Abstract
We have used laser-capture microdissection and microarray hybridization to characterize gene expression in the three principal neuron layers of rat hippocampus. Correlative and clustering analyses revealed all three layers to be easily differentiated from one another based on gene expression profile alone. A greater disparity in gene expression exists between dentate granule and pyramidal cell layers, reflecting phenotypic and ontological differences between those cell populations. Remarkably, the level of more than 45% of expressed transcripts was significantly different among the three neuron populations, with more than a third of those (>1,000 transcripts) being at least 2-fold different between layers. Even CA1 and CA3 pyramidal cell layers were dramatically different on a transcriptional level with a separate analysis indicating that nearly 20% of transcripts are differentially expressed between them. Only a small number of transcripts were specific for a given hippocampal cell layer, suggesting that functional differences are more likely secondary to wide-ranging expression differences of modest magnitude rather than very large disparities in a few genes. Categorical analysis of transcript abundance revealed concerted differences in gene expression among the three cell layers referable to specific cellular pathways. For instance, transcripts encoding proteins involved in glucose metabolism are most highly expressed in the CA3 pyramidal layer, which may reflect an underlying greater metabolic rate of these neurons and partially explain their exquisite vulnerability to seizure-induced damage. Conversely, transcripts related to MAP kinase signaling pathways and transcriptional regulator activity are prominent in the dentate granule cell layer, which could contribute to its resistance to damage following seizure activity by positioning these neurons to respond to external stimuli by altering transcription. Taken together, these data suggest that unique physiological characteristics of major cell layers, such as neuronal activity, neuronal plasticity, and vulnerability to neurodegeneration, are reflected in substantial transcriptional heterogeneity within the hippocampus.
Keywords: laser capture microdissection, pyramidal neuron, dentate gyrus, granule cell, microarray
Introduction
Pathophysiology in the hippocampus underlies abnormal neurological function in many human diseases, including epilepsy and stroke. Abnormal hippocampal morphology and aberrant neuronal excitability are well-described in temporal lobe epilepsy and thought to underlie epileptogenesis. The hippocampus is exquisitely vulnerable to hypoxic and ischemic insults leading to cognitive and psychiatric disturbances in stroke patients. Interestingly, in both settings, pathophysiology in the hippocampus is not uniform, but heterogeneous among the principal hippocampal cell layers. For example, granule cells in the dentate gyrus are remarkably resistant to neuronal damage caused by most insults, including hypoxia/ischemia and seizures (Borges et al., 2003; Mathern et al., 1995; Ordy et al., 1993). Conversely, pyramidal neurons in the CA3 region of Ammon's horn are extremely vulnerable to seizure-induced or trauma-induced damage, and CA1 pyramidal neurons are sensitive to both hypoxia/ischemia- and seizure-induced neurodegeneration (Borges et al., 2003; Mathern et al., 1995; Maxwell et al., 2003; Ordy et al., 1993).
Differential susceptibility to dysfunction and degeneration is most likely due to the distinctive anatomical and physiological characteristics of neurons in the principal hippocampal cell layers, the circuitry and neurochemistry of which is well-described. Less defined is the library of mRNA transcripts available to hippocampal cells. Identification of hippocampal gene expression profiles is important not only to determine what cellular pathways may be affected by the unique characteristics of each cell layer, but also to determine if differential gene expression is in part responsible for the manifestation of those properties. A few investigators have begun to explore this issue. A “molecular atlas” of the hippocampus is beginning to be defined based primarily on in situ hybridization studies targeted by a microarray experiment performed on microdissected hippocampal regions (Lein et al., 2004; Zhao et al., 2001). These data are convincing and intriguing, but only genes with dramatic differences between the cell layers have been investigated. More recently, laser capture microdissection (LCM) was used to compare gene expression between dentate gyrus and CA3 to demonstrate technical feasibility, but that study was performed on only a single rat (Datson et al., 2004).
We have taken a broader approach to exploring the transcriptional neuroanatomy of the hippocampus by focusing less on the magnitude of individual transcript differences and more on concerted, broad-based differences between three main cell layers in the rat hippocampal formation: dentate gyrus granule cells (DG) and pyramidal cells from CA1 and CA3. We have found that gene expression profiles in different hippocampal cell populations are widely disparate, not only on a gene-by-gene basis, but also based on concerted differences in a restricted number of cellular pathways.
Methods
Animals and tissue preparation
All animal procedures were performed in accordance with NIH guidelines and were approved by the Emory University IACUC. The animals used in this study served as “sham-preconditioned” rats in our previous study (Borges et al., 2007). Briefly, on two consecutive days adult male Sprague Dawley rats (200-270 g) obtained from Charles River were intraperitoneally injected with approximately 1 ml phosphate buffered saline (PBS, pH 7.4) followed 90 minutes later by an injection of pentobarbital (40 mg/kg, i.p.). On the third day rats were decapitated after isoflurane anesthesia. Under RNase-free conditions, brains were removed and immediately frozen on dry ice. Fourteen micron frozen sections through the hippocampus were collected onto uncoated microscope slides, refrozen on dry ice, and placed in a −80 °C freezer. For staining, sections were fixed in ice-cold 70% ethanol for 2 min, rinsed with water, dipped in cresyl violet for Nissl stain, and dehydrated to xylene. Sections were dried in a fume hood and LCM performed within 24 hours (Goldsworthy et al., 1999; Greene et al., 2005).
LCM, RNA isolation, and RNA amplification
Laser capture microdissection was performed using an Arcturus Pixcell IIe system with transmission illumination (Arcturus, CA) and the following parameters: spot size = 30 μm; power = 85 mW; and duration = 750-1200 μs (Emmert-Buck et al., 1996). The three major hippocampal cell layers (DG, CA1, and CA3) were harvested from 2-3 sections from each animal (3.5-4.5 mm caudal to bregma) onto separate LCM HS Caps (Arcturus) (Fig 1A). Cells near the boundary between the regions were not dissected to ensure anatomical distinction. CA2 neurons were not collected due to inability to clearly differentiate this small subregion reliably in Nissl-stained sections. Total RNA was immediately extracted using the Extractsure adapter and PicoPure Isolation Kit (Arcturus) with DNase digestion (Qiagen RNase-free DNase Set), and stored at −80 °C until use.
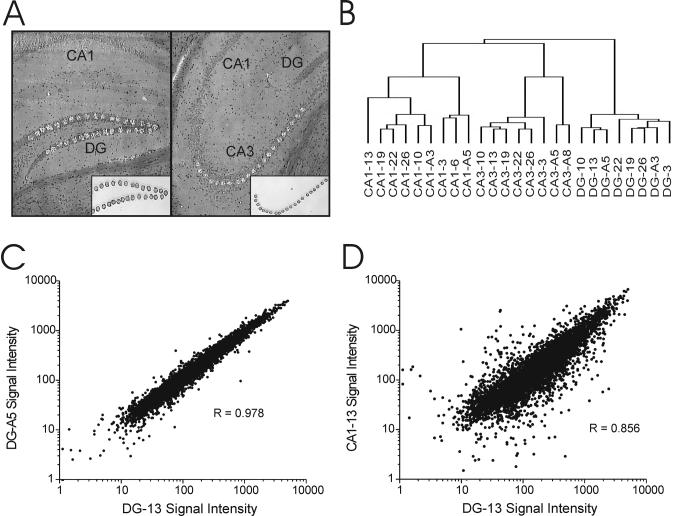
Figure 1. Differential gene expression between hippocampal cell layers.
A. Photomicrographs depicting coronal hippocampal sections after LCM of DG granule neurons (left) and CA3 pyramidal neurons (right). Corresponding inserts show isolated neuronal layers. B. Unsupervised hierarchical clustering using all expressed genes differentiates the three main hippocampal cell layers based on gene expression profile alone. C. Close correlation of gene expression between two DG samples, despite the fact that they are from two different animals from different sacrifice days. D. Lower correlation between DG and CA1 samples from the same animal.
