24 h, viruses were removed and replaced with medium with or without EGF (10 μg/mL). After an additional 24 h, cells were treated with either 0.1% DMSO or TCDD (10 nM) for 24 h.
Keratinocyte Differentiation.
The formation of CEs was quantified by using published procedures (16). At the concentration used in this study, α-naphthoflavone (1 μM) has been shown to act as an AHR antagonist (43).
Additional Reagents.
TGFα was from Sigma, EGFR Ab LA1 was from Upstate Tech, ammonium pyrrolidinedithiocarbamate was from Calbiochem, and SN50 was from Biomol.
Supplementary Material
Acknowledgments.
We acknowledge the expert technical assistance of Ying Li, Xui Fen Liu, and Sufang Zhang; the helpful discussions with members of the lab and our colleagues; the critical reviews of this manuscript by Drs. Paul Talalay and Pierre Coulombe; the careful editing of this manuscript by Deborah Hernandez, James M. Kovarik, and Engrid Roy; and Dr. Gary Perdew (Pennsylvania State University, University Park, PA) and Dr. Rick Pollenz (University of South Florida, Tampa, FL) for providing antibodies to the AHR and ARNT proteins. This work was supported in part by National Institutes of Health Grants ES06071, ES03819, and ES07141; the American Forest and Paper Association; the Johns Hopkins Center for Alternatives to Animal Testing; and the W. Harry Feinstone Center for Genomic Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900874106/DCSupplemental.
References
- 1.Steenland K, Bertazzi P, Baccarelli A, Kogevinas M. Dioxin revisited: Developments since the 1997 IARC classification of dioxin as a human carcinogen. Environ Health Perspect. 2004;112:1265–1268. doi: 10.1289/ehp.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hafferty AJ, Pavlou SP, Hom W. Release of polychlorinated biphenyls (PCB) in a salt-wedge estuary as induced by dredging or contaminated sediments. Sci Total Environ. 1977;8:229–239. doi: 10.1016/0048-9697(77)90023-7. [DOI] [PubMed] [Google Scholar]
- 3.Nishijo M, et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin in maternal breast milk and newborn head circumference. J Expo Sci Environ Epidemiol. 2008;18:246–251. doi: 10.1038/sj.jes.7500589. [DOI] [PubMed] [Google Scholar]
- 4.Yao G, Harstad EB, Bradfield CA. The Ah Receptor. In: Crews ST, editor. PAS Proteins: Regulators and Sensors of Development and Physiology. Norwell, MA: Kluwer; 2003. pp. 149–182. [Google Scholar]
- 5.Panteleyev AA, Bickers DR. Dioxin-induced chloracne–reconstructing the cellular and molecular mechanisms of a classic environmental disease. Exp Dermatol. 2006;15:705–730. doi: 10.1111/j.1600-0625.2006.00476.x. [DOI] [PubMed] [Google Scholar]
- 6.Wilke MS, Hsu BM, Wille JJ, Jr, Pittelkow MR, Scott RE. Biologic mechanisms for the regulation of normal human keratinocyte proliferation and differentiation. Am J Pathol. 1988;131:171–181. [PMC free article] [PubMed] [Google Scholar]
- 7.Dotto GP. Signal transduction pathways controlling the switch between keratinocyte growth and differentiation. Crit Rev Oral Biol Med. 1999;10:442–457. doi: 10.1177/10454411990100040201. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs E. Epidermal differentiation and keratin gene expression. J Cell Sci Suppl. 1993;17:197–208. doi: 10.1242/jcs.1993.supplement_17.28. [DOI] [PubMed] [Google Scholar]
- 9.Osborne R, Greenlee WF. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) enhances terminal differentiation of cultured human epidermal cells. Toxicol Appl Pharmacol. 1985;77:434–443. doi: 10.1016/0041-008x(85)90183-8. [DOI] [PubMed] [Google Scholar]
