 The efficient synthesis of a range of stable SAM mimetics, and their ability to promote the binding of the E. coli methionine repressor (MetJ) to its operator DNA, is described. Active analogues functionalised with bioorthogonal tags were identified.
The efficient synthesis of a range of stable SAM mimetics, and their ability to promote the binding of the E. coli methionine repressor (MetJ) to its operator DNA, is described. Active analogues functionalised with bioorthogonal tags were identified.
Abstract
The efficient synthesis of a range of stable SAM mimetics, and their ability to promote the binding of the E. coli methionine repressor (MetJ) to its operator DNA, is described.
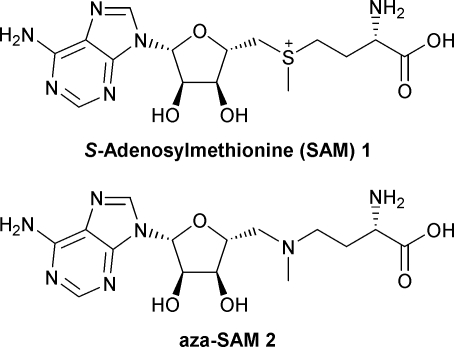
S-Adenosylmethionine or SAM, 1 (also known as AdoMet), is a co-factor with a wide range of biological functions, including methylation, regulation of gene expression in the bacterial methionine biosynthetic pathway, catalysis of some biological radical reactions1,2 and control of a number of riboswitch elements.3 In total, SAM, 1, has been identified as a ligand for fifteen protein superfamilies.1 The use of SAM, 1, to study these processes has been hampered by its inherent instability.4,5 Thus, stable SAM analogues, such as aza-SAM, 2, which is a titratable, isostructural mimic, have been developed6 and shown to be useful tools in the analysis of SAM-dependent processes.7,8
Here, we describe an improved synthesis of aza-SAM, 2, which we have adapted to the preparation of a range of novel derivatives. We have also determined the ability of the analogues to promote the binding of the E. coli methionine repressor, the MetJ dimer, to its operator DNA. MetJ represses at least nine genes in the methionine biosynthetic pathway9 following activation by the non-cooperative binding of two molecules of SAM, 1, which increases the affinity of MetJ for its operator DNA by a factor of 100–1000.2,8,10
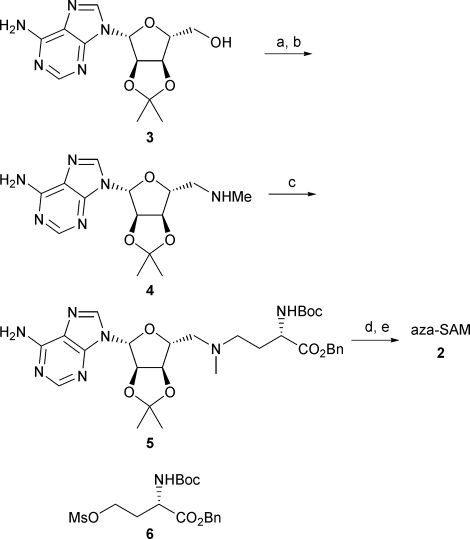
Fukuyama–Mitsunobu reaction11 of the protected adenosine derivative12 3 with MeNNsH as the nucleophile gave the corresponding crystalline sulfonamide in 60% yield; deprotection gave the key amine intermediate 4.13 Our approach avoided the synthesis of the 5′-tosyl derivative of 3, which was prone to cyclisation.14 Furthermore, the approach avoided the use of methylamine in a high pressure reaction vessel, which was required for conversion of the tosylate into 4.15 The amine 4 was reacted with the mesylate 6, prepared from the corresponding aspartic acid derivative,16 to give 5. Deprotection gave aza-SAM, 2, in 44% yield (Scheme 1).
