Abstract
The N-end rule pathway targets protein degradation through the identity of the amino-terminal residue of specific protein substrates. Two components of this pathway in Arabidopsis thaliana, PROTEOLYSIS6 (PRT6) and arginyl-tRNA:protein arginyltransferase (ATE), were shown to regulate seed after-ripening, seedling sugar sensitivity, seedling lipid breakdown, and abscisic acid (ABA) sensitivity of germination. Sensitivity of prt6 mutant seeds to ABA inhibition of endosperm rupture reduced with after-ripening time, suggesting that seeds display a previously undescribed window of sensitivity to ABA. Reduced root growth of prt6 alleles and the ate1 ate2 double mutant was rescued by exogenous sucrose, and the breakdown of lipid bodies and seed-derived triacylglycerol was impaired in mutant seedlings, implicating the N-end rule pathway in control of seed oil mobilization. Epistasis analysis indicated that PRT6 control of germination and establishment, as exemplified by ABA and sugar sensitivity, as well as storage oil mobilization, occurs at least in part via transcription factors ABI3 and ABI5. The N-end rule pathway of protein turnover is therefore postulated to inactivate as-yet unidentified key component(s) of ABA signaling to influence the seed-to-seedling transition.
Keywords: abscisic acid, aminoacyl tRNA protein transferase, lipid bodies, targeted protein degradation
The N-end rule pathway of protein degradation dictates the half-life of proteins containing destabilizing amino-terminal residues (1, 2). In Arabidopsis, several of the components of this pathway have been identified (3–5) (Fig. 1). Amino-terminal destabilizing residues of proteins (N-degrons) are recognized by N-recognin E3 ligases (N-recognins) and are targeted for proteolysis via the 26S proteasome (2). In yeast and mammals, 2 classes of N-recognins (types I and II) recognize either basic or hydrophobic amino-terminal amino acids, respectively. Yeast UBR1 was the first characterized E3 ligase with N-degron activity, and in mammals there are at least 7 proteins that contain the characteristic UBR box (2). In contrast, the Arabidopsis UBR box-containing E3 ligase PROTEOLYSIS6 (PRT6) recognizes only basic amino acids, and PRT1, which does not contain a UBR box, recognizes aromatic amino acids (3, 4, 6). Other N-recognins must also exist in Arabidopsis, because other N-degrons are unstable in prt6 prt1 double mutants (3). N-degrons containing primary destabilizing amino-terminal amino acids can also be created through the conversion of pre-N-degron tertiary or secondary destabilizing residues (Fig. 1). Tertiary destabilizing residues can be converted to secondary through the action of amino-terminal aminohydrolase (NTAN) or through chemical modification of cysteine, and secondary to primary through the action of arginyl-tRNA:protein arginyltransferase (ATE1; Fig. 1). Arabidopsis contains 2 ATE genes, one of which (ATE1) has been implicated in leaf senescence (5). Model substrates starting with Asp or Glu were specifically stabilized in protoplasts of the ate1 mutant, confirming a biochemical role in the N-end rule pathway (5). Key to the formation of N-degrons is the activity of specific endopeptidases that expose primary, secondary, or tertiary destabilizing residues (Fig. 1). Degradation usually requires prior modification of substrates. For instance, Drosophila IAP 1 is cleaved by caspase to expose a destabilizing residue, a process required for the correct regulation of apoptosis (7). Mammalian G protein signaling components RGS 4, 5, and 16 become substrates of the N-end rule pathway through oxidation and subsequent arginylation of Cys-2 exposed following methionine aminopeptidase (MetAP) activity (8), and Arabidopsis protein RIN4 is cleaved by a bacterial pathogenicity factor before rapid degradation (9).
Fig. 1.