Amplification of poly-A RNA was performed independently on each sample (Greene et al., 2005). Total RNA was used as template in a reverse transcription reaction at 37 °C using the Superscript II cDNA Synthesis Kit (Invitrogen) and an oligo-dT24 primer containing the T7 promoter (Proligo, LLC). Second strand synthesis was performed at 15 °C with E. Coli DNA polymerase. Ends were polished with T4 DNA polymerase, and the product was isolated using the Qiaquick PCR purification Kit (Qiagen). In vitro transcription of the template was performed overnight at 37 °C using the Megascript T7 Transcription Kit (Ambion). aRNA was isolated using the RNeasy Kit (Qiagen) and used as template for a second reverse transcription reaction with random hexamers (5 ng/μl) for priming. Following RNaseH digestion of the parent strand, second strand synthesis was performed at 15 °C with E. Coli DNA polymerase and the oligo-dT24 primer. Ends were polished with T4 DNA polymerase, and the product was isolated using the Qiaquick PCR purification Kit (Qiagen). Product quality was assessed after both rounds of cDNA synthesis using endpoint PCR for neuron specific enolase. The degree of amplification using this procedure was on the order of 3 × 105 fold, and products of amplification were from 250-2000 bases long.
Microarray hybridization
Sample labeling, microarray hybridization, and preliminary analyses were performed by the NINDS NIMH Microarray Consortium at the Translational Genomics Institute in Phoenix, AZ (TGEN; http://arrayconsortium.tgen.org). Briefly, we sent the Consortium second round cDNA, which was used to produce biotinylated cRNA using the EnzoBioArray High Yield RNA Transcript Labeling Kit (Affymetrix, CA). Samples (10 μg) were hybridized to Affymetrix Rat RAE230A Gene Chips. The RAE230A is a high-density microarray that surveys more than 10,000 unique transcripts. Chips were developed, scanned, and normalized by global scaling. Visual inspection was performed to identify arrays with production defects or uneven hybridization. Image files and data from all hybridizations are available online at the TGEN website.
The relative abundance of each probe set and an evaluation of whether a particular transcript was expressed above background were calculated using Microarray suite (MAS 5.0, Affymetrix). The assignment of each probe pair on the Rat RAE230A GeneChip to a gene was originally based by Affymetrix on the sequences available in Unigene build #99. The probe pair assignments have not been updated by Affymetrix, and approximately 11% of the original accession numbers assigned to probe sets on the Rat RAE230A chip either match fewer than half of the probe pairs in the corresponding set or are retired from current databases. Dai et al. created a custom CDF file based on Unigene build 154 (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp) that can be read by the MAS 5.0 program to assign signal intensities of each probe pair to genes (Dai et al., 2005). All probe pairs for a particular transcript are pooled into a single probe set, which eliminates duplicate or triplicate instances of genes on the Chip. Moreover, probes hybridizing to the non-coding strand of a transcript are deleted from analysis, which greatly reduces the number of expressed sequence tags (ESTs) called. Discrimination scores of the signal intensities for each spot on an individual chip were determined to be significantly different from background (i.e., present, marginally present, or absent calls) using a one-sided Wilcoxon's Sign Ranked test. We selected genes for subsequent analyses if signal intensities were significantly above background in 65% of arrays from at least one region.
When necessary, conversion from Unigene ID to other public ID types (e.g., gene symbol or GenBank Accession number) was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID), available online at NIAID (http://david.abcc.ncifcrf.gov/) (Dennis et al., 2003).
Statistical determination of differential expression
Genes that were differentially expressed among the three regions were determined by one way ANOVA with Benjamini-Hochberg (B-H) correction for multiple comparisons (Hochberg and Benjamini, 1990) using a false discover rate (FDR) = 1%. Separate unpaired, two-tailed t-tests with B-H correction (FDR = 1%) were performed to examine differences between dentate granule and pyramidal cell layers, and separately between CA1 and CA3 pyramidal neuron layers. Input for all analyses was a list of all expressed genes with corresponding log2 transformed signal intensities from every sample.
Unsupervised hierarchical clustering analysis
Unsupervised hierarchical clustering was performed using GenePattern (http://www.broad.mit.edu/cancer/software/genepattern/index.html) using Pearson's correlation (Reich et al., 2006). Input was a list of all expressed genes with corresponding log2 transformed signal intensities from every sample.
Gene set enrichment analysis (GSEA)
Gene set enrichment analysis was performed using the GSEA-P software available at http://www.broad.mit.edu/gsea/ (Subramanian et al., 2005). This method does not require prior statistical determination of which genes are differentially expressed, but instead evaluates all transcripts expressed above background. For this reason, GSEA provides a very sensitive approach to detecting broad expression differences in functional cellular pathways. Using GSEA, we compared the cell layer predominance of 522 predefined functional groups of transcripts or “gene sets” originally described by Subramanian et al (curated “c2” gene set from MSigDB 1.0) (Subramanian et al., 2005). Input was a list of all genes expressed in at least one region with corresponding signal intensities for every sample. Default program settings were used for analysis, including a minimum gene set size of 15 to exclude very small sets. Gene sets with a nominal p-value < 0.05 and false discovery rate (FDR) < 0.3 were considered significantly different between groups, as suggested by Subramanian et al (Subramanian et al., 2005).
Categorical analysis of differentially expressed genes using eGOn
Categorical differences between different layers were further examined using the web-based application eGOn (Explore Gene ONtology; http://www.genetools.microarray.ntnu.no/common/intro.php) (Beisvag et al., 2006). eGOn automatically categorizes genes into gene ontology classifications under three main GO headings: biological process, molecular function, and cellular compartment, making it an excellent complement to GSEA. Two separate analyses were performed. First, transcripts that were significantly more abundant in the dentate granule cell layer were compared to those more abundant in pyramidal layers. Second, transcripts more abundant in CA1 were compared to those more abundant in CA3. All four lists of transcripts were determined by t-test as described above. Fisher's exact test was used to evaluate the statistical significance of categorical differences between layers. GO categories were considered significantly different from the whole if there was at least a 2-fold difference in abundance between layers and the p-value was < 0.01.
Results
LCM was performed on hippocampi from 10 rats (Fig 1A). All samples (CA1, CA3, and DG) from 7 rats met minimum standards for RNA and microarray hybridization quality. In addition, we obtained reliable data from CA1 and DG from rat number A3, CA1 from rat 6, and CA3 from rat A8. As such, we analyzed 9 CA1 samples and 8 samples each from CA3 and DG.
A total of 5982 genes (59% of transcripts assessed by the microarray) were expressed above background in at least one region of the hippocampus (Table 1). The full dataset is available as Supplementary Table 1. Unsupervised hierarchical clustering based on all expressed transcripts differentiated CA1, CA3, and DG by expression profile alone indicating consistent differences in gene expression among the cell layers (Fig 1B). CA1 and CA3 pyramidal cell layers more closely resembled one another than the dentate granule cell layer. Furthermore, although there was an excellent correlation between samples from the same region across different animals (Fig 1C), the correlation between regions, even from the same animal, was much less robust (Fig 1D). Lower interregional correlation resulted from the combination of a scattered few outlying transcripts exhibiting large differences between layers and a large number of transcripts exhibiting more modest differences.
Table 1.
Gene expression in CA1, CA3, and DG.
| Number of unique probe sets on chip | 10179 | (% of total) |
| Number of probe sets expressed in at least one region | 5982 | 58.8 |
| Number of differentially expressed probe sets | 2838 | 27.9 |
| Number of differentially expressed probe sets (> 2-fold) | 1027 | 10.1 |
| Number of differentially expressed probe sets (> 8-fold) | 110 | 1.1 |
| Number of differentially expressed probe sets (> 32-fold) | 17 | 0.2 |
| Mean difference between highest and lowest region | 2.1 fold | |
| Median difference between highest and lowest region | 1.8 fold | |
| Largest significant difference between highest and lowest region | 337 fold | |
| Smallest significant difference between highest and lowest region | 1.1 fold |
Transcripts called ‘present’ in > 65% of animals in any region were considered expressed. Differential expression was determined by one-way ANOVA with Benjamini-Hochberg correction (FDR < 1%).
