Summary
The relationship between cell growth (cell mass increase over time) and cell division is poorly understood in animal stem cells. Recent studies in several Drosophila stem cell types have provided the tools to interrogate this relationship. In several cases (brat, mei-P26, pros, bam, lethal giant larvae, polo), mutations have been defined that trigger tumorous overproliferation of progenitor cells and reveal how unrestricted self-renewing capacity is controlled. Moreover, microRNAs have been discovered as essential regulators of stem cell division rate and identity, suggesting that stem cell self-renewal depends on protein translational control. Biosynthetic capacity has also been found to be limiting for stem cell division rates. Finally, asymmetric cell division can impose dominant differentiation signals in a stem cell’s daughter, and this can inhibit the stem cell specific proliferation signature and lock in cell cycle exit.
Introduction
Invertebrate research has been instrumental in formulating basic concepts of stem cell biology. Indeed the term “Stammzelle”, german for stem cell, was coined during experimentation with Crustacean primordial germ cells by Valentin Haecker around 1890. ”Stem cell” is still today an operational definition, defined by continued self-renewal and the potential to produce daughter cells that can commit to lineage specific differentiation. The intense current interest in understanding and manipulating “stemness”, has focused largely on cell identity – i.e. pluripotency- but the significance of growth potential for self-renewal is beginning to be appreciated.
Stem cell growth maintains constant stem cell size over many rounds of self-renewing division. Protein biosynthesis is one critical factor that is limiting for growth rate. Although it is generally the case that cell growth can become limiting for rates of cell division in proliferating cells, it is not clear how growth and cell cycle progression are coordinated (for review see: [1]). In Drosophila, boosting the speed of the cell division cycle by expressing the limiting core cell cycle regulators for G2/M (stg/Cdc25) and G1/S progression (cyclin E or E2f1/Dp) does not activate growth in imaginal disc, larval progenitors of the adult ectoderm. Conversely, the important question of whether growth is sufficient to drive cell cycle progression has not been definitively addressed in most stem cell types, including those found in Drosophila. A growth controlled stem cell cycle could elegantly restrict self-renewing proliferation to undifferentiated stem cells while avoiding division in differentiating daughters. Growth activation could also help to overcome stem cell quiescence, a phase with reduced growth and G1 DNA content that is thought to assure long-term self-renewing capacity. Because of the technical effort to purify stem cells from tissues and derive quantitative parameters of cell growth, little is known about the significance of growth for the stem cell cycle in multicellular systems in vivo. Here, we discuss recent studies that begin to reveal the reciprocal regulation between growth, cell cycle and cellular polarity in Drosophila stem cells.
Proliferation control in Drosophila stem cells
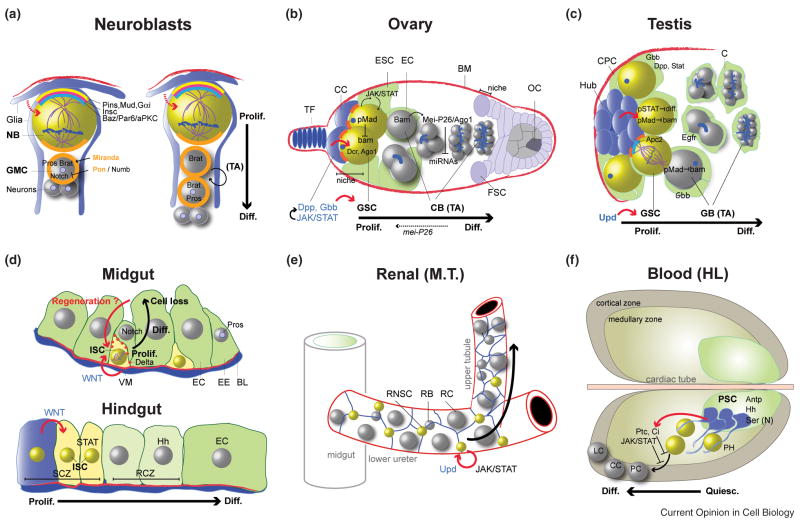
Recent work in Drosophila has provided the systems and tools to dissect how stem cellintrinsic factors are integrated with signalling events in the microenvironment of the “niche”, in order to maintain stem cell identity and proliferation capacity. So far, mechanistic insight into stem cell growth control has been obtained mostly from adult male and female germline stem cells (GSCs) and from postembryonic neural stem cell-like neuroblasts (NBs) (Figure 1a–c). NBs are a transient, heterogeneous population that accomplishes most if not all of their proliferation during larval development (for review: [2,3]). Other insects such as crickets, however, show robust neuroblast proliferation (BrdU incorporation) in the mushroom body of adults, which is dependent on juvenile hormone and on sensory nerve input [4]. Growth regulation has also been described in imaginal disc cells, and these cells can exhibit features of stemness under regenerating conditions. Disc cells obtain the potential to regenerate an injured disc tissue, display developmental autonomy, and can be kept by in vivo culture for years, maintaining their differentiation potential [5,6]. Recently, adult epithelial stem cells that homeostatically maintain the intestine (posterior midgut and hindgut intestinal stem cells-ISCs) and lower tubules and ureters of the malpighian tubules (renal-nephric stem cells-RNSCs) have been described (Figure 1d, e) [7–9]. Finally stem cell-like, slowly proliferating haematopoietic progenitors (HSCs) reside in the medullary zone of the larval lymph gland. These have been shown to control haemocyte homeostasis (Figure 1f) [10,11]. To summarize, stem cells can reside in cellular niches (GSC, HSC) or in non-cellular niches on basement membranes (ISCs and RNSCs) implying a role for the underlying circular muscle (ISCs, figure 1d). As described for GSCs, Drosophila stem cells can perform polarized mitotic divisions that position differentiating daughter cells away from the source of short-range (1 cell diameter) mitogenic and anti-differentiation signals (Figure 1b). Consequently, upon physical displacement, differentiation pathways in daughter cells become derepressed. Analogously, NBs or ISCs employ asymmetric mitotic partitioning of cell fate determinants or Notch/Delta signalling components, respectively, in order to activate differentiation specifically in one daughter cell (Figure 1a, d) [2,3]. Janus-Kinase/Signal-Transducer-and-Activator-of-Transcription (JAK/STAT) signalling by leptin-like cytokines (Unpaired, Upd) and signalling via the bone morphogenetic protein (BMP) ligand decapentaplegic (Dpp) represent major pathways for activating self-renewing proliferation in the different stem cell systems (Figure 1b–f).