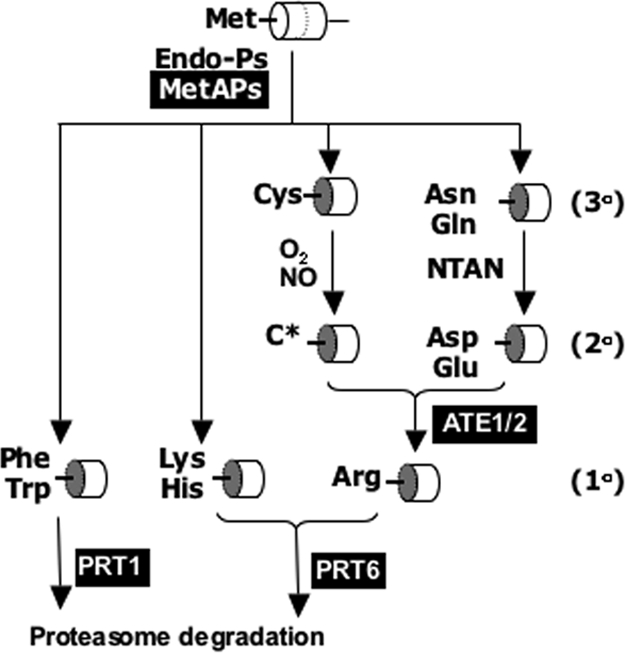
Diagrammatic representation of the N-end rule pathway associated with PRT6 function. Components for which orthologous Arabidopsis genes have been identified are highlighted. Protein substrate (represented as a cylinder) is cleaved by a methionine aminopeptidase (MetAP) or a specific endopeptidase (Endo-P) at a unique amino acid sequence (dotted curve). Cleavage reveals destabilizing amino acid residues, which may be further modified by enzymatic (NTAN, amino-terminal amidohydrolase; ATE1/2, Arg-tRNA-protein transferase) or chemical (O2, NO) action. 1°, 2°, and 3° indicate primary, secondary, and tertiary destabilizing amino-terminal residues, respectively. Generation of a primary destabilizing residue (1°) leads to targeting by E3 ubiquitin ligases PROTEOLYSIS6 (PRT6) or PRT1, depending on the nature of the destabilizing residue. C*, oxidized derivative of cysteine.
In this article, we investigate the role of the N-end rule pathway in seed germination and establishment. Arabidopsis seed germination is regulated by intrinsic hormonal pathways and external environmental signals, which influence whether an imbibed seed completes germination or remains dormant (10–12). Dry Arabidopsis seeds exhibit physiologically non-deep dormancy, and following shedding they go through a period of after-ripening, through which the capacity for dormancy is lost (10, 13). The Arabidopsis embryo is surrounded by 2 structures: a single endosperm cell layer and an external, dead testa layer that is maternally derived (11). Germination proceeds through 2 stages: initially, the testa layer ruptures, and subsequently the endosperm ruptures opposite the emerging embryo radicle. Germination is said to be complete when the radicle has penetrated the endosperm (10, 11). The phytohormone abscisic acid (ABA) represses germination and is presumed to function to stabilize the dormant state. Catabolism of ABA through ABA 8′-hydroxylase is required to reduce the dormancy of imbibed seeds (14, 15), and many loci have been identified that enhance the desensitization of seeds and seedlings to ABA or enhance the effect of ABA (11). In particular, the transcription factors ABI3, ABI4, and ABI5 enhance sensitivity to ABA (12). ABI3 has been shown to act upstream of ABI5 (16), and ABI4 has been implicated in the repression of seedling lipid degradation (17). The mechanisms through which ABA desensitization occurs are not well understood. However, specific proteolysis is important during the seed-to-seedling transition, because E3 ligases (not associated with the N-end rule pathway) have been shown to have a small influence on germination potential in the presence of ABA, or to regulate seedling establishment following germination (18, 19). In addition, there is a relationship between ABA and sugar signaling and sensitivity during seedling establishment, such that ABA synthesis/signal transduction mutants are insensitive to exogenous sugars for establishment, although the physiological significance of these observations is still not clear (20, 21). In this paper, we show that ATE and PRT6 are major regulators of seed ABA sensitivity and seedling establishment, indicating a key role for the N-end rule pathway in the seed-to-seedling transition.
Results
Influence of Components of the N-End Rule Pathway on Key Aspects of Germination and Establishment.
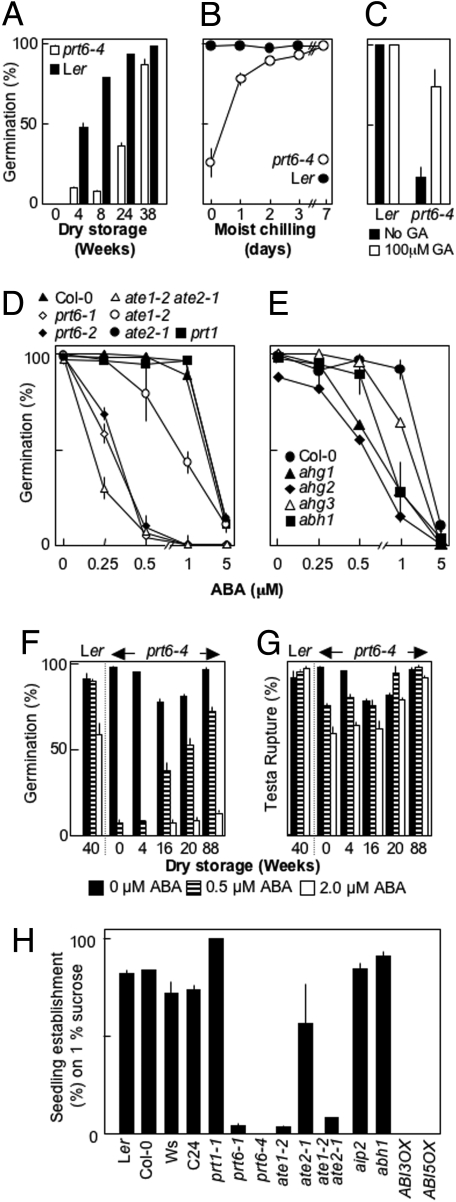