Based on anatomy (Fig 1A), these samples are expected to be highly enriched, but not completely homogenous, populations of excitatory pyramidal or granule neurons. Gene expression data revealed the probable inclusion of some inhibitory neurons by detection of GAD1 (GAD67) mRNA in all cell layers (Table 2). Several genes typically associated with glial cells were also expressed in these samples (Table 2). As such, while the samples contain predominantly principal neurons, a portion of the gene expression signal is derived from other cell types.
Table 2.
Genes expressed by minor cell populations in hippocampal cell layers.
| Gene Symbol |
Gene Name | CA1 | CA3 | DG | ANOVA | Cell type |
|---|---|---|---|---|---|---|
| GAD1 | Glutamic acid decarboxylase, 67kDa | 205 | 199 | 232 | NS | GABAergic neurons |
| CD81 | CD 81 antigen | 1553 | 1313 | 1488 | NS | Microglia |
| GFAP | Glial fibrillary acidic protein | 532 | 203 | 631 | SIG | Astrocytes |
| GLUL | Glutamine synthase | 115 | 118 | 130 | NS | Astrocytes |
| SLC1A3 | Glial high affinity glutamate transporter | 148 | 130 | 122 | NS | Astrocytes |
| MBP | Myelin basic protein | 112 | 250 | 141 | NS | Oligodendrocytes |
| MOG | Myelin oligodendrocyte glycoprotein | 46 | 53 | 36 | NS | Oligodendrocytes |
| PLP | Proteolipid protein | 912 | 1399 | 478 | SIG | Oligodendrocytes |
Mean expression levels in different cell layers of selected genes known to be expressed by GABAergic interneurons or glial cells. SIG, significantly different by ANOVA, P < 0.01; NS, not significant.
We determined which transcripts were significantly different among layers using a one-way ANOVA with B-H correction. This analysis revealed a different level of expression between regions for 2838 transcripts (47% of expressed genes). At a false discovery rate (FDR) of 1%, only 29 of these are expected to be false positive differences. The median difference in transcript level between the highest and lowest expressing region was 1.8 fold. Data in Table 1 indicate a broad-based difference in gene expression between CA1, CA3, and DG, supporting the impression obtained from Fig 1. A large population of transcripts is enriched in certain hippocampal cell layers, including over 1,000 that are at least 2-fold different between layers. A very small proportion of transcripts were nearly specific for CA1, CA3, or DG (at least 32-fold different).
While this study was not primarily focused on detecting high-magnitude differences in individual transcripts, examination of transcripts at least 8-fold different among layers was helpful for validation purposes. Sixty-three were found in the Allen Brain Atlas, which is a compilation of serial mouse brain sections stained for nearly 20,000 individual transcripts created by the Allen Institute for Brain Science (Lein et al., 2007). Given species and technical differences, the concordance between the methods was quite high. Our microarray data were qualitatively confirmed by the Atlas for 45 transcripts (71%). Ten transcripts (14%) appeared by visual inspection to be more homogeneous in the Atlas (Table 3). Eight out of 54 (13%) appeared undetectable in the Atlas, suggesting that the microarray platform is somewhat more sensitive than in situ hybridization.
Table 3.
Comparison of array results with the Allen Brain Atlas.
| Gene Symbol | CA1 | CA3 | DG | ABA Pattern |
|---|---|---|---|---|
| GRIK4 | 5 | 174 | 45 | Same |
| KCND3 | 17 | 102 | 152 | Same |
| TRPC6 | 14 | 22 | 130 | Same |
| DCN | 347 | 35 | 25 | Same |
| FOXO1A | 7 | 24 | 56 | Same |
| LPHN2 | 64 | 6 | 3 | Same |
| PRSS23 | 33 | 619 | 35 | Same |
| SOCS2 | 21 | 79 | 8 | Same |
| PTN | 817 | 197 | 86 | Same |
| DDIT4L | 22 | 35 | 489 | Same |
| TLE3 | 423 | 174 | 46 | Same |
| EPHA5 | 366 | 1484 | 175 | Same |
| MEF2C | 98 | 57 | 510 | Same |
| INHBB | 150 | 23 | 3 | Same |
| RASL11B | 774 | 295 | 40 | Same |
| LPL | 508 | 863 | 21 | Same |
| TIAM1 | 5 | 8 | 233 | Same |
| CALB1 | 258 | 39 | 1099 | Same |
| SULF2 | 275 | 963 | 84 | Same |
| NEGR1 | 141 | 250 | 1667 | Same |
| DUSP6 | 815 | 416 | 31 | Same |
| SIPA1L2 | 14 | 15 | 133 | Same |
| KCNN2 | 240 | 172 | 23 | Same |
| NNAT | 90 | 2465 | 233 | Same |
| SV2B | 231 | 1346 | 62 | Same |
| PLAGL1 | 1 | 72 | 27 | Same |
| PKIG | 126 | 15 | 336 | Same |
| KLK8 | 144 | 51 | 1 | Same |
| PCBD1 | 5 | 72 | 16 | Same |
| BID3 | 5 | 24 | 84 | Same |
| SLC6A7 | 47 | 156 | 19 | Same |
| GRM2 | 20 | 23 | 240 | Same |
| NTF3 | 26 | 57 | 379 | Same |
| GPC3 | 2 | 2 | 120 | Same |
| PCP4 | 71 | 725 | 2085 | Same |
| KCNA1 | 24 | 390 | 243 | Same |
| CCK | 1099 | 340 | 127 | Same |
| PERP_PREDICTED | 3 | 3 | 86 | Same |
| AUTS2_PREDICTED | 79 | 146 | 755 | Same |
| FZD7_PREDICTED | 99 | 21 | 8 | Same |
| TP53I11_PREDICTED | 26 | 18 | 183 | Same |
| MAN1A_PREDICTED | 898 | 412 | 31 | Same |
| RREB1_PREDICTED | 25 | 21 | 322 | Same |
| LATS2_PREDICTED | 470 | 1052 | 89 | Same |
| SEMA6D_PREDICTED | 88 | 59 | 524 | Same |
| NOV | 461 | 179 | 34 | CA1>>CA3,DG |
| KHDRBS2 | 14 | 8 | 92 | DG,CA1>>CA3 |
| RGS10 | 210 | 152 | 2085 | DG,CA3>CA1 |
| GRP | 14 | 98 | 12 | Diffuse |
| SLC17A6 | 407 | 159 | 28 | Diffuse |
| JMJD3_PREDICTED | 16 | 33 | 134 | Diffuse |
| S100A10 | 674 | 133 | 33 | Diffuse |
| GDF10 | 4 | 17 | 89 | Diffuse |
| JUN | 31 | 134 | 299 | Diffuse |
| HCRTR2 | 9 | 73 | 8 | Diffuse |
| ARG1 | 29 | 28 | 409 | n.d. |
| NELL1 | 917 | 625 | 108 | n.d. |
| DSC2 | 2 | 21 | 21 | n.d. |
| CTSK | 190 | 33 | 3 | n.d. |
| CYP27A1 | 20 | 274 | 14 | n.d. |
| CST6 | 24 | 399 | 74 | n.d. |
| PLSCR1 | 4 | 41 | 90 | n.d. |
| PKIB | 5 | 37 | 67 | n.d. |
Transcripts with an 8-fold difference between two hippocampal regions were visually compared against in situ hybridization pictures in the Allen Brain Atlas. Relative expression levels were confirmed for the majority of transcripts. n.d., not detected.
A direct comparison of dentate granule to pyramidal layers (CA1 and CA3) by t-test supported the results from the ANOVA, showing that 1683 transcripts (28%) were different between dentate granule neurons and pyramidal neurons. A separate analysis specifically of CA1 versus CA3 pyramidal neuron layers revealed that 19% of expressed transcripts were different by t-test even between those two similar populations.