Figure 1. Proliferative control in Drosophila stem cells.<.
Stem cells (yellow) usually divide asymmetrically by receiving short-range signals (red arrows) from niche cells (blue). Reduction in adhesion (orange rectangles) to niche support (blue stromal cells or red basement membranes) contributes to displacement of differentiating daughter cells (grey) from the niche. The direction of cell “flow” (major black arrow) reflects spatial organization into zones of stem cell proliferation and differentiation. Tumorous proliferation can arise by progenitor cell expansion or de-differentiation of weakly specified stem cell daughters.
1a. Proliferation in different larval neuroblast (NB) lineages
Left: NBs (yellow) divide asymmetrically in a presumptive niche-like environment of glial cells (blue), and bud off intermediate progenitors (ganglion mother cells - GMC), which will divide only once more into two differentiating neurons. During asymmetric NB division, apical restriction of cortical aPKC/Par, Inscuteable and Pins/Gαi complexes causes cell fate determinants Numb, Prospero and Brat to be partitioned basally to the GMC with the help of their adaptors Partner of Numb (Pon) and Miranda. Right: In contrast to other central brain NBs, posterior-asense-negative (PAN) neuroblasts (yellow) display an intermediate transit-amplifying (TA) lineage by generating asymmetrically dividing secondary neuroblasts. Mutation of brat causes tumor-like overproliferation arising from uncommitted secondary neuroblasts.
1b. Female germline stem cells in the stromal cell niche at the tip of the ovariole.
Niche cap cells (CC) are in close contact with germline stem cells (GSCs, yellow) that divide in coordination with somatic escort stem cells (ESC). ESCs produce squamous nondividing daughter escort cells (EC) that encyst the differentiating GSC daughters (cystoblasts-CB). After 4 transit-amplifying (TA) division cycles with incomplete cytokinesis, one CB is determined as oocyte (OC). JAK/STAT activated BMP signalling via the ligands decapentaplegic (Dpp) and glass-bottom-boat (Gbb) triggers transcriptional inhibition of the differentiation factor bag-of-marbles (bam). During differentiation Bam de-repression in cystocytes activates the microRNA inhibitor Mei-P26. Thereby, microRNAs responsible for balanced growth and cell cycle progression in stem cells are inactivated during cyst differentiation. Mutation of mei-P26 (dashed arrow, bottom) causes cystocyte tumors. Terminal filament (TF), basement membrane (BM), follicle stem cell with epithelial niche (FCS).
1c. Male germline stem cell niche at the testis tip.
Producing transit-amplifying gonialblast daughters (GBs), male GSCs (yellow) proliferate due to secretion of the JAK/STAT ligand unpaired (upd) from the somatic hub cells (niche). GSCs divide asymmetrically based on astral microtubule capture by cortical localization of Apc2 at the interphase between hub and GSCs. GB differentation involves Egfr signal reception in the cyst cells. Drosophila male germline stem cells do not exhibit a quiescent reserve stem cell pool.
1d. Adult Intestinal stem cells (ISCs).
Top: ISCs of the adult midgut epithelium (yellow) proliferate at a basal position, sandwiched between the differentiated midgut epithelial cells and a basement membrane (BM) adjacent to visceral muscle (VM). ISC daughters, called enteroblasts (EB), are more apical and directly differentiate either into enterocytes (EC) or enteroendocrine cells (EE). Lineage choice towards EC fate is achieved by high levels of Delta signalling from the ISC to activate Notch for EB differentiation. ISC self-renewal is regulated by paracrine Wnt signalling (Drosophila Wingless) from the niche of circular visceral muscle, which antagonizes Notch activation. Bottom: Slowly cycling ISCs (yellow) in the adult hindgut proliferation zone (HPZ) self-renew in the spindle cell zone (SCZ) due to short-range Wingless release from niche cells (blue) in the anterior SCZ. Hindgut ISCs produce fast cycling hindgut progenitors, which divide in the round cell zone (RCZ, light green), and finally exit cell cycle and differentiate to enterocytes (dark green).