A genetic screen was carried out to identify Arabidopsis loci involved in the activation of processes associated with germination. Mutants were identified that showed reduced germination potential when after-ripening of wild-type (WT) seeds was complete (22). Positional cloning and complementation analysis with previously published alleles identified the PROTEOLYSIS6 (PRT6) gene as one of these loci (allele prt6-4, At5g02310; Fig. S1) (3). After-ripening of prt6 seeds was substantially delayed, although moist chilling or exogenous gibberellic acid (GA) removed this block to germination (Fig. 2 A–C). The prt6 alleles exhibited extreme ABA-hypersensitive inhibition of germination in comparison with previously published ABA-hypersensitive mutants or with transgenics ectopically expressing ABI3 and ABI5, positive components of the ABA signal transduction pathway (Fig. 2 D and E, Fig. 4A, and Fig. S2D). Endogenous ABA levels were not altered in prt6-4 or changed with after-ripening status (Fig. S2A). ABA sensitivity of WT seeds did not change during after-ripening (Fig. 2 F and G and Fig. S2C). In contrast, sensitivity to exogenous ABA decreased with dry storage time of mutant seeds (Fig. 2F). Previous studies have shown that Arabidopsis germination is composed of 2 phases: testa rupture followed by endosperm rupture (reviewed in ref. 11). Decreased germination potential of prt6 seeds resulted from a defect subsequent to testa rupture, because this was unaffected by storage time or in ABA sensitivity (Fig. 2G). Germination was also analyzed by using mutants of 2 other components of the N-end rule pathway: PRT1 (4) and ATE (Fig. 1). Arabidopsis contains 2 ATE genes: ATE1 (At5g05700) and ATE2 (At3g11240) (5) (Fig. S1). Neither prt1-1 nor the single ate1-2 or ate2-1 mutants demonstrated the extreme ABA hypersensitivity shown by prt6 alleles, although ate1-2 was as hypersensitive as some previously characterized mutants (Fig. 2 D and E). However, ate1-2 ate2-1 double-mutant seed showed hypersensitivity similar to prt6 alleles. In contrast to ABA hypersensitivity of germination, root growth of prt6 was not hypersensitive to ABA inhibition of root elongation, as was also the case for another ABA-hypersensitive mutant, abh1 (Fig. S3).
Fig. 2.
ABA sensitivity of germination, after-ripening, and sucrose sensitivity of establishment. (A) After-ripening of WT (Ler) and prt6-4 seeds assayed on water agarose after 7 days. (B) Germination potential of seeds imbibed for 7 days on water-agarose media after increasing periods of cold, moist chilling. (C) Germination potential of seeds imbibed for 7 days on agarose media containing 1/2MS in the absence (black bars) and presence (white bars) of 100 μM GA3. (D and E) Germination potential after 7 days of WT, mutant, and transgenic seeds assayed on 1/2MS media in the presence of exogenous ABA following 2 days of moist chilling. Seeds were stored for 2 months before assay. Respective WT and mutant seeds are: Col-0, prt1-1, prt6-1, prt6-2, ate1-2, ate2-1, abh1, ahg1, 2, 3 (3–5 and 36–39). Data points for ate2-1 are slightly obscured by those of prt1. Results obtained for ABI3OX, ABI5OX, and aip2 are presented in Fig. S2D. (F and G) Change in sensitivity of endosperm rupture (F) and testa rupture (G) to exogenous ABA, with time of dry storage of cold, chilled WT (Ler) and prt6-4 seeds, assayed on 1/2MS media and measured at 7 days following 2 days of moist chilling. (H) Sucrose sensitivity of establishment (greening of cotyledons) of WT, transgenic, and mutant seedlings grown on water-agarose supplemented with 1% (wt/vol) sucrose measured at 7 days, following 2 days of moist chilling. Dose–response curves are presented in Fig. S4. Data represent means ± SE of the mean.