Scheme 1. Improved synthesis of aza-SAM. Conditions: (a) MeNNsH, DEAD, PPh3, THF, 18 h, 60%; (b) PhSH, Cs2CO3, MeCN, 18 h, 83%; (c) 6, DIPEA, TBAI, MeCN, 65 °C, 13 h, 75%; (d) 2 M NaOH, MeCN, 3 h; (e) 5 M HCl, 1 h, 44% over two steps.
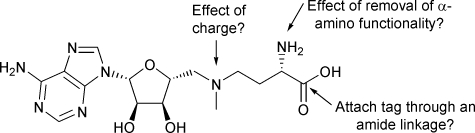
The terminal carboxyl group of SAM is exposed to solvent in the structure of its complex with the MetJ dimer,2 suggesting that it may be possible to attach a bioorthogonal tag at this position (Fig. 1). In addition, we wanted to explore the effect of (a) removal of the α-amino group in order to simplify the structure of aza-SAM; and (b) quaternisation of the 5′-tertiary amine in order to determine the role of the charge.
Fig. 1. Identification of a position for attaching a bioorthogonal tag.
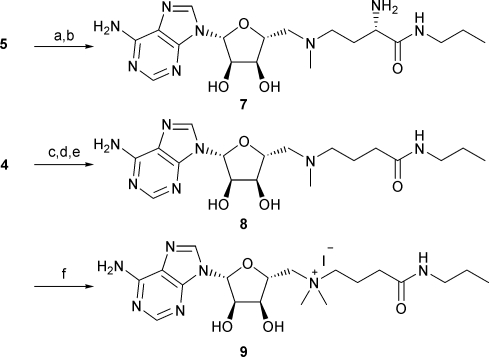
The amide analogue 7 was prepared by aminolysis of the ester 5, followed by removal of the protecting groups (Scheme 2). Similarly, alkylation of the amine 4 with methyl 4-iodobutyrate was followed by aminolysis with propylamine and deprotection to yield the analogue 8 lacking the α-amino group. Treatment of 8 with methyl iodide gave the corresponding quaternised derivative 9.
Scheme 2. Conditions: (a) propylamine, 43 °C, 23 h, 99%; (b) 5 M HCl, 5 min, 91%; (c) methyl 4-iodobutyrate, DIPEA, MeCN, 70 °C, 6.5 h, 98%; (d) propylamine, 47 °C, 5 d, 93%; (e) TFA, 1:1 MeOH–H2O, 5 min, 98%; (f) MeI, 3:1 MeCN–H2O, 97%.
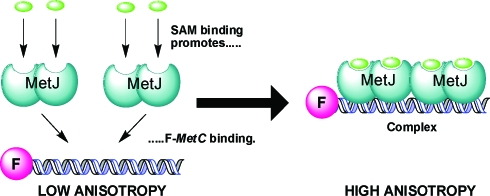
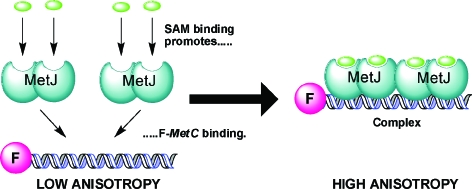
Fluorescence anisotropy17 was used to measure the ability of the SAM analogues to act as co-repressors. Two 18-base oligonucleotides, one of which had a covalently-linked terminal fluorescein group, were annealed to yield the fluorescently-labelled duplex (F-metC). F-metC contained the shortest naturally occurring operator sequence for MetJ: metC. 18 MetJ was titrated into a solution of 10 nM F-metC in 50 mM Trizma hydrochloride pH 7.6 and 100 mM KCl, in the presence or absence of 2 mM ligand, and the increase in fluorescence anisotropy due to DNA binding was recorded (Fig. 2). The assay thus allowed us to determine the effect of modification of the SAM analogues on their relative ability to promote complex formation.
Fig. 2. Cartoon illustrating the fluorescence anisotropy assay. Binding of SAM molecules promotes formation of a SAM–F-metC–protein complex, with two MetJ dimers bound to the 18 base-pair DNA duplex.