1e. Adult Renal-nephric stem cell (RNSC)
Adult RNSCs (yellow) proliferate in the lower tubules and ureters of the malpighian tubules (M.T.) and produce renalblast daughters (RB), which will differentiate into renalcytes (RC) in the lower tubule/ureters or into type I and type II cells in the regions of the upper tubules. Adult stem cell proliferation rate could be determined by autocrine unpaired secretion and activation of JAK/STAT signalling in the stem cells.
1f. Haematopoiteic stem cells in the larval lymph gland.
Slowly cycling larval haematopoietic progenitors (yellow) are contacted by cytoplasmic extensions from cells of the posterior signalling center (PSC, niche), which is specified by expression of the homeotic gene Antennapedia (Antp). An undifferentiated prohaemocyte (PH) pool in the medullary zone is maintained by Hedgehog (Hh) and JAK/STAT activation. Differentiated cell types comprise phagocytic plasmatocytes (PC), crystal cells (CC), involved in innate immune responses and wound healing, and lamellocyte (LC), which engulf and neutralize eggs of parasitizing wasps in the larva. At the begin of metamorphosis the gland disintegrates, releasing cells into the open circulatory system (haemolymph-HL).
The cell size limit hypothesis and neuroblast proliferation
Neural development provides a unique system to address the critical question of whether growth potential causally determines cell cycle progression in a stem cell type. During most differentiative divisions, neuroblasts (NBs) divide asymmetrically into a large NB that has continued self-renewing growth potential, and a small ganglion mother cell (GMC), which will only divide terminally into a pair of differentiated neurons or glia (Figure 1a, left).
With the exception of mushroom body and lateral neuroblasts, central nervous system embryonic NBs first enlarge, and then perform a series of stereotyped rapid apico-basally oriented divisions. Due to progressive mass loss, embryonic NBs decrease their size over time until they are small and enter quiescence [12]. Larval neuroblasts increase their cell mass before cell cycle reactivation. Reentry into S-phase is mediated by non-autonomous growth factor signalling (Hedgehog and FGF), which is modulated by glial heparin sulfate proteoglycan (trol) function. Cell mass increase is most likely dependent upon feeding on a protein diet during larval stage L1, which is required for resumed NB divisions. Likewise, imaginal disc cells increase their size 6-fold before overcoming their developmental larval G1 quiescence [13–15]. After a larval neuroblast initiates its proliferative program, it then becomes independent of ongoing nutritional supplies from the diet. Therefore, relative to differentiated cells in the larva, which arrest cell cycle progression upon food withdrawal, NBs appear to have an enhanced ability to take up recycled nutrients from the haemolymph (blood), grow, and divide. Nevertheless, NBs do reduce their division rate if the larva is starved [14]. Although not yet clarified by genetic mosaic analysis or cell ablation experiments at progressive timepoints during postembryonic development, glia-NB contacts have been reported to exhibit features of a stem cell niche-like microenvironment (Figure 1a, red dashed arrow). Reception of Activin signalling seems to maintain NB proliferation rates in the optic centers and central larval brain, and TGF-β type ligands are indeed expressed in surface glial cells [16]. Elegant evidence corroborates, however, the central role of Prospero (Pros) as an output of a temporal transcription factor cascade that terminates most postembryonic NB divisions [17**].
To summarize, staying above a limiting small size (mass) has been correlated with the ability to continue self-renewing stem cell divisions. Studies have so far, however, failed to determine whether cell growth is the critical limiting factor for stem cell proliferation or not.
Asymmetric cell division maintains a cell size difference without differential growth
Independent of actual growth regulation, the big Drosophila NB displays an intriguing strategy to stay big by budding off small GMC daughters. Thereby, the difference in cell size between a self-renewing NB and its GMC daughter occurs instantaneously at mitosis. This size asymmetry is dependent on the activity of two redundant regulatory pathways that are active in the short period of mitosis, and does not necessarily require differential growth. One pathway, depending on the evolutionarily conserved Baz/aPKC/PAR-6 complex, which resides on the apical NB cortex and spatially instructs processes of cell polarization, displaces the cortical cleavage plane off-center in the basal GMC hemisphere (Figure 1a). The other results in much shorter mitotic spindle fibers on the future GMC through a Pins/Galphai/Cno and Loco/Galphai pathway proposed to sustain a feedback loop with Gbeta13F/Ggamma1 activity, hence spatially organizing heterotrimeric G protein signalling and microtubule polymerization regulators (Figure 1a) [18]. Whether whole “growth organelles” (mitochondria, ribosomes) might be actively partitioned in morphologically symmetric stem cell divisions, remains unknown. Relevant pathways for symmetric growth factor signal distribution over mitosis have, however, been described in wing imaginal cells, where endocytic vesicles transport the Dpp/TGF-β receptor complex [19]. Moreover, pericentrosomal proteins destined for proteasomal degradation are asymmetrically inherited over mitosis even in seemingly symmetric somatic divisions [20].