1027 genes that were differentially expressed and at least 2-fold different between the highest and lowest layer (Table 1) underwent unsupervised hierarchical clustering using GenePattern 2.0. Similar to the clustering results obtained using every expressed gene (Fig 1B), clustering using transcripts with high magnitude differences between hippocampal cell layers consistently grouped samples from the same cell layer together, with pyramidal transcriptomes being more alike than their counterparts from the dentate (Fig 2). Every permutation of anatomical gene expression pattern (e.g., DG>CA1>CA3; CA1=CA3>DG; etc.) was represented by numerous transcripts in the rat hippocampus.
Figure 2. Hierarchical clustering of differentially expressed genes.
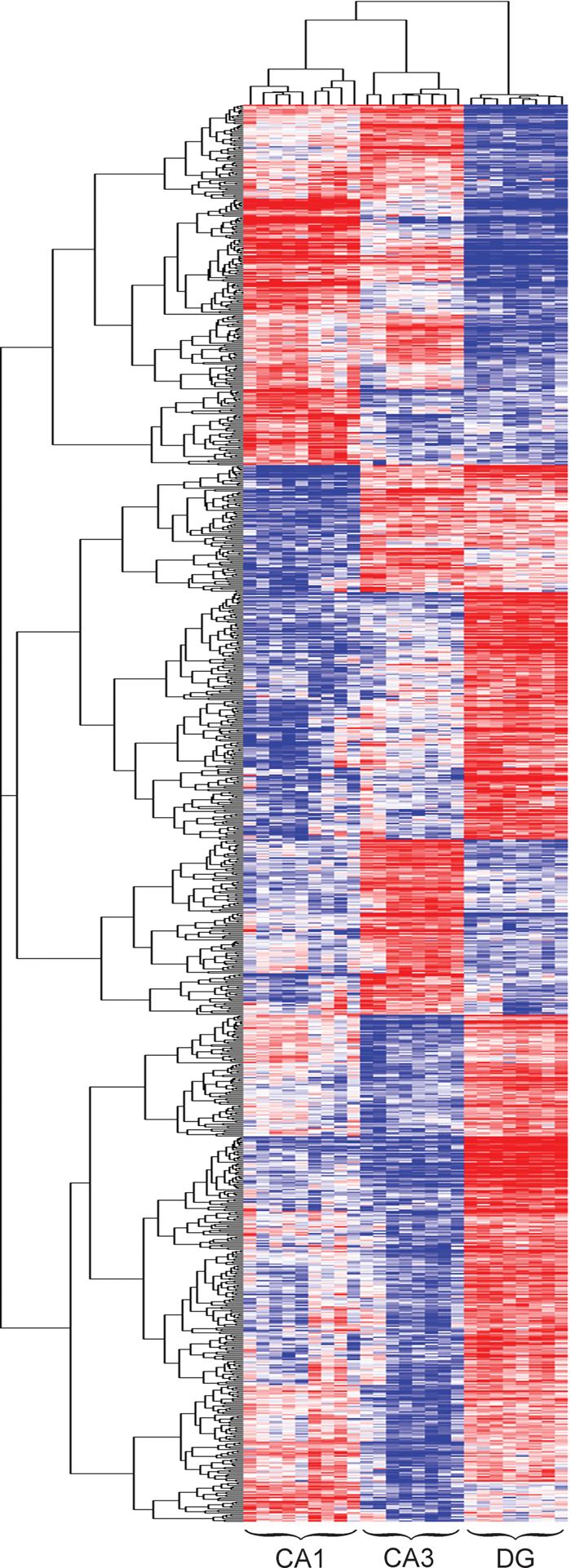
Each cell in the heat map represents the expression level of a gene (row) in a single sample (column). Note that samples cluster together based on gene expression profile and that genes cluster together into groups based on similar regional expression profiles. Every potential pattern of differential expression between the major hipocampal cell layers is apparent.
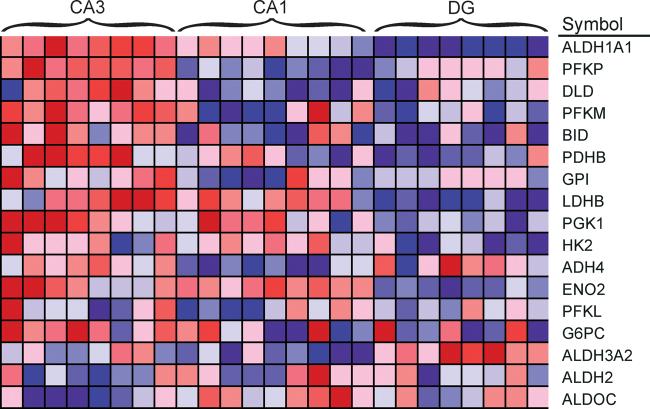
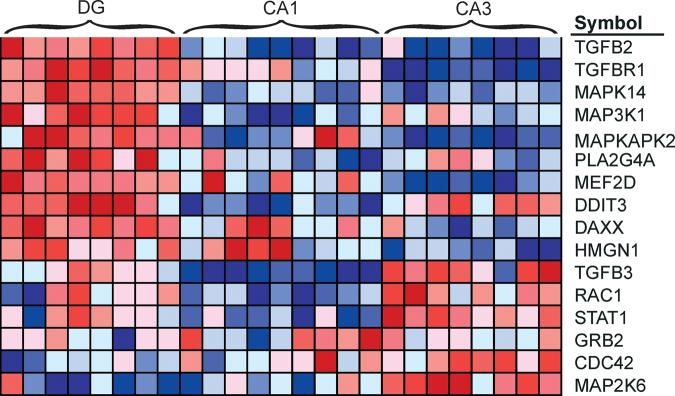
Gene set enrichment analysis (GSEA) based on all expressed genes indicated that differences in gene expression profile between the three cell layers are based in part on concerted differences in a limited number of cellular processes (Table 4). The most prominent enrichments were in gene sets related to glucose metabolism in CA3 and an abundance of transcripts related to MAPK signaling and transcription factors in DG. Consistent CA3 enrichment of transcripts involved in glucose metabolism is apparent in a heatmap depicting relative expression of transcripts across cell layers (Fig 3). A similar heatmap of the p38 MAP kinase pathway shows not only an overrepresentation of the pathway in the dentate granule layer, but also a qualitative difference between the two pyramidal populations (Fig 4).
Table 4.
Gene sets enriched in hippocampal cell layers.
| GENE SET NAME | SIZE | CORE | p-value | FDR | CLASS |
|---|---|---|---|---|---|
| Enriched in CA1 | |||||
| ANDROGEN_GENES_FROM_NETAFFX | 15 | 8 | 0.006 | 0.11 | |
| RAR_UP | 15 | 5 | 0.018 | 0.06 | |
| VEGFPATHWAY | 15 | 5 | 0.017 | 0.08 | |
| AR_ORTHOS_MAPPED_TO_U133_VIA_NETAFFX | 17 | 4 | 0.035 | 0.14 | |
| AR_MOUSE | 17 | 4 | 0.035 | 0.11 | |
| Enriched in CA3 | |||||
| CR_PROTEIN_MOD | 45 | 11 | 0.002 | 0.24 | |
| ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | 15 | 6 | 0.018 | 0.16 | |
| PPARAPATHWAY | 21 | 5 | 0.004 | 0.21 | Met |
| KRAS_TOP100_KNOCKDOWN | 23 | 4 | 0.023 | 0.17 | |
| INSULIN_2F_UP | 71 | 21 | 0.000 | 0.14 | Met |
| SIG_INSULINRECEPTORPATHWAYINCARDIACMYOCYTES | 23 | 7 | 0.014 | 0.19 | Met |
| MAP00010_GLYCOLYSIS_GLUCONEOGENESIS | 17 | 14 | 0.039 | 0.21 | Met |
| Enriched in DG | |||||
| HOXA9_DOWN | 17 | 3 | 0.002 | 0.09 | Tr |
| CR_CELL_CYCLE | 24 | 6 | 0.004 | 0.19 | |
| P38MAPKPATHWAY | 16 | 9 | 0.015 | 0.20 | MAP |
| HUMAN_CD34_ENRICHED_TF_JP | 41 | 22 | 0.002 | 0.15 | Tr |
| MAPKPATHWAY | 34 | 17 | 0.031 | 0.26 | MAP |
| DNA_DAMAGE_SIGNALLING | 21 | 4 | 0.039 | 0.23 | Tr |
GSEA of hippocampal cell layers. Geneset name is from MSigDB 1.0. Size is the number of genes from the geneset that are expressed in our dataset. Core is the number of genes in the ‘leading edge’ of the gene set; a higher ratio of core to total genes indicates a more robust enrichment. P-value and False Discovery Rate are measures of the significance of the enrichment in one region as compared to the other two. Seceral broad classes of gene sets were prominently represented: Transcription and MAPK signaling in DG and Metabolism of glucose in CA3.