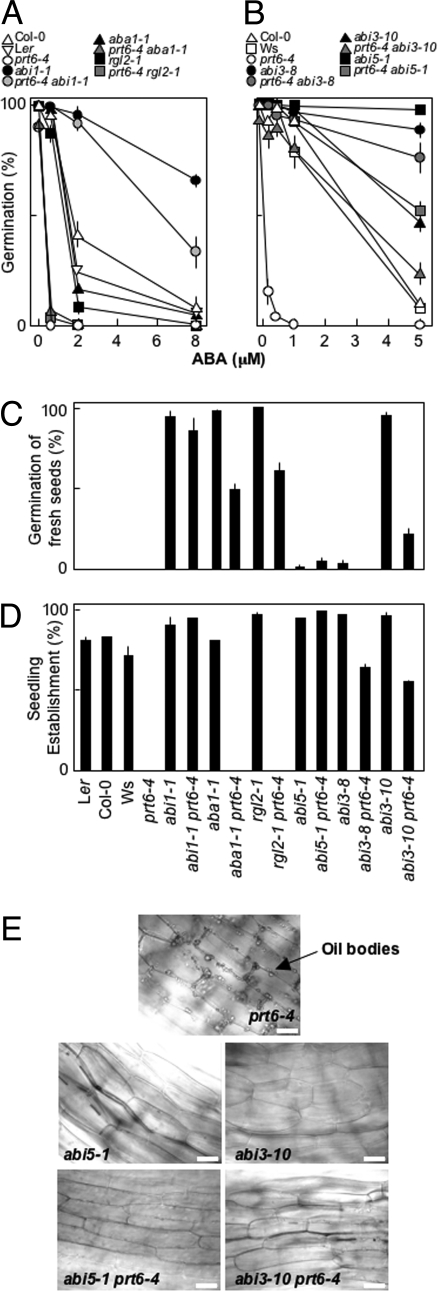
Fig. 4.
Genetic interactions of PRT6 with components of ABA and GA biosynthesis and signaling. (A and B) Germination potential after 7 days of WT mutant and transgenic seeds on 1/2MS media in the presence of exogenous ABA, following 2 days of moist chilling. abi1-1 (Ler background) (26), aba1-1 (Ler) (25), rgl2-1 (Ler) (24), abi3-8 (Col-0), abi3-10 (Col-0) (28), abi5-1 (Ws) (27). (C) Germination potential, after 7 days, of freshly harvested seeds on water-agarose. (D) Seedling establishment, after 7 days, on water-agarose containing 1% (wt/vol) sucrose, following 2 days of moist chilling (dose–response curves are presented in Fig. S4.). (E) Light micrographs of hypocotyl epidermis of single- and double-mutant seeds showing retention of lipid bodies in prt6-4, but not in single- or double-mutant combinations with abi3-10 or abi5-1, on 1/2MS media after 5 days, following 2 days of moist chilling. (Scale bar: 20 μm.). Data represent means ± SE of the mean.
Numerous genetic studies have indicated a role for ABA in sugar signaling during seedling establishment (20, 21). Therefore, we analyzed sugar sensitivity of establishment for N-end rule mutants in comparison with ABA signaling mutants (Fig. 2H and Fig. S4). prt6-4 was hypersensitive to inhibition of establishment by sugars but not to sugar alcohols (Fig. 2H and Fig. S2B), and sucrose sensitivity was greater on water agarose compared with media supplemented with half-strength MS salts (1/2MS), with IC50 values of 0.27% and 1.4%, respectively (Fig. 3A).
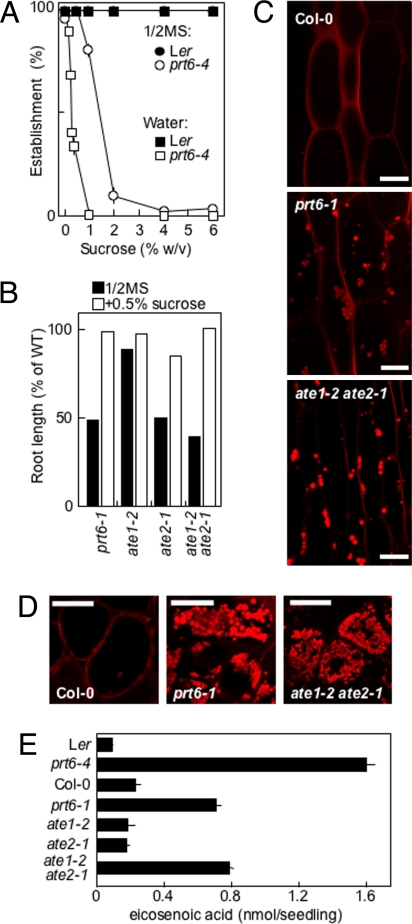
Fig. 3.
Influence of PRT6 and ATE1/2 on lipid degradation during establishment. (A) Establishment of prt6-4 and WT (Ler) seedlings after 7 days of growth on increasing concentrations of sucrose either in the presence (1/2MS) or absence (water) of 1/2 MS following 2 days of moist chilling. (B) Influence of exogenous sucrose on root length of prt6 and ate1 and ate2 alleles in relation to WT, measured at 7 days of imbibition on 1/2MS media following 2 days of moist chilling. (C and D) Representative confocal microscopy images of hypocotyl epidermal (C) and endosperm (D) cells (5 days of imbibition on 1/2MS, no added sucrose, following 2 days of moist chilling) stained with Nile Red to reveal oil bodies, which can be seen as red-staining spherical inclusions in prt6-1 and ate1 ate2, but not in Col-0. (Scale bar: 20 μm.) Brightfield images are presented in Fig. S5. (E) Eicosenoic acid (20:1) content of 5-day-old seedlings grown on 1/2MS media following 2 days of moist chilling. Data represent means ± SE of the mean.
prt6 and ate1-2 ate2-1 Mutant Seeds Retain Oil Bodies Following Germination.