Mutual regulation of stem cell polarity, cell cycle and growth
Regulation of asymmetric cell division by cell cycle factors
Neuroblasts reveal another interesting aspect of growth control by partitioning protein translation regulators during mitosis. Firstly, asymmetric mitotic division restricts active growth and cell division to stem cells and distributes proliferation inhibitors and differentiation factors (segregating determinants) to daughters (Figure 1a). Interestingly, cell cycle factors directly control the asymmetric cell division machinery and vice versa (for review see: [21,22]). For example, sufficiently high activity of the mitotic regulator Cdk1/Cyclin B and B3 activity, or Aurora A and Polo kinase function, are required for maintenance of asymmetric protein localization during NB division [23,24**]. The two kinases are known to regulate centrosomes, spindle assembly, cohesion and cleavage furrow ingression. Mitosis in polo kinase mutants, however, proceeds with aberrantly symmetrical splitting of unphosphorylated Pon and its binding partner, the segregating determinant Numb, between both daughters (Figure 1a) [24**]. Consequently, insufficient Numb activity fails to antagonize Notch, and both daughters continue rapid growth and division as self-renewing NB-like cells. The “stem cell expansion” caused by exponential NB proliferation can continue, resulting in tumorous overgrowth. Since partial rescue of Numb asymmetry in polo mutants does not abolish overproliferation, a parallel impact of spindle orientation and aPKC kinase localization (which affects Numb phosphorylation and activity) can be expected [24**,25].
Regulation of the stem cell cycle by cellular polarity
Reciprocally, the polarity machinery can also directly impact the cell cycle. Combined loss of lethal (2) giant larvae (lgl), a cytoskeletal tumor suppressor and regulator of cell fate determinant localization, and partner of inscuteable (pins), increases the amount of cortical aPKC activity, and this triggers symmetric self-renewing neuroblast overproliferation [26]. Net overproliferation of neuroblasts that produce daughters of equal size is also observed in mud mutant mushroom body neuroblasts [27]. The mud gene product regulates mitotic spindle orientation by binding Pins and enhancing microtubule polymerization (Figure 1a) [28]. We learn that defective daughter cell determination can cause tumors, defective spindle orientation can result in aberrant partitioning of determinants, and centrosome malfunction may deregulate spindle orientation. This does, however not address whether the stem cell cycle is actually accelerated, or which pathways account for self-sufficient growth and division in stem cells and NB tumors.
Partitioning transcription and translation regulators
The cross-regulation of mitosis and symmetry is intriguing, because factors that are asymmetrically distributed by the adaptor protein Miranda into the GMC include the cell cycle regulatory transcription factor Prospero (Pros), as well as the putative translation inhibitor Brat (Figure 1a) [29**,30**,31]. Due to apically restricted auto-inhibition of Lgl upon phosphorylation by aPKC kinase, Lgl directs cortical Miranda to the basal cortex. Therefore, it looks like two critical regulators, one suppressing growth and the other suppressing the cell cycle, are delivered to the GMC. Indeed, both the prospero and brat mutants get neuroblast stem cell-like derived tumors [30**,31].
The homeodomain transcription factor Pros inhibits progression of the cell cycle by directly binding and repressing promoter regions of G1/S (e2f1, cyclin E) and G2/M transition (string/cdc25) as well as NB-type asymmetric cell division and renewal genes [29**]. By interaction with coactivators, Pros also activates differentiation genes in GMCs, locking in the switch to differentiate [29**]. Pros overexpression can terminate most of overproliferation that occurs in brat mutant neuroblasts [32], suggesting that the transcriptional cell cycle exit program imposed by Pros is dominant over permissive growth driven by loss of brat [29**].
A mechanistic paradigm for understanding how cell growth and the cell cycle are co-regulated in stem cells comes from the recent analysis of Drosophila Trim-NHL domain proteins including the tumor suppressor brat. Homology and genetic studies in C. elegans as well as Drosophila suggest that Brat may suppress cell growth by repressing general translation rather directly. In addition, or perhaps as part of this function, Brat posttranscriptionally inhibits dMyc, a transcription factor that stimulates many processes required for growth, including ribosome production and translation [30**]. brat mutation causes metastatic tumorous proliferation of certain types of NBs [30**]. These tumors arise from continued self-renewing growth and division of transient amplifying secondary neuroblasts that are unable to differentiate, and do not express the differentiation factor Prospero (Figure 1a, right panel) [30**,31,33*]. Whether brat mutant NBs overproliferate due simply to increased protein synthesis and growth, however, is not so clear. brat mutations only cause overproliferation of specific classes on NBs, and known translational regulators such as dMyc and the TOR activator Rheb have not yet been shown to transform even these NBs. Nevertheless, the extant data has been interpreted to indicate that deregulation of growth by translational control might be the trigger for tumorous NB self renewal and cell cycle progression by never allowing the balance to tip towards differentiation associated cell cycle exit, at least in especially sensitive cell states [30**,33*]. A strategy to translationally control the production of limiting mitotic (S. pombe Cdc25) or G1/S cell cycle regulators (S. cerevisiae Cln3) by motifs in the 5′ untranslated (UTR) mRNA regions allows cells to couple general growth and cell cycle rate [34,35]. Although these mechanisms are likely to apply in Drosophila and other animal cells, they have not yet been assessed experimentally in any depth. Since vertebrate microRNAs have recently been shown not only to inhibit, but also to stimulate protein production by binding to 5′UTRs, translational regulation of cell cycle regulators must be reevaluated as a novel and potentially important mode of growth control [36*,37].