Figure 3. Higher expression of glycolytic genes in CA3 pyramidal cells.
Each cell in the heatmap represents the expression level of a gene (row) in a single sample (column). N=8 for both DG and CA3 samples and N=9 for CA1 samples. Deep red reflects higher expression of the transcript, whereas deep blue represents lower expression.
Figure 4. Higher expression of p38 MAPK pathway genes in dentate granule cells.
Each cell in the heatmap represents the expression level of a gene (row) in a single sample (column). N=8 for both DG and CA3 samples and N=9 for CA1 samples. Deep red reflects higher expression of the transcript, whereas deep blue represents lower expression. Note the primary distinction between DG and pyramidal neurons, but also quantitative differences between CA1 and CA3.
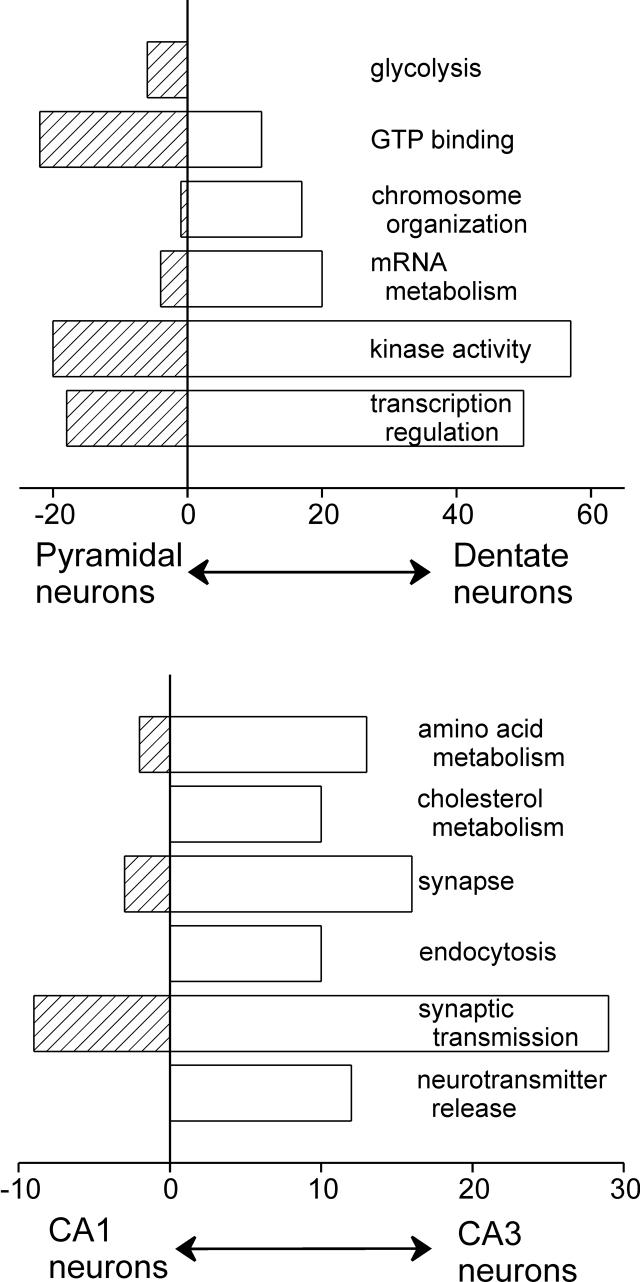
To complement the GSEA and further characterize the broad transcript differences between the hippocampal cell populations, direct comparison was made between dentate granule cell and pyramidal neuron layers and then between CA1 and CA3. Figure 5 summarizes categorical analysis of differentially expressed genes completed using eGOn. Transcripts related to transcription, DNA packaging, and kinase activity are consistently higher in DG. In particular, the p38 MAP kinase pathway is overrepresented. All categories were concordant with the results from GSEA. Glycolytic transcripts were significantly enriched in pyramidal neurons, as were transcripts encoding GTP-binding proteins.
Figure 5. Concerted and categorical differences in transcript abundance between hippocampal cell layers.
A. GO classifications different between dentate and pyramidal neuron layers (CA1 and CA3) B. GO classifications different between CA1 and CA3. All categories are significantly different with p < 0.01 by Fisher's exact test. X-axis labels represent numbers of transcripts higher in each layer.
The transcriptional differences between CA1 and CA3 pyramidal neuron layers are also broad-based. In particular, transcripts related to synaptic transmission and neurotransmitter release are dramatically overrepresented in CA3. Mirroring results from GSEA, no large categories were enriched in the CA1 layer.
Seventeen transcripts nearly specific for CA1, CA3, or DG, as defined by at least a 32-fold difference between regions, are presented in Table 5. Several transcripts on this list have been previously described to be different between the cell layers (Chen et al., 1995; Datson et al., 2004; Lein et al., 2004; Paradis et al., 2004; Werner et al., 1991), but most are novel. The consistency and magnitude of specificity is reflected in the representative graphs in Fig 6.
Table 5.
Transcripts highly specific tor certain cell layers.
| Accession | Gene Name | CA1 | CA3 | DG |
|---|---|---|---|---|
| H31456 | EST | 2438 | 302 | 43 |
| AI408583 | SIMILAR TO BM150J22.1 (NOVEL PROTEIN (ORTHOLOG OF HUMAN C22ORF1)) (PREDICTED) |
2055 | 94 | 31 |
| AA943310 | SIMILAR TO OCIA DOMAIN CONTAINING 2 | 1189 | 559 | 37 |
| M72711 | POU DOMAIN, CLASS 3, TRANSCRIPTION FACTOR 1 | 933 | 41 | 11 |
| XM_230449 | MEIS1, MYELOID ECOTROPIC VIRAL INTEGRATION SITE 1 HOMOLOG 2 (PREDICTED) |
569 | 455 | 2 |
| AI009639 | LIPOPROTEIN LIPASE | 508 | 863 | 21 |
| BF413126 | EST | 318 | 9 | 2 |
| AW435360 | CATHEPSIN K | 190 | 33 | 3 |
| CF112064 | INHIBIN BETA-B | 150 | 23 | 3 |
| BI282567 | KALLIKREIN 8 (NEUROPSIN/OVASIN) | 144 | 51 | 1 |
| CK845349 | PLEIOMORPHIC ADENOMA GENE-LIKE 1 | 1 | 72 | 27 |
| NM_012572 | GLUTAMATE RECEPTOR, IONOTROPIC, KAINATE 4 | 5 | 174 | 45 |
| BE099933 | GLYPICAN 3 | 2 | 2 | 120 |
| AA892022 | LOC363020 (PREDICTED) | 5 | 247 | 158 |
| BM389265 | T-CELL LYMPHOMA INVASION AND METASTASIS 1 | 5 | 8 | 233 |
| AW918391 | SIMILAR TO RIKEN CDNA 6330406115 (PREDICTED) | 15 | 215 | 608 |
| CK840869 | EST | 55 | 50 | 1755 |
Significantly different transcripts (ANOVA p < 0.05 with Benjamini-Hochberg correction) with at least a 32-fold difference between two hippocampal cell layers. Values represent mean signal intensity across all animals for each region.
Figure 6. Transcripts highly specific to individual hippocampal cell layers.