Roots of single prt6 and double ate1-2 ate2-1 mutants were shorter than WT on 1/2MS media but exhibited enhanced growth in the presence of 0.5% sucrose (a concentration noninhibitory to establishment; Fig. 3A), indicating that seedlings may be energetically challenged (Fig. 3B). Hypocotyls and endosperms of these mutants retained oil bodies well beyond the time that these had disappeared in the respective WT seedlings (Fig. 3 C and D and Fig. S5). Accordingly, analysis of fatty acid composition demonstrated that mutant seedlings retained eicosenoic acid (20:1), a fatty acid considered to be diagnostic of Arabidopsis seed triacylglycerol (23) (Fig. 3E). However, the capacity for β-oxidation of the model substrate, 2,4-dichlorophenoxybuytric acid (2,4-DB), was not markedly reduced in these mutants, as judged by root elongation inhibition assays (Fig. S6), suggesting that the affected step occurs before peroxisome import and β-oxidation of fatty acids.
Genetic Interactions of PRT6 with Seed ABA and GA Signal Transduction Components.
Both ABA and GA have been shown to play antagonistic roles in the regulation of germination. Therefore, we analyzed the genetic interactions between PRT6 and genes associated with signaling and synthesis of these hormones. The RGL2 DELLA protein is a negative regulator of germination (24). We observed that rgl2-1 seeds did not exhibit marked sensitivity to ABA, whereas rgl2-1 prt6-4 seeds were hypersensitive (Fig. 4A). Freshly harvested rgl2-1 prt6-4 seeds showed a reduction in germination potential in comparison with rgl2-1 seeds (Fig. 4C), and seeds were also hypersensitive to sucrose for establishment (Fig. 4D and Fig. S4).
Mutations in ABA biosynthesis (aba1-1) and signal transduction (abi1-1) reduce seed dormancy and sensitivity of establishment to exogenous sugar, and in addition, abi1-1 reduces sensitivity of germination to exogenous ABA (25, 26). Whereas aba1-1 seeds were similar in ABA sensitivity to WT, aba1-1 prt6-4 double-mutant seeds exhibited hypersensitivity similar to prt6-4. Both abi1-1 and abi1-1 prt6-4 seeds showed highly ABA-insensitive germination potential (Fig. 4A). Although freshly harvested abi1-1 prt6-4 seeds were nondormant (Fig. 4C), aba1-1 prt6-4 seeds did retain some dormancy in comparison with aba1-1. Both abi1-1 and aba1-1 showed sucrose insensitivity of establishment (Fig. 4D); however, only abi1-1 seeds retained this phenotype in the prt6 background.
The transcription factors ABI5 and ABI3 participate in germination-related ABA signaling (27, 28). We used 2 alleles of ABI3 (abi3-8, missense mutation in the B1 domain; abi3-10, missense mutation in the B2 domain) and 1 allele of ABI5, all of which show highly reduced sensitivity of germination to ABA (27, 28). In all cases, double mutants containing prt6-4 also showed ABA-insensitive germination (Fig. 4B). Freshly harvested seeds of both abi5-1 and abi3-8 exhibited high levels of primary dormancy, whereas abi3-10 seeds were nondormant (Fig. 4C), and the abi3-10 prt6-4 double-mutant seeds showed reduced dormancy. Although prt6-4 partially increased sucrose hypersensitivity for abi3-8 and abi3-10 seeds, it did not affect sensitivity in the abi5-1 background (Fig. 4D). Light microscopic analysis demonstrated that oil bodies were not retained in establishing seedlings of abi3-10 prt6-4 and abi5-1 prt6-4, in contrast to prt6-4 (Fig. 4E).
Discussion
In this paper we describe the identification of a major function for the N-end rule pathway of protein degradation in the regulation of the seed-to-seedling transition. We show that this pathway is required for key aspects of seed after-ripening, ABA sensitivity, lipid degradation, and seedling establishment.
The PRT6 gene was identified as a regulator of germination potential. Mutant seeds exhibited a greatly increased after-ripening time (Fig. 2), suggesting that PRT6 function is required to potentiate after-ripening. Prolonged after-ripening may be due to enhanced ABA signaling, because germination was shown to be highly sensitive to exogenous ABA, and ABA sensitivity of prt6-4 reduced with seed storage time, as the germination potential of untreated seeds increased. ABA sensitivity occurred following testa rupture, indicating action late in phase II of germination, although ABA levels in imbibed prt6 seeds were similar to those of WT. The prt6 alleles exhibited an extreme sensitivity of germination to ABA, suggesting that this protein is key to the removal of ABA sensitivity immediately before germination. In agreement with a role in germination, analysis of publicly available expression data indicated that all 3 components are expressed at low levels in the embryo and endosperm of germinating seeds (Fig. S7).