Together, available data suggest that known translation/growth pathways can sometimes become dominant over cell cycle promoting transcription factors in limiting proliferation. This may be particularly true in contexts where nutrient availability limits proliferation or when dropping intracellular ATP levels trigger an AMP-activated protein kinase based metabolic checkpoint, downregulating cyclin E protein [38–41]. Such a strategy makes sense, because translational responses are fast for unstable proteins (Cyclin E, String), and do not necessarily reset the general proliferative transcription program of stem cells [41]. But pros-like transcriptional regulators may be dominant over translational activity, imposing cell cycle exit by direct repression of cell cycle factors and redundant activation of differentiation factors that further inhibit proliferation [42]. Direct repression of rRNA transcription by vertebrate MyoD and Runx2 during lineage specification is another example [29**,43]. Important exceptions do, however, exist, and productive polysome levels are actually increased in differentiating mouse stem cells [44].
Cell cycle and growth: where is the pattern?
The notion that a mere increase in general protein biosynthesis after dmyc derepression may trigger cell cycling in brat mutant NBs is not supported by experimental evidence in other Drosophila cell types. dMyc specifically enhances ribosome biogenesis and protein translation by coordinately facilitating PolI rRNA transcription and expression of many Pol II and III targets including ribosomal components, rRNA processing and translation initiation factors [45]. In fact, forced expression of dmyc or other growth regulators that are known to directly control metabolism, like PI3K (insulin signalling) or Rheb (a TOR activator), does not trigger overproliferation of Drosophila adult intestinal stem cells (H. Jiang, B.A.Edgar personal communication), or imaginal disc cells. Such factors can posttranscriptionally increase Cyclin E levels and accelerate G1/S progression in imaginal discs, but mitotic entry remains limiting, effectively not speeding up the cell cycle [46]. On the other hand many of the signalling systems that regulate organ patterning and cell specification can also effect G2/M transitions and overall proliferation rates, at least in imaginal discs [8,16,46,47]. Since a transient G2/M arrest precedes overproliferation in brat mutant NBs, the prime reason that tumors arise could potentially lie in de-repression of patterning growth factor signal transduction, activating mitotic entry. Interestingly, microRNAs could be involved in growth factor signal interpretation, as has been elegantly demonstrated in the Xenopus Spemann organizer, where a graded TGF-β/Nodal response is quantitatively determined by a mirror microRNA gradient inhibiting the corresponding growth factor receptor [48].
Coordinating growth and cell cycle control: a place for microRNAs
Since Ncl-1, the C. elegans ortholog of brat, has been implicated in RNAi function, the TRIM-NHL family protein Brat has also been suggested to inhibit mRNA targets via regulation of small regulatory RNAs [30**]. This is especially interesting in the light of the finding that both the dicer-1/microRNA and the dicer-2 dependent RNAi-mediated chromatin regulation pathway were shown to affect Drosophila female and male germline stem cell division rate and stem cell identity (Figure 1b, c). Moreover, a recent study reveals that Mei-P26, another TRIM/NHL growth regulator, directly binds the argonaute microRNA effector Ago1 via its NHL domain and represses microRNAs including Bantam, which has been implicated as a positive regulator of self-renewing growth and cell cycle progression. Mei-P26 function is activated by the differentiation factor bag of marbles (Bam) in ovarian stem cell daughters only, and its mutation results in tumor initiation in the transit amplifying cystocyte lineage due to loss of microRNA inhibition, and consequently enhanced rRNA biogenesis, self-renewing growth and cell cycle competence (Figure 1b) [49**]. Since Brat is analogously segregated in the differentiating daughters, it will be interesting to see whether microRNAs and translation control also affect proliferation in transit-amplifying NB lineages (Figure 1a, right) [33*].
Indeed, a number of core microRNA biogenesis factors (Dicer-1, Loquacious) as well as argonaute family effector proteins used in the microRNAs and RNA interference (RNAi) pathways (Ago1, Piwi) and a microRNA itself (bantam) have previously been implicated as essential cell autonomous components that determine germ line stem cell (GSC) division rate and cell fate maintenance (Figure 1b) [50,51,52,53*] (for review see: [54]). Variations exist for example in that Piwi can also function in the female somatic GSC niche to effect stem cell mitotic activity. In dicer mutant GSCs, S-phase and mitotic index are reduced, while Cyclin E and Dacapo/p27 expression is enhanced, suggesting a CKI-based G1/S transition block with stable but inactive cycE/CDK2 complexes. Indeed, reducing dacapo dosage can rescue the division defect in dicer mutants [50]. Importantly, loss of the single bantam miRNA, an activator of balanced growth and cell division, recapitulates the growth, division and maintenance defects seen in dicer mutant GSCs [49**,53*,55,56]. Whether bantam is used to promote the cell cycle in other stem cell types, or whether different stem cell types share a common microRNA expression profile is not known at this point. Likewise, it remains unknown which microRNA target proteins must be inhibited to maintain stem cell growth and cell cycle progression, but an miRNA network might underlie the elusive parallelization of growth and cell cycle control in higher eukaryotes.