Fitting the data to a sigmoidal binding model allowed extraction of the EC50, the concentration of protein required to promote half-maximal formation of the complex. Although the EC50 values obtained (Table 1) allow the relative effect of the ligands to be compared, the assay set-up may mean that the data has little physiological relevance. The EC50 in the presence of 2 mM SAM, 1, was 96 ± 3 nM. In the absence of SAM, 1, the affinity of MetJ for the DNA was about 30-fold lower (EC50 = 2900 ± 20 nM). The effect of SAM was smaller than previously reported estimates, which vary considerably.19,20 Aza-SAM, 2, was a less effective ligand than SAM, 1, although it did still promote complex formation (EC50 650 ± 40 nM). The pK a of aza-SAM, 2, has been shown to be 7.18,12 and so will only be around 25% protonated under the assay conditions (pH 7.6 buffer).
Table 1. EC50 values, i.e. the concentration of MetJ required to promote half-maximal formation of its complex with the F-metC DNA, in the presence and absence of ligands.
| Entry | Liganda | EC50/nM |
| 1 | SAM, 1 | 96 ± 3 |
| 2 | aza-SAM, 2 | 650 ± 40 |
| 3 | 7 | 910 ± 10 |
| 4 | 8 | 1000 ± 100 |
| 5 | 9 | 150 ± 10 |
| 6 | no ligand | 2900 ± 20 |
aThe concentration of the ligands was 2 mM.
The effects of the novel SAM analogues were also investigated. Replacing the carboxylic acid of aza-SAM, 2, with the propyl amide of 7 resulted in a higher EC50 (compare entry 3 with entry 2, Table 1) though removal of the α-amino functionality, as in 8, did not have a significant effect (entry 4). However, quaternisation of the tertiary amine of 8, to give 9, resulted in an EC50 that was much lower than with aza-SAM, and comparable to that of SAM itself (compare entry 5 with entry 1). Thus, amide formation and removal of the α-amino group had a relatively small effect on ligand function. However, the charge of the amine can be deduced to be important for promoting the formation of the MetJ–DNA complex. It was therefore decided to attach bioorthogonal tags through an amide linkage formed from the terminal carboxyl group.
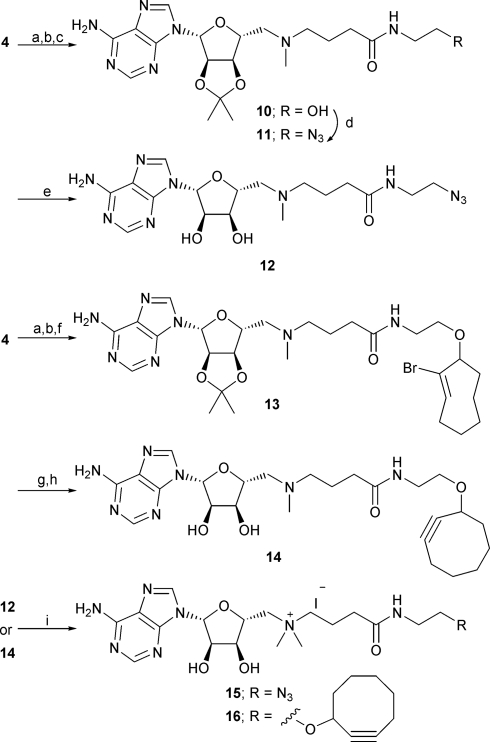
With this information in hand, we prepared ligands which were functionalised with the bioorthogonal azide and cyclooctyne tags. The preparation of these analogues was, however, problematic. Alkylation of 4 with methyl 4-iodobutyrate was followed by ester hydrolysis and amide formation with ethanolamine to yield the amide 10 (Scheme 3). Activation of 10 by treatment with methanesulfonic anhydride, however, triggered cyclisation to yield the corresponding oxazoline; nonetheless, this intermediate could be opened to yield the required azide 11 (yield: 56% from 10). Similarly, coupling with 2-(cyclooct-2-ynoxy)-ethylamine did give the required amide; however attempted deprotection resulted in hydration of the strained alkyne. We therefore decided to delay the formation of the cyclooctyne until after deprotection of the ligand. Thus, 2-((Z)-2-bromocyclooct-2-enyloxy) ethylamine was reacted to yield the amide 13; after removal of the acetonide protecting group, treatment with five equivalents of sodium hydride in THF–DMF resulted in elimination to give the required cyclooctyne 14 in 95% yield. Remarkably, under these conditions, the elimination proceeded more smoothly than has been previously reported21 and clean elimination was possible in the presence of five potentially acidic protons. Treatment of the tagged analogues 12 and 14 with methyl iodide yielded the corresponding quaternised derivatives 15 and 16.