Other components of the N-end rule pathway have been identified in Arabidopsis, including PRT1 (4, 6) and ATE (5). It is unlikely that this pathway acts through bulk degradation of proteins in imbibed seeds, because we did not observe any influence of the prt1 mutant on ABA or sucrose sensitivity. However, germination of double-mutant ate1-2 ate2-1 seeds was highly sensitive to exogenous ABA. This observation confirms the importance of the N-end rule pathway in seed ABA signaling, because ATE provides arginylation of secondary destabilizing residues for PRT6. The similarity of the phenotypes of prt6 and ate1-2 ate2-1 mutants demonstrates that arginylation of a secondary destabilizing residue is required for the substrate(s), and also indicates that the relevant substrate(s) continue to be functional throughout processing to their arginylated form (Fig. 1). In common with some other ABA-hypersensitive mutants (but not aip2 or abh1), prt6 and ate1-2 ate2-1 were also hypersensitive to sucrose for seedling establishment (as were transgenic seedlings ectopically expressing ABI3 and ABI5). Sugar sensitivity of establishment was specific to endogenous monosaccharides and disaccharides but not to sugar alcohols, discounting an osmotic effect.
Remarkably, the N-end rule mutants retained oil bodies after they had been degraded in WT seedlings, indicating that this pathway influences many aspects of development, from imbibition to establishment. The inhibition of lipid degradation in these mutants was an unexpected finding, because prt6 and ate1-2 ate2-1 seedlings are able to complete establishment in the absence of exogenous sucrose, a process that is considered to require seed oil mobilization (29). β-Oxidation of the model substrate 2,4-DB was unaffected, consistent with an early block in lipid mobilization or in an associated pathway (30). Further studies are required to characterize the precise mechanism by which this pathway regulates lipid breakdown and to explore its biotechnological potential.
Analysis of the genetic relationship between PRT6 and components of ABA/GA pathways indicated that PRT6 function is closely associated with ABA but not GA signaling. Assay for the effect of exogenously applied ABA on the repression of early endosperm rupture in moist prechilled seeds indicated an interaction between PRT6 and the ABA signaling components ABI1, ABI3, and ABI5. The abi1-1 mutation dominantly represses ABA signaling, thereby bypassing the requirement for PRT6, although the dominant-negative nature of this mutation means that it is difficult to provide a straightforward explanation of the associated signaling hierarchy. In the absence of activity of the ABI3 or ABI5 transcription factors, PRT6 function is not required because ABA sensitivity is removed. RGL2 is one member of the DELLA family of proteins that has been shown to negatively regulate germination (24, 31). Removal of RGL2 activity is required for activation of GA-regulated responses (24, 32). Mutation of RGL2 did not alter ABA hypersensitivity of prt6-4, suggesting that PRT6 function to remove ABA sensitivity is required before that of GA signaling. Assay for the effect of genetic interactions on primary dormancy showed that the dominant abi1-1 mutation bypasses a requirement for PRT6. Interestingly, both abi5-1 and abi3-8 exhibited high levels of primary dormancy, whereas abi3-10 did not. Dormancy in abi3-10 prt6-4 seeds was increased, suggesting that PRT6 function may be required before that of ABI3 in the removal of ABA signal transduction. Alternatively, abi3-10 may represent a weaker allele that may still provide some protein function in the absence of PRT6 activity in the double mutant. Although aba1-1 seeds are in a low-dormancy state, dormancy is increased in combination with prt6-4. This may be due to the failure to degrade PRT6 substrate(s) in a prt6 background, an effect that could be enhanced through low levels of remaining ABA. Similarly, rgl2 seeds exhibited a low-dormancy state, in this case due to lack of repression of GA signaling. Stabilization of PRT6 substrate in the double mutant may partially overcome this reduced dormancy, as suggested above for aba1-1 prt6-4, by increasing the strength of ABA signaling, in opposition to GA activation of germination. Assay for the sensitivity of seedling establishment to exogenous sucrose also demonstrated a genetic interaction between PRT6 and components of ABA signaling.
Double mutants abi3 prt6 and abi5 prt6 did not retain oil bodies, suggesting a role for ABA signaling in the repression of lipid degradation. Previous work has shown that ABI4, not ABI5, is required for lipid breakdown in seedlings (17); however, the experimental system used was different from that reported here, involving application of very high levels (20 μM) of ABA to mutant seeds.