Recent experiments in the imaginal discs provide a conceptual framework for how the bantam microRNA could regulate both the cell cycle and growth. Dependent on repression of an inhibitory Notch signal, a self-enforcing cell cycle – growth positive feedback loop builds up between bantam and dMyc which stimulates E2F activity and promotes G1/S transition [57*].
Organismal growth control of stem cell proliferation
Finally, how a homeostatically growing tissue instructs the described growth and cell cycle control of its stem cell population is a question of great complexity. Recent studies in Drosophila intestinal, renal and haematopoietic systems have begun to address the largely unknown dynamic coupling between organ cellularity and stem cell proliferation (Figure 1d–f). Absorptive differentiated cell types of the midgut epithelium (enterocytes) turn over weekly for example (Figure 1d, black arrow), suggesting that the midgut epithelium is homeostatically regenerated. Paracrine canonical Wnt signalling from the underlying visceral muscle and the antagonizing Notch activity in differentiating stem cell daughters have been implicated in balancing ISC self-renewal and differentiation respectively (Figure 1d, red arrow) [7,58]. Consistently, clonal deletion of Notch results in excess progenitors (ISCs/EBs) referred to as tumors. Conceptually very similar, adult hindgut stem cells self-renew under the influence of short-range paracrine Wnt signalling. Leaving the Wnt signal range, fast cycling hindgut progenitors finally exit the cell cycle and differentiate to enterocytes due to Hedgehog activity (Figure 1d, bottom). Secondly, but much less clear, high and low signal strengths of autocrine JAK/STAT activity are proposed to instruct self-renewal and differentiation of the renal stem cells respectively (Figure 1e) [8]. Thirdly, a niche-like microevironment in the lymph gland appears required and limiting for maintenance of slowly cycling, uncommitted haematopoietic progenitors implicating Hedgehog signalling (Figure 1f) [10,11]. Although long-term repopulation ability in analogy to the vertebrate haematopoietic system is still unclear, the progenitor pool as already been shown essential for an efficient production of lamellocytes in the case of infestation by parasitoid wasps laying eggs into Drosophila larvae [11]. Forthly, Drosophila female and male germline stem cell (GSC) niches offer the cellularly best understood systems to investigate stem cell programming by animal physiology and organismal growth cues [2]. Interestingly, when protein levels in the diet are low, reduced brain derived insulin like peptide (DILP) signalling leads to a GSC cell-autonomous activation of the transcription factor Foxo and a G2/M delay. According to richness of diet, redundant factors modulate the cell cycle in G1/S and G2/M independent of the niche [38,39*]. Consistent with the growth dependence of female germline stem cells, cell cycle progression in regenerating imaginal disc cells, which exhibit stem cell-like features, does not occur without a growth activating amino-acid supply [6,59].
Conclusions and outlook
On a molecular level the role of RNAs as central transcriptional and translational regulators of stem cell function is emerging. This includes poised chromatin states that leave open the potential for activation or inhibition during lineage specification. These bivalent states are determined by the rate and loci where RNA polymerases transcribe genomes, and by RNA molecules that instruct chromatin function [60–62]. Secondly, recent work in Drosophila puts forward single molecular species (e.g. microRNAs, growth/mitogen cues) that co-regulate both stem cell growth and the cell cycle. Thereby, the undifferentiated cell state self-sufficiently replicates, as long as short range growth factors or even physical cues (anchoring in the niche) prevent differentiation factors from locking-in cell cycle exit. Work in Drosophila also reveals how the asymmetric cell division machinery distributes information, in the form of growth and cell cycle regulators and cell fate determinants, during mitosis. Robustness in distinguishing cell growth and cell cycle control is further conferred by putting stem cells and daughters in different spatial contexts relative to external pro- and anti-differentiative signals in the cellular microenvironment. This leads to self-renewal of the stem cell but not its differentiating daughter. Therefore, although paradoxical, asymmetry might be the driving force in bringing order, as has been recognized a long time ago (“C’est la dissymétrie qui crée le phénomène”. Pierre Curie.)