- 10.Sutter TR, Guzman K, Dold KM, Greenlee WF. Targets for dioxin: Genes for plasminogen activator inhibitor-2 and interleukin-1 beta. Science. 1991;254:415–418. doi: 10.1126/science.1925598. [DOI] [PubMed] [Google Scholar]
- 11.Choi EJ, Toscano DG, Ryan JA, Riedel N, Toscano WA., Jr Dioxin induces transforming growth factor-alpha in human keratinocytes. J Biol Chem. 1991;266:9591–9597. [PubMed] [Google Scholar]
- 12.Ray SS, Swanson HI. Alteration of keratinocyte differentiation and senescence by the tumor promoter dioxin. Toxicol Appl Pharmacol. 2003;192:131–145. doi: 10.1016/s0041-008x(03)00277-1. [DOI] [PubMed] [Google Scholar]
- 13.Greenlee WF, Poland A. An improved assay of 7-ethoxycoumarin O-deethylase activity: Induction of hepatic enzyme activity in C57BL/6J and DBA/2J mice by phenobarbital, 3-methylcholanthrene and 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Pharmacol Exp Ther. 1978;205:596–605. [PubMed] [Google Scholar]
- 14.Taketani Y, Oka T. Biological action of epidermal growth factor and its functional receptors in normal mammary epithelial cells. Proc Natl Acad Sci USA. 1983;80:2647–2650. doi: 10.1073/pnas.80.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hankinson O. Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor. Arch Biochem Biophys. 2005;433:379–386. doi: 10.1016/j.abb.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 16.Sun TT, Green H. Differentiation of the epidermal keratinocyte in cell culture: Formation of the cornified envelope. Cell. 1976;9:511–521. doi: 10.1016/0092-8674(76)90033-7. [DOI] [PubMed] [Google Scholar]
- 17.Poumay Y, Pittelkow MR. Cell density and culture factors regulate keratinocyte commitment to differentiation and expression of suprabasal K1/K10 keratins. J Invest Dermatol. 1995;104:271–276. doi: 10.1111/1523-1747.ep12612810. [DOI] [PubMed] [Google Scholar]
- 18.Takagi S, et al. Alteration of the 4-sphingenine scaffolds of ceramides in keratinocyte-specific Arnt-deficient mice affects skin barrier function. J Clin Invest. 2003;112:1372–1382. doi: 10.1172/JCI18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng S, et al. Targeted ablation of Arnt in mouse epidermis results in profound defects in desquamation and epidermal barrier function. J Cell Sci. 2006;119:4901–4912. doi: 10.1242/jcs.03282. [DOI] [PubMed] [Google Scholar]
- 20.Sogawa K, et al. Repression of cytochrome P-450c gene expression by cotransfection with adenovirus E1a DNA. Eur J Biochem. 1989;181:539–544. doi: 10.1111/j.1432-1033.1989.tb14757.x. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi A, Numayama-Tsuruta K, Sogawa K, Fujii-Kuriyama Y. CBP/p300 functions as a possible transcriptional coactivator of Ah receptor nuclear translocator (Arnt) J Biochem. 1997;122:703–710. doi: 10.1093/oxfordjournals.jbchem.a021812. [DOI] [PubMed] [Google Scholar]
- 22.Taylor RT, Wang F, Hsu EL, Hankinson O. Roles of coactivator proteins in dioxin induction of CYP1A1 and CYP1B1 in human breast cancer cells. Toxicol Sci. 2009;107:1–8. doi: 10.1093/toxsci/kfn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Ge K, Roeder RG, Hankinson O. Role of mediator in transcriptional activation by the aryl hydrocarbon receptor. J Biol Chem. 2004;279:13593–13600. doi: 10.1074/jbc.M312274200. [DOI] [PubMed] [Google Scholar]