Mechanisms regulating after-ripening are not well understood (11, 13). Previously, it has been shown that catabolism of ABA in imbibed seeds is an important regulator of germination potential, and that seeds lacking ABA 8′-hydroxylase activity exhibit enhanced dormancy (14, 15). However, ABA catabolism alone cannot explain reduced functioning of ABA pathways in the repression of germination, because there is only a slight increase in dormancy of these mutant seeds. Mechanisms for the reduction of sensitivity to ABA must also be important. Many proteins, representing different biochemical pathways, are involved in the desensitization of seeds to ABA (11), but the function of these proteins in after-ripening has not been reported. The N-end rule components PRT6 and ATE play key roles in the removal of ABA sensitivity. After-ripening of prt6-4 was delayed and sensitivity to ABA reduced with after-ripening time, suggesting that the N-end rule pathway may be responsible for desensitizing seeds during a “window” of ABA responsiveness (10, 12). This pathway may function through the ABA signaling components ABI3 and ABI5, although it is not possible from genetic evidence to state whether these are direct targets or whether they function upstream or downstream of the N-end rule pathway. Germination potential following imbibition is signaled by after-ripening, and it is likely that both ABA catabolism (through ABA 8′-hydroxylase activity) and desensitization (through the N-end rule pathway and also through other, more minor components) play important roles in removing the capacity for ABA to repress germination and maintain a dormant state. Because after-ripening is a key ecological trait, it will be interesting to understand the influence of the environment on these 2 components. Recent work has shown that light influences ABA 8′-hydroxylase function (33).
The discovery of new roles for the N-end rule pathway in plants poses many intriguing questions. For example, nitric oxide (NO) can remove dormancy in imbibed seeds (34), and we note that NO has been shown to function within the N-end rule pathway (2), raising the possibility that NO operates through this pathway. The pathway requires a specific endopeptidase–substrate interaction or action of MetAPs (Fig. 1), and it has been shown that many peptidases (including MetAPs) are induced following after-ripening (13). Although ABI3 and/or ABI5 are possible substrates, ectopic expression of these transcription factors did not result in the extreme ABA sensitivity observed for prt6 and ate1/2 mutants (Fig. S2D). The future identification of the substrate(s) for this pathway in imbibed seeds should shed further light on the regulation of ABA desensitization, the mechanism(s) through which after-ripening signals germination potential, and the regulation of lipid degradation.
Materials and Methods
Plant Material and Physiological Analysis.
Arabidopsis genetic resources were obtained from NASC or individual researchers. Plant growth and germination/seedling establishment assays were carried out as before (13, 35). Unless otherwise stated, seed germination/establishment was assayed at 7 days' imbibition at 22 °C: radicle protrusion through the endosperm was used as the criterion for germination, and cotyledon greening was used for establishment [establishment assayed on water agarose containing 1% (wt/vol) sucrose]. Where indicated in figure legends, seeds were exposed to 2 days of moist chilling at 4 °C before transfer to 22 °C to remove residual dormancy.
Lipid Analysis.
Observation of lipid bodies was carried out following staining with the lipophilic dye Nile Red by using a Nikon eC1/TiU inverted confocal microscope, and seedling images (Fig. 4) were obtained by differential interference contrast microscopy using a Nikon OptiPhot 2 Upright Microscope. Fatty acids were analyzed by gas chromatography following conversion to methyl esters (23) adapted for small tissue samples and using 17:0 as an internal standard.
Supplementary Material
Acknowledgments.
T.J.H. was supported by a PhD grant from the Lawes Trust, P.D.J. by a Biotechnology and Biological Sciences Research Council (BBSRC) studentship, L.R. by a National Fruit and Cider Institute PhD studentship, and A.M. and H.S. by fellowship grants from the University of Nottingham (Nottingham, U.K.). Rothamsted Research receives grant-aided support from the BBSRC. Financial assistance was provided to S.Ú.T. and M.J.H. by a BBSRC Centre for Plant Integrative Biology (CPIB) award to the University of Nottingham, and to A.B. by the Deutsche Forschungsgemeinschaft (Grant BA 1158/3/1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810280106/DCSupplemental.