Acknowledgments
A.K. wishes to thank Norman Zielke and Mihaela Zigman for critical comments, and is supported by a Human Frontier Science Program Long-Term postdoctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Edgar BA, Nijhout HF. Growth and Cell Cycle Control in Drosophila. In: Hall MN, Raff M, Thomas G, editors. Cell Growth: Control of Cell Size. Cold Spring Harb. Lab. Press; 2004; 2004. pp. 23–83. [Google Scholar]
- 2.Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- 3.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Cayre M, Malaterre J, Scotto-Lomassese S, Aouane A, Strambi C, Strambi A. Hormonal and sensory inputs regulate distinct neuroblast cell cycle properties in adult cricket brain. J Neurosci Res. 2005;82:659–664. doi: 10.1002/jnr.20672. [DOI] [PubMed] [Google Scholar]
- 5.Karpen GH, Schubiger G. Extensive regulatory capabilities of a Drosophila imaginal disk blastema. Nature. 1981;294:744–747. doi: 10.1038/294744a0. [DOI] [PubMed] [Google Scholar]
- 6.Sustar A, Schubiger G. A transient cell cycle shift in Drosophila imaginal disc cells precedes multipotency. Cell. 2005;120:383–393. doi: 10.1016/j.cell.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 8.Singh SR, Liu W, Hou SX. The adult Drosophila malpighian tubules are maintained by multipotent stem cells. Cell Stem Cell. 2007;1:191–203. doi: 10.1016/j.stem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takashima S, Mkrtchyan M, Younossi-Hartenstein A, Merriam JR, Hartenstein V. The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signalling. Nature. 2008;454:651–655. doi: 10.1038/nature07156. [DOI] [PubMed] [Google Scholar]
- 10.Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, Banerjee U. A Hedgehogand Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446:320–324. doi: 10.1038/nature05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzemien J, Dubois L, Makki R, Meister M, Vincent A, Crozatier M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature. 2007;446:325–328. doi: 10.1038/nature05650. [DOI] [PubMed] [Google Scholar]
- 12.Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 13.Barrett AL, Krueger S, Datta S. Branchless and Hedgehog operate in a positive feedback loop to regulate the initiation of neuroblast division in the Drosophila larval brain. Dev Biol. 2008;317:234–245. doi: 10.1016/j.ydbio.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- 15.Madhavan V. Histological analysis of the dynamics of growth of imaginal discs and histoblast nests during the larval development of Drosophila melanogaster. Roux’s Archives Dev Biol. 1977;183:269–305. doi: 10.1007/BF00848459. [DOI] [PubMed] [Google Scholar]
- 16.Zhu CC, Boone JQ, Jensen PA, Hanna S, Podemski L, Locke J, Doe CQ, O’Connor MB. Drosophila Activin- and the Activin-like product Dawdle function redundantly to regulate proliferation in the larval brain. Development. 2008;135:513–521. doi: 10.1242/dev.010876. [DOI] [PubMed] [Google Scholar]
- 17**.Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. This study reveals for the first time how larval neuroblast stem cell-like divisions are terminated. The transcriptional outputs from transient consecutive expression bursts of transcription factors are integrated over time (Hb, Kr, Pdm, Cas, Svp). This culminates in timed nuclear Prospero activity to enforce cell cycle exit after the final mitosis. [DOI] [PubMed] [Google Scholar]
- 18.Yu F, Wang H, Qian H, Kaushik R, Bownes M, Yang X, Chia W. Locomotion defects, together with Pins, regulates heterotrimeric G-protein signaling during Drosophila neuroblast asymmetric divisions. Genes Dev. 2005;19:1341–1353. doi: 10.1101/gad.1295505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bokel C, Schwabedissen A, Entchev E, Renaud O, Gonzalez-Gaitan M. Sara endosomes and the maintenance of Dpp signaling levels across mitosis. Science. 2006;314:1135–1139. doi: 10.1126/science.1132524. [DOI] [PubMed] [Google Scholar]
- 20.Fuentealba LC, Eivers E, Geissert D, Taelman V, De Robertis EM. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc Natl Acad Sci U S A. 2008;105:7732–7737. doi: 10.1073/pnas.0803027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prokopenko SN, Chia W. When timing is everything: role of cell cycle regulation in asymmetric division. Semin Cell Dev Biol. 2005;16:423–437. doi: 10.1016/j.semcdb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol. 2008;180:267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 2007;449:96–100. doi: 10.1038/nature06056. Based on biochemical and genetic evidence, this paper highlights the important link how a central cell cycle regulator (Polo kinase) imposes asymmetric partitioning of a cell fate determinant (the Notch inhibitor Numb) during mitosis of larval neuroblast stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. Embo J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- 27.Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- 28.Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 29**.Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. The authors combine mRNA microarray expression profiling, DamID technology on oligonucleotide DNA tiling arrays and genetic analysis to reveal the central tumor suppressive function of the cell cycle regulatory transcription factor Prospero in embryonic NBs in vivo. Binding to 1000 putative regulatory regions in the genome, prospero exerts a dual role in assuring differentiation associated cell cycle exit in a context-dependent fashion. Thereby, prospero both inhibits the NB signature (cell cycle genes, symmetry regulators and the temporal cascade) and activates differentiation genes. [DOI] [PubMed] [Google Scholar]
- 30**.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. This study identifies the putative translational regulator and NHL-TRIM protein family member Brat as novel segregating determinant in Drosophila neuroblasts. Brat mutation triggers unrestricted growth and cell division of specific larval stem cell-like neuroblasts in the central nervous system. Although the mRNA targets remain unclear, a function for translational repression is suggested in stem cell differentiation, proposing that dedifferentiation can initiate growth-sufficient and self-replicating tumorous proliferation. [DOI] [PubMed] [Google Scholar]