- 24.Hottiger MO, Felzien LK, Nabel GJ. Modulation of cytokine-induced HIV gene expression by competitive binding of transcription factors to the coactivator p300. EMBO J. 1998;17:3124–3134. doi: 10.1093/emboj/17.11.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1:2005. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 27.Arora S, et al. Egr1 regulates the coordinated expression of numerous EGF receptor target genes as identified by ChIP on chip. Genome Biol. 2008;9:R166. doi: 10.1186/gb-2008-9-11-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YJ, Wang YN, Chang WC. ERK2-mediated C-terminal serine phosphorylation of p300 is vital to the regulation of epidermal growth factor-induced keratin 16 gene expression. J Biol Chem. 2007;282:27215–27228. doi: 10.1074/jbc.M700264200. [DOI] [PubMed] [Google Scholar]
- 29.Ke S, Rabson AB, Germino JF, Gallo MA, Tian Y. Mechanism of suppression of cytochrome P-450 1A1 expression by tumor necrosis factor-alpha and lipopolysaccharide. J Biol Chem. 2001;276:39638–39644. doi: 10.1074/jbc.M106286200. [DOI] [PubMed] [Google Scholar]
- 30.Obata H, et al. NF-kappa B is induced in the nuclei of cultured rat aortic smooth muscle cells by stimulation of various growth factors. Biochem Biophys Res Commun. 1996;224:27–32. doi: 10.1006/bbrc.1996.0979. [DOI] [PubMed] [Google Scholar]
- 31.Drozdoff V, Pledger WJ. Commitment to differentiation and expression of early differentiation markers in murine keratinocytes in vitro are regulated independently of extracellular calcium concentrations. J Cell Biol. 1993;123:909–919. doi: 10.1083/jcb.123.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peus D, Hamacher L, Pittelkow MR. EGF-receptor tyrosine kinase inhibition induces keratinocyte growth arrest and terminal differentiation. J Invest Dermatol. 1997;109:751–756. doi: 10.1111/1523-1747.ep12340759. [DOI] [PubMed] [Google Scholar]
- 33.Kolev V, et al. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol. 2008;10:902–911. doi: 10.1038/ncb1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sando GN, Howard EJ, Madison KC. Induction of ceramide glucosyltransferase activity in cultured human keratinocytes. Correlation with culture differentiation. J Biol Chem. 1996;271:22044–22051. doi: 10.1074/jbc.271.36.22044. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez-Salguero PM, Ward JM, Sundberg JP, Gonzalez FJ. Lesions of aryl-hydrocarbon receptor-deficient mice. Vet Pathol. 1997;34:605–614. doi: 10.1177/030098589703400609. [DOI] [PubMed] [Google Scholar]
- 36.Loertscher JA, Sattler CA, Allen-Hoffmann BL. 2,3,7,8-Tetrachlorodibenzo-p-dioxin alters the differentiation pattern of human keratinocytes in organotypic culture. Toxicol Appl Pharmacol. 2001;175:121–129. doi: 10.1006/taap.2001.9202. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ausubel FM, et al. Current Protocols in Molecular Biology. New York: Wiley; 1990. pp. 12.11.11–12.11.13. [Google Scholar]
- 40.Beischlag TV, et al. Recruitment of thyroid hormone receptor/retinoblastoma-interacting protein 230 by the aryl hydrocarbon receptor nuclear translocator is required for the transcriptional response to both dioxin and hypoxia. J Biol Chem. 2004;279:54620–54628. doi: 10.1074/jbc.M410456200. [DOI] [PubMed] [Google Scholar]
- 41.Hines RN, Mathis JM, Jacob CS. Identification of multiple regulatory elements on the human cytochrome P450IA1 gene. Carcinogenesis. 1988;9:1599–1605. doi: 10.1093/carcin/9.9.1599. [DOI] [PubMed] [Google Scholar]
- 42.Tang YM, et al. Isolation and characterization of the human cytochrome P450 CYP1B1 gene. J Biol Chem. 1996;271:28324–28330. doi: 10.1074/jbc.271.45.28324. [DOI] [PubMed] [Google Scholar]
- 43.Gasiewicz TA, Rucci G. Alpha-naphthoflavone acts as an antagonist of 2,3,7, 8-tetrachlorodibenzo-p-dioxin by forming an inactive complex with the Ah receptor. Mol Pharmacol. 1991;40:607–612. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.