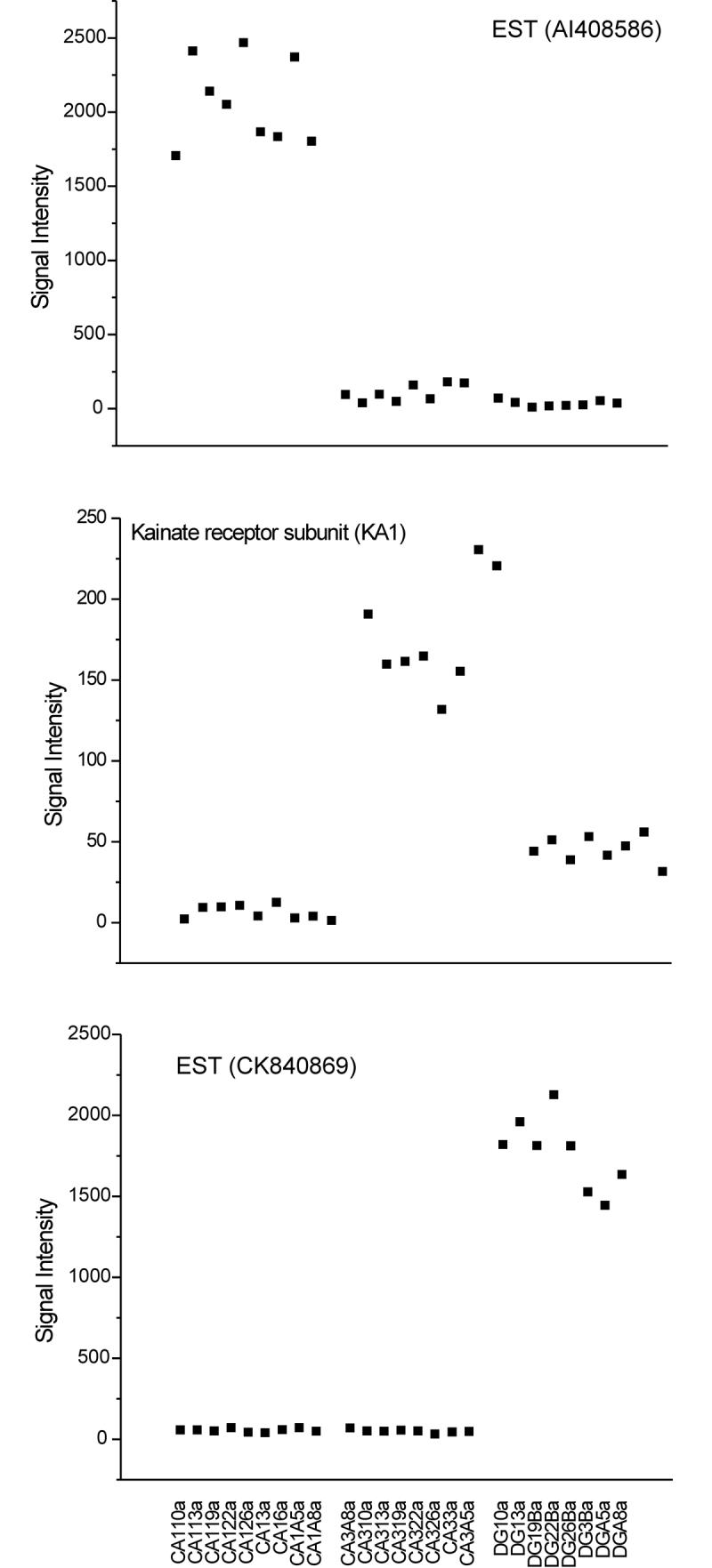
A. Example of a transcript specific for CA1. B. Example of a transcript enriched in CA3. C. Example of a transcript specific for DG. Transcript level is strikingly different between the cell layers and consistent within a given layer.
Discussion
Understanding the normal transcriptional neuroanatomy of the brain is vital to generate deeper insight into CNS function in both normal and disease states. The hippocampus is an ideal structure in which to begin investigations due to its well-defined anatomy and extensively-studied physiology and pathophysiology. Using approaches for data acquisition and analysis that generate results more sophisticated than the simple distribution of individual transcripts, we have discovered a striking disparity in gene expression among hippocampal principal cell layers that is evident in the large proportion of individual genes that are expressed differently between the cell layers. More than 45% of genes expressed in at least one region are differentially expressed, and greater than 17% show at least 2-fold difference between regions. Furthermore, categorical analysis reveals that these differences are concerted in nature and encompass such fundamental areas of neuronal function as cell signaling, metabolism, transcriptional regulation, and neurotransmission. These data suggest that unique physiological characteristics of major cell layers, such as neuronal activity, neuronal plasticity, and vulnerability to neurodegeneration are reflected in, and likely caused by, substantial transcriptional heterogeneity of the hippocampus.
Previous expression profiling experiments of the hippocampus have not reported the same level of differential expression between principal cell layers (Datson et al., 2004; Lein et al., 2004; Zhao et al., 2001). Datson et al. (2004) reported a differential expression rate between dentate and CA3 of about 17%, although that dataset was limited because it was derived from only one rat. However, the seeming disparity between our data and previous reports is most likely artificial, since prior analyses have highlighted high magnitude differences between the regions as opposed to taking into account the breadth of the observed differences. Comparison of our results to the Allen Brain Atlas, a library of nearly 20,000 in situ hybridization probes in the mouse, revealed good agreement between the two methods and was thus beneficial for validating both sets of results (Lein et al., 2007). However, it should be stressed that as opposed to earlier hippocampal expression profiling efforts, our analysis is primarily centered on evaluating cellular pathways, instead of individual genes, and that analysis of expression data in this manner has been shown to be more robust than the more traditional method of ‘validating’ individual genes of interest using other techniques that are less sensitive, such as real-time PCR, Northern blotting, or in situ hybridization. Pathways analysis dramatically lowers false positive findings, increases power to detect true differences, and eliminates bias associated with arbitrarily highlighting individual transcript differences (Greene, 2006; Subramanian et al., 2005; Toronen, 2004; Ye and Eskin, 2007). Since these results are dependent on accurate classification of genes based on function, it is important that comparison of our dataset to both a manually curated group of gene sets (GSEA) and the Gene Ontology hierarchy (eGOn) gave qualitatively similar findings.
Due to the LCM technique used for isolation of cell layers, the gene expression signal from these samples is derived primarily from principal excitatory cells (CA1 and CA3 pyramidal and dentate granule neurons). However, some inhibitory GABAergic interneurons were likely collected. GAD1 expression in the samples would tend to support that idea, but GAD1 is also expressed at low levels in DG neurons and processes (Sloviter et al., 1996). Also relevant are transcript contributions from minor populations of glial cells interwoven with principal neurons in the three layers, particularly astrocytes and oligodendrocytes (Borges et al., 2006; Borges et al., 2007; Jorgensen et al., 1993; Ong and Levine, 1999; Shapiro et al., 2008). As such, while these are highly enriched samples of principal neurons, they are not homogeneous, and the results should be interpreted with that in mind.
The results indicate that the dentate gyrus is substantially different from CA1 or CA3 pyramidal cell layers with regard to basal gene expression in several fundamental cellular pathways. For example, transcripts encoding members of the MAP kinase cascade are particularly prominent in the dentate. More specifically, transcripts involved in transforming growth factor beta (TGF-β) signaling through the p38 MAP kinase pathway account for a considerable fraction of that expression profile. Signaling via TGF-β has been implicated in the regulation of apoptosis, neurogenesis, and neuronal survival (Bravo et al., 2006; Lu et al., 2005; Zhu et al., 2004; Zhu et al., 2002), and these data suggest the TGF-β pathway is an important part of the basal physiology of the dentate gyrus and its response to injury or insult. In addition, several members of this pathway, including TGF-β, MAPK, and MAPKAPK2, have previously been described to be induced in dentate granule cells by seizure activity (Garrido et al., 1998; Kim et al., 2002; Kodama et al., 2005; Vician et al., 2004).