References
- 1.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 2.Tasaki T, Kwon YT. The mammalian N-end rule pathway: New insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Garzon M, et al. PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 2007;581:3189–3196. doi: 10.1016/j.febslet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Potuschak T, et al. PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc Natl Acad Sci USA. 1998;95:7904–7908. doi: 10.1073/pnas.95.14.7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida S, Ito M, Callis J, Nishida I, Watanabe A. A delayed leaf senescence mutant is defective in arginyl-tRNA: Protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 2002;32:129–137. doi: 10.1046/j.1365-313x.2002.01407.x. [DOI] [PubMed] [Google Scholar]
- 6.Stary S, et al. PRT1 of Arabidopsis is a ubiquitin protein ligase of the plant N-end rule pathway with specificity for aromatic amino-terminal residues. Plant Physiol. 2003;133:1360–1366. doi: 10.1104/pp.103.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ditzel M, et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 8.Lee MJ, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takemoto D, Jones DA. Membrane release and destabilization of Arabidopsis RIN4 following cleavage by Pseudomonas syringae AvrRpt2. Mol Plant Microbe Interact. 2005;18:1258–1268. doi: 10.1094/MPMI-18-1258. [DOI] [PubMed] [Google Scholar]
- 10.Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 11.Holdsworth MJ, Soppe WJJ, Bentsink L. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 12.Kucera B, Cohn MA, Leubner-Metzger G. Plant hormone interactions during seed dormancy release and germination. Seed Sci Res. 2005;15:281–307. [Google Scholar]
- 13.Carrera E, et al. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 2008;53:214–224. doi: 10.1111/j.1365-313X.2007.03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kushiro T, et al. The Arabidopsis cytochrome P450CYP707A encodes ABA 8′- hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto M, et al. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Molina L, Mongrand B, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- 17.Penfield S, Li Y, Gilday AD, Graham S, Graham IA. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell. 2006;18:1887–1899. doi: 10.1105/tpc.106.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang XR, Garreton V, Chua NH. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dekkers BJW, Schuurmans J, Smeekens SCM. Interaction between sugar and abscisic acid signalling during early seedling development in Arabidopsis. Plant Mol Biol. 2008;67:151–167. doi: 10.1007/s11103-008-9308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finkelstein RR, Gibson SI. ABA and sugar interactions regulating development: Cross-talk or voices in a crowd? Curr Opin Plant Biol. 2002;5:26–32. doi: 10.1016/s1369-5266(01)00225-4. [DOI] [PubMed] [Google Scholar]
- 22.Russell L, Larner V, Kurup S, Bougourd S, Holdsworth MJ. The Arabidopsis COMATOSE locus regulates germination potential. Development. 2000;127:3759–3767. doi: 10.1242/dev.127.17.3759. [DOI] [PubMed] [Google Scholar]
- 23.Lemieux B, Miquel M, Somerville C, Browse J. Mutants of Arabidopsis with alterations in seed lipid fatty-acid composition. Theor Appl Genet. 1990;80:234–240. doi: 10.1007/BF00224392. [DOI] [PubMed] [Google Scholar]
- 24.Lee SC, et al. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002;16:646–658. doi: 10.1101/gad.969002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koornneef M, Jorna ML, Brinkhorst Van der Swan DLC, Karssen CM. The isolation of Abscisic Acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L) Heynh. Theor Appl Genet. 1982;61:385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- 26.Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61:377–383. [Google Scholar]
- 27.Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nambara E, et al. A screen for genes that function in abscisic acid signaling in Arabidapsis thaliana. Genetics. 2002;161:1247–1255. doi: 10.1093/genetics/161.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker A, Graham IA, Holdsworth M, Smith SM, Theodolou FL. Chewing the fat: Beta-oxidation in signalling and development. Trends Plant Sci. 2006;11:124–132. doi: 10.1016/j.tplants.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Eastmond PJ. MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell. 2007;19:1376–1387. doi: 10.1105/tpc.106.043992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrera E, et al. Gene expression profiling reveals defined functions of the ABC transporter COMATOSE late in phase II of germination. Plant Physiol. 2007;143:1669–1679. doi: 10.1104/pp.107.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyler L, et al. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004;135:1008–1019. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo M, et al. Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006;48:354–366. doi: 10.1111/j.1365-313X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- 34.Bethke PC, et al. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 2007;143:1173–1188. doi: 10.1104/pp.106.093435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Footitt S, et al. Analysis of the role of COMATOSE and peroxisomal beta-oxidation in the determination of germination potential in Arabidopsis. J Exp Bot. 2006;57:2805–2814. doi: 10.1093/jxb/erl045. [DOI] [PubMed] [Google Scholar]
- 36.Hugouvieux V, Kwak JM, Schroeder JI. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- 37.Nishimura N, et al. Isolation and characterization of novel mutants affecting the abscisic acid sensitivity of Arabidopsis germination and seedling growth. Plant Cell Physiol. 2004;45:1485–1499. doi: 10.1093/pcp/pch171. [DOI] [PubMed] [Google Scholar]
- 38.Parcy F, Giraudat J. Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J. 1997;11:693–702. doi: 10.1046/j.1365-313x.1997.11040693.x. [DOI] [PubMed] [Google Scholar]
- 39.Brocard-Gifford I, Lynch TJ, Garcia ME, Malhotra B, Finkelstein RR. The Arabidopsis thaliana ABSCISIC ACID-INSENSITIVE8 locus encodes a novel protein mediating abscisic acid and sugar responses essential for growth. Plant Cell. 2004;16:406–421. doi: 10.1105/tpc.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.