- 31.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 32.Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Develop. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. Detailed cell biological investigation shows that loss of growth regulation by Brat initiates self-renewing tumorous proliferation selectively in transit-amplifying NB lineages in the larval brain. Overcoming a transient G2 block, immature neuroblasts are unable to complete differentiation. Instead of eventually exiting the cell cycle, not fully committed brat or numb mutant secondary neuroblasts continue to proliferate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polymenis M, Schmidt EV. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daga RR, Jimenez J. Translational control of the cdc25 cell cycle phosphatase: a molecular mechanism coupling mitosis to cell growth. J Cell Sci. 1999;112(Pt 18):3137–3146. doi: 10.1242/jcs.112.18.3137. [DOI] [PubMed] [Google Scholar]
- 36*.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. In contrast to the classical view that microRNAs negatively regulate mRNA translation by binding to regulatory regions in the 3′UTR, this study in vertebrate cells shows that miR-10a stimulates translation of target mRNAs encoding ribosomal proteins by a novel undescribed mechanism. MiR-10a binds downstream of a TOP (5′-terminal oligopyrimidine tract) motif in the 5′UTR. The TOP motif or 5′ untranslated open reading frames (uORFs) are known to confer reduction of translation rate of the corresponding downstream ORF under poor growth conditions with low translation rates. Repression can be bypassed during fast growth or by miR-10a based translational stimulation. [DOI] [PubMed] [Google Scholar]
- 37.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 38.LaFever L, Drummond-Barbosa D. Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science. 2005;309:1071–1073. doi: 10.1126/science.1111410. [DOI] [PubMed] [Google Scholar]
- 39*.Hsu HJ, LaFever L, Drummond-Barbosa D. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev Biol. 2008;313:700–712. doi: 10.1016/j.ydbio.2007.11.006. The authors show how Drosophila germline stem cell proliferation responds to diet and brain derived insulin-like peptides. When protein levels in the diet are low, activation of the transcription factor Foxo contributes to a G2/M delay. Additionally, rapamycin (Tor) kinase signalling, a major integrator of nutrient availability, could be involved, because PI3-K and Tor signalling can converge into common regulation of myc and 4E-BP, controlling ribosome biogenesis and translation initiation respectively. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- 43.Ali SA, Zaidi SK, Dacwag CS, Salma N, Young DW, Shakoori AR, Montecino MA, Lian JB, van Wijnen AJ, Imbalzano AN, et al. Phenotypic transcription factors epigenetically mediate cell growth control. Proc Natl Acad Sci U S A. 2008;105:6632–6637. doi: 10.1073/pnas.0800970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- 46.Prober DA, Edgar BA. Ras1 promotes cellular growth in the Drosophila wing. Cell. 2000;100:435–446. doi: 10.1016/s0092-8674(00)80679-0. [DOI] [PubMed] [Google Scholar]
- 47.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 48.Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, Dupont S, et al. MicroRNA control of Nodal signalling. Nature. 2007;449:183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 49**.Ralph A, Neumüller JB, Fischer Anja, Bushati Natascha, Poernbacher Ingrid, Mechtler Karl, Cohen Stephen M, Knoblich Juergen A. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008 doi: 10.1038/nature07014. Published online 4 June 2008. This study is of general importance and provides novel mechanistic insights into the significance of translation control for coordination of cell cycle and growth in stem cells lineages. The authors show that Drosophila TRIM-NHL family proteins interact with the microRNA effector Argonaute-1 in order to inhibit miRNA function. MiRNA/translational control by Mei-P26 is a novel tumor suppressor mechanism that ensures termination of self-renewing proliferative capacity during differentiation of transit-amplifying cystocyte lineages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 51.Yang L, Chen D, Duan R, Xia L, Wang J, Qurashi A, Jin P, Chen D. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development. 2007;134:4265–4272. doi: 10.1242/dev.009159. [DOI] [PubMed] [Google Scholar]
- 52.Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- 53*.Shcherbata HR, Ward EJ, Fischer KA, Yu JY, Reynolds SH, Chen CH, Xu P, Hay BA, Ruohola-Baker H. Stage-specific differences in the requirements for germline stem cell maintenance in the Drosophila ovary. Cell Stem Cell. 2007;1:698–709. doi: 10.1016/j.stem.2007.11.007. The authors highlight how Drosophila germline stem cell proliferation and maintenance is instructed by bantam function. Interestingly, there appears to be a Dcr-2 dependent safeguard mechanism that maintains GSCs before adult stages when Mad or dicer/bantam microRNA function is compromised. A robust Dcr-2 mediated RNAi-templated chromatin control is a likely explanation for this effect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 55.Raisin S, Pantalacci S, Breittmayer JP, Leopold P. A new genetic locus controlling growth and proliferation in Drosophila melanogaster. Genetics. 2003;164:1015–1025. doi: 10.1093/genetics/164.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 57*.Herranz H, Perez L, Martin FA, Milan M. A Wingless and Notch double-repression mechanism regulates G1-S transition in the Drosophila wing. Embo J. 2008 doi: 10.1038/emboj.2008.84. Both the bantam microRNA and myc individually are rate limiting for G1/S progression in Drosophila wing imaginal discs. But upon derepression of bantam, a positive feedback loop between the bantam microRNA and myc becomes effective. Bantam stimulates dMyc protein levels and dMyc stimulates bantam transcriptionally, to overcome a cell cycle block, which further involves bantam in stimulating E2F1 activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008 doi: 10.1038/nature07329. Published online 21 Sept. [DOI] [PubMed] [Google Scholar]
- 59.McClure KD, Schubiger G. Transdetermination: Drosophila imaginal disc cells exhibit stem cell-like potency. Int J Biochem Cell Biol. 2007;39:1105–1118. doi: 10.1016/j.biocel.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]