Concurrently, these data show that transcripts encoding proteins with transcriptional regulator activity are nearly three times more prominent in the dentate than in the pyramidal neuron layers. Relative abundance of transcripts related to mRNA metabolism and chromosome organization is even more lopsided toward the dentate. Transcription has been previously mentioned as a category that may be different between hippocampal layers, but this had not been previously demonstrated statistically (Lein et al., 2004). In conjunction with the abundance of MAP kinase signaling transcripts, a wealth of transcription factor mRNAs suggests that dentate granule cells at baseline are poised to respond transcriptionally to external insults and stimuli. This is particularly interesting given our recent results which indicate that hippocampal neuroprotection following seizure preconditioning is associated primarily with a dramatic alteration in DG gene expression (Borges et al., 2007).
The hippocampal pyramidal neuron layers in general and CA3 in particular express an abundance of transcripts related to glucose metabolism, confirming hints from other profiling studies of the hippocampus (Datson et al., 2004; Lein et al., 2004). This is interesting given the exquisite vulnerability of pyramidal neurons to damage caused by extended seizures and ischemia (Pulsinelli et al., 1982; Schreiber and Baudry, 1995), both of which place dramatic metabolic stress on neurons. Even at baseline, high spontaneous firing rates in pyramidal neurons result in high metabolic demand (Biscoe and Duchen, 1985), and metabolic activation is tightly associated with states of high activity frequently seen in hippocampal pyramidal neurons (Csicsvari et al., 2000; Csicsvari et al., 2003; Huchzermeyer et al., 2008). We and others have previously demonstrated that an abundance of energy metabolism transcripts is a marker for high susceptibility to metabolic insults in midbrain dopamine neurons, suggesting that high metabolic activity may be a generalizable marker for neuronal susceptibility to seizures, ischemia, and other neurodegenerative insults (Chung et al., 2005; Greene et al., 2005).
CA1 and CA3 pyramidal neurons differ anatomically and biophysically from each other as well as from dentate granule cells (Carnevale et al., 1997). This is mirrored by differential expression of numerous transcripts involving cell shape and ion channels (Supplementary Table 1). For example, the α1C and α1B Ca2+ channel subunits are more prominent in CA3 than CA1, as are the KCNMB4 and KCNA1 potassium channel transcripts, and KA1, GluR3 and mGluR1 glutamate receptor transcripts. As mentioned above, the extensive differences in gene expression between CA1 and CA3 observed in this study were somewhat unexpected since both layers consist mainly of pyramidal neurons, which use glutamate as their primary neurotransmitter, and are derived from similar precursors in the ammonic neuroepithelium (Altman and Bayer, 1990). Nevertheless, nearly 20% of transcripts were expressed differently between them, with nearly 400 different by a factor more than two. Obvious differences in the transcriptional profiles of hippocampal pyramidal neurons have recently been described (Lein et al., 2004); however, the current data indicate that expression differences are much more pervasive than previously suspected.
The most striking area of distinction between the CA1 and CA3 neuron layers was that transcripts encoding proteins involved in synaptic function and transmitter release were more abundant in CA3. This difference likely contributes to the extensive synaptic remodeling capacity inherent in the Schaffer-CA1 synapse insofar as the plastic potential of the presynaptic terminals is supported by possession of the machinery necessary to induce such synaptic changes. For example, numerous transcripts encoding synaptic vesicle proteins or proteins involved in exocytosis are more highly expressed in CA3 than CA1 (e.g., CPLX1, CPLX2, NSF, SYN2, SV2A, SNAP25, STX1A, SYNJ1, CADPS). Neurexin 1 (NRXN1) and neurotrophin 3 (NTF3), which promote synapse stabilization, are also overexpressed in CA3 relative to CA1. High levels of synapse-related transcripts are likely a marker for the high level of synaptic neuronal activity observed in CA3 pyramidal neurons and correlate with their vulnerability to seizure-induced damage.
In conclusion, this study provides new and complex anatomical knowledge about the organization of the rodent hippocampus and supports a novel framework for interpreting regional differences in hippocampal neuron function and vulnerability to insults.
Supplementary Material
Acknowledgements
This work was supported by NIH K08NS048858 (JGG) and Citizens United for Epilepsy (KB). The authors wish to thank Renee Shaw for expert technical assistance.
References
- Altman J, Bayer SA. Mosaic organization of the hippocampal neuroepithelium and the multiple germinal sources of dentate granule cells. J Comp Neurol. 1990;301(3):325–42. doi: 10.1002/cne.903010302. [DOI] [PubMed] [Google Scholar]
- Beisvag V, Junge FK, Bergum H, Jolsum L, Lydersen S, Gunther CC, Ramampiaro H, Langaas M, Sandvik AK, Laegreid A. GeneTools--application for functional annotation and statistical hypothesis testing. BMC Bioinformatics. 2006;7:470. doi: 10.1186/1471-2105-7-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscoe TJ, Duchen MR. An intracellular study of dentate, CA1 and CA3 neurones in the mouse hippocampal slice. Q J Exp Physiol. 1985;70(2):189–202. doi: 10.1113/expphysiol.1985.sp002902. [DOI] [PubMed] [Google Scholar]
- Borges K, Gearing M, McDermott DL, Smith AB, Almonte AG, Wainer BH, Dingledine R. Neuronal and glial pathological changes during epileptogenesis in the mouse pilocarpine model. Exp Neurol. 2003;182(1):21–34. doi: 10.1016/s0014-4886(03)00086-4. [DOI] [PubMed] [Google Scholar]
- Borges K, McDermott D, Irier H, Smith Y, Dingledine R. Degeneration and proliferation of astrocytes in the mouse dentate gyrus after pilocarpine-induced status epilepticus. Exp Neurol. 2006;201(2):416–27. doi: 10.1016/j.expneurol.2006.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges K, Shaw R, Dingledine R. Gene expression changes after seizure preconditioning in the three major hippocampal cell layers. Neurobiol Dis. 2007;26(1):66–77. doi: 10.1016/j.nbd.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo JA, Parra CS, Arancibia S, Andres S, Morales P, Herrera-Marschitz M, Herrera L, Lara HE, Fiedler JL. Adrenalectomy promotes a permanent decrease of plasma corticoid levels and a transient increase of apoptosis and the expression of Transforming Growth Factor beta1 (TGF-beta1) in hippocampus: effect of a TGF-beta1 oligoantisense. BMC Neurosci. 2006;7:40. doi: 10.1186/1471-2202-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale NT, Tsai KY, Claiborne BJ, Brown TH. Comparative electrotonic analysis of three classes of rat hippocampal neurons. J Neurophysiol. 1997;78(2):703–20. doi: 10.1152/jn.1997.78.2.703. [DOI] [PubMed] [Google Scholar]
- Chen ZL, Yoshida S, Kato K, Momota Y, Suzuki J, Tanaka T, Ito J, Nishino H, Aimoto S, Kiyama H. Expression and activity-dependent changes of a novel limbic-serine protease gene in the hippocampus. J Neurosci. 1995;15(7 Pt 2):5088–97. doi: 10.1523/JNEUROSCI.15-07-05088.1995. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Seo H, Sonntag KC, Brooks A, Lin L, Isacson O. Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum Mol Genet. 2005;14(13):1709–25. doi: 10.1093/hmg/ddi178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csicsvari J, Hirase H, Mamiya A, Buzsaki G. Ensemble patterns of hippocampal CA3-CA1 neurons during sharp wave-associated population events. Neuron. 2000;28(2):585–94. doi: 10.1016/s0896-6273(00)00135-5. [DOI] [PubMed] [Google Scholar]
- Csicsvari J, Jamieson B, Wise KD, Buzsaki G. Mechanisms of gamma oscillations in the hippocampus of the behaving rat. Neuron. 2003;37(2):311–22. doi: 10.1016/s0896-6273(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datson NA, Meijer L, Steenbergen PJ, Morsink MC, van der Laan S, Meijer OC, de Kloet ER. Expression profiling in laser-microdissected hippocampal subregions in rat brain reveals large subregion-specific differences in expression. Eur J Neurosci. 2004;20(10):2541–54. doi: 10.1111/j.1460-9568.2004.03738.x. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID:Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274(5289):998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Garrido YC, Sanabria ER, Funke MG, Cavalheiro EA, Naffah-Mazzacoratti MG. Mitogen-activated protein kinase is increased in the limbic structures of the rat brain during the early stages of status epilepticus. Brain Res Bull. 1998;47(3):223–9. doi: 10.1016/s0361-9230(98)00075-6. [DOI] [PubMed] [Google Scholar]
- Goldsworthy SM, Stockton PS, Trempus CS, Foley JF, Maronpot RR. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog. 1999;25(2):86–91. [PubMed] [Google Scholar]
- Greene JG. Gene expression profiles of brain dopamine neurons and relevance to neuropsychiatric disease. J Physiol. 2006;575(Pt 2):411–6. doi: 10.1113/jphysiol.2006.112599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JG, Dingledine R, Greenamyre JT. Gene expression profiling of rat midbrain dopamine neurons: implications for selective vulnerability in parkinsonism. Neurobiol Dis. 2005;18(1):19–31. doi: 10.1016/j.nbd.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Huchzermeyer C, Albus K, Gabriel HJ, Otahal J, Taubenberger N, Heinemann U, Kovacs R, Kann O. Gamma oscillations and spontaneous network activity in the hippocampus are highly sensitive to decreases in pO2 and concomitant changes in mitochondrial redox state. J Neurosci. 2008;28(5):1153–62. doi: 10.1523/JNEUROSCI.4105-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen MB, Finsen BR, Jensen MB, Castellano B, Diemer NH, Zimmer J. Microglial and astroglial reactions to ischemic and kainic acid-induced lesions of the adult rat hippocampus. Exp Neurol. 1993;120(1):70–88. doi: 10.1006/exnr.1993.1041. [DOI] [PubMed] [Google Scholar]
- Kim HC, Bing G, Kim SJ, Jhoo WK, Shin EJ, Bok Wie M, Ko KH, Kim WK, Flanders KC, Choi SG. Kainate treatment alters TGF-beta3 gene expression in the rat hippocampus. Brain Res Mol Brain Res. 2002;108(1-2):60–70. doi: 10.1016/s0169-328x(02)00514-4. others. [DOI] [PubMed] [Google Scholar]
- Kodama M, Russell DS, Duman RS. Electroconvulsive seizures increase the expression of MAP kinase phosphatases in limbic regions of rat brain. Neuropsychopharmacology. 2005;30(2):360–71. doi: 10.1038/sj.npp.1300588. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–76. doi: 10.1038/nature05453. others. [DOI] [PubMed] [Google Scholar]
- Lein ES, Zhao X, Gage FH. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci. 2004;24(15):3879–89. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wu Y, Sousa N, Almeida OF. SMAD pathway mediation of BDNF and TGF beta 2 regulation of proliferation and differentiation of hippocampal granule neurons. Development. 2005;132(14):3231–42. doi: 10.1242/dev.01893. [DOI] [PubMed] [Google Scholar]
- Mathern GW, Babb TL, Vickrey BG, Melendez M, Pretorius JK. The clinical-pathogenic mechanisms of hippocampal neuron loss and surgical outcomes in temporal lobe epilepsy. Brain. 1995;118(Pt 1):105–18. doi: 10.1093/brain/118.1.105. [DOI] [PubMed] [Google Scholar]
- Maxwell WL, Dhillon K, Harper L, Espin J, MacIntosh TK, Smith DH, Graham DI. There is differential loss of pyramidal cells from the human hippocampus with survival after blunt head injury. J Neuropathol Exp Neurol. 2003;62(3):272–9. doi: 10.1093/jnen/62.3.272. [DOI] [PubMed] [Google Scholar]
- Ong WY, Levine JM. A light and electron microscopic study of NG2 chondroitin sulfate proteoglycan-positive oligodendrocyte precursor cells in the normal and kainate-lesioned rat hippocampus. Neuroscience. 1999;92(1):83–95. doi: 10.1016/s0306-4522(98)00751-9. [DOI] [PubMed] [Google Scholar]
- Ordy JM, Wengenack TM, Bialobok P, Coleman PD, Rodier P, Baggs RB, Dunlap WP, Kates B. Selective vulnerability and early progression of hippocampal CA1 pyramidal cell degeneration and GFAP-positive astrocyte reactivity in the rat four-vessel occlusion model of transient global ischemia. Exp Neurol. 1993;119(1):128–39. doi: 10.1006/exnr.1993.1014. [DOI] [PubMed] [Google Scholar]
- Paradis E, Clavel S, Julien P, Murthy MR, de Bilbao F, Arsenijevic D, Giannakopoulos P, Vallet P, Richard D. Lipoprotein lipase and endothelial lipase expression in mouse brain: regional distribution and selective induction following kainic acid-induced lesion and focal cerebral ischemia. Neurobiol Dis. 2004;15(2):312–25. doi: 10.1016/j.nbd.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Pulsinelli WA, Brierley JB, Plum F. Temporal profile of neuronal damage in a model of transient forebrain ischemia. Ann Neurol. 1982;11(5):491–8. doi: 10.1002/ana.410110509. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Schreiber SS, Baudry M. Selective neuronal vulnerability in the hippocampus--a role for gene expression? Trends Neurosci. 1995;18(10):446–51. doi: 10.1016/0166-2236(95)94495-q. [DOI] [PubMed] [Google Scholar]
- Shapiro LA, Wang L, Ribak CE. Rapid astrocyte and microglial activation following pilocarpine-induced seizures in rats. Epilepsia. 2008;49(Suppl 2):33–41. doi: 10.1111/j.1528-1167.2008.01491.x. [DOI] [PubMed] [Google Scholar]
- Sloviter RS, Dichter MA, Rachinsky TL, Dean E, Goodman JH, Sollas AL, Martin DL. Basal expression and induction of glutamate decarboxylase and GABA in excitatory granule cells of the rat and monkey hippocampal dentate gyrus. J Comp Neurol. 1996;373(4):593–618. doi: 10.1002/(SICI)1096-9861(19960930)373:4<593::AID-CNE8>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toronen P. Selection of informative clusters from hierarchical cluster tree with gene classes. BMC Bioinformatics. 2004;5:32. doi: 10.1186/1471-2105-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vician LJ, Xu G, Liu W, Feldman JD, Machado HB, Herschman HR. MAPKAP kinase-2 is a primary response gene induced by depolarization in PC12 cells and in brain. J Neurosci Res. 2004;78(3):315–28. doi: 10.1002/jnr.20251. [DOI] [PubMed] [Google Scholar]
- Werner P, Voigt M, Keinanen K, Wisden W, Seeburg PH. Cloning of a putative high-affinity kainate receptor expressed predominantly in hippocampal CA3 cells. Nature. 1991;351(6329):742–4. doi: 10.1038/351742a0. [DOI] [PubMed] [Google Scholar]
- Ye C, Eskin E. Discovering tightly regulated and differentially expressed gene sets in whole genome expression data. Bioinformatics. 2007;23(2):e84–90. doi: 10.1093/bioinformatics/btl315. [DOI] [PubMed] [Google Scholar]
- Zhao X, Lein ES, He A, Smith SC, Aston C, Gage FH. Transcriptional profiling reveals strict boundaries between hippocampal subregions. J Comp Neurol. 2001;441(3):187–96. doi: 10.1002/cne.1406. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Culmsee C, Klumpp S, Krieglstein J. Neuroprotection by transforming growth factor-beta1 involves activation of nuclear factor-kappaB through phosphatidylinositol-3-OH kinase/Akt and mitogen-activated protein kinase-extracellular-signal regulated kinase1,2 signaling pathways. Neuroscience. 2004;123(4):897–906. doi: 10.1016/j.neuroscience.2003.10.037. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Yang GY, Ahlemeyer B, Pang L, Che XM, Culmsee C, Klumpp S, Krieglstein J. Transforming growth factor-beta 1 increases bad phosphorylation and protects neurons against damage. J Neurosci. 2002;22(10):3898–909. doi: 10.1523/JNEUROSCI.22-10-03898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.