Scheme 3. Conditions: (a) methyl 4-iodobutyrate, DIPEA, MeCN, 70 °C, 6.5 h, 98%; (b) NaOH, MeOH, 2 d, 57%; c) ethanolamine, PyBOP, DIPEA, DMF, 30 min, 93%; (d) Ms2O, NEt3, CH2Cl2, 0 °C, 30 min then NaN3, DMSO, 70 °C, 2 h, 56%; (e), 5 M HCl, 20 min, 94%; (f) 2-((Z)-2-bromocyclooct-2-enyloxy) ethylamine, PyBOP, DIPEA, DMF, 30 min, 80%; (g) 5 M HCl, 6 min, 75%; (h) NaH, 1:1 DMF–THF, 30 min, 95%; (i) MeI, 3:1 MeCN–H2O, 15: 88%, 16: 86%.
The ability of the tagged analogues to promote the binding of MetJ dimer to DNA was also investigated using fluorescence anisotropy. The assay allowed us to determine the effect of attachment of the bioorthogonal tags on the activity of the SAM analogues. Both tagged analogues 12 and 14 were more efficient at binding to MetJ than the model propyl amide 8 (compare entry 4, Table 1 with entries 3 and 4, Table 2). Once more, however, quaternised analogues were significantly more effective than the corresponding uncharged derivatives (compare entries 5 and 6, with entries 3 and 4, Table 2). The most potent compound was the quaternised analogue, 16, that had been derivatised with the cyclooctyne tag. Furthermore, 16 was a more effective ligand than SAM itself (EC50 96 ± 3 nM).
Table 2. EC50 values, i.e. the concentration of MetJ required to promote half-maximal formation of its complex with the F-metC DNA, in the presence and absence of ligands.
| Entry | Liganda | EC50/nM |
| 1 | SAM, 1 | 96 ± 3 |
| 2 | aza-SAM, 2 | 650 ± 40 |
| 3 | 12 | 540 ± 20 |
| 4 | 14 | 650 ± 40 |
| 5 | 15 | 154 ± 6 |
| 6 | 16 | 65 ± 3 |
| 7 | no ligand | 2900 ± 20 |
aThe concentration of the ligands was 2 mM.
In summary, we have described an improved synthesis of aza-SAM which we have adapted to the synthesis of a range of other stable analogues of the co-factor SAM. We have investigated the ability of these analogues to promote the binding of the MetJ dimer to its DNA operator. In particular, we have shown that quaternisation enhances the activity of aza-SAM analogues, and that it is possible to append a bioorthogonal tag through conversion of the terminal carboxyl group into an amide. SAM analogues which have been derivatised with a bioorthogonal tag may be useful chemical tools for isolating, identifying and modulating other SAM-binding macromolecules such as methyltransferases, fluorinases, radical SAM proteins and riboswitch domains.
Acknowledgments
We thank the Wellcome Trust, EPSRC and BBSRC for funding, and Dr Bruce Turnbull for discussions.
Footnotes
References
- Loenen W. A. M. Biochem. Soc. Trans. 2006;34:330–333. doi: 10.1042/BST20060330. [DOI] [PubMed] [Google Scholar]
- Rafferty J. B., Somers W. S., Saint-Girons I., Phillips S. E. V. Nature. 1989;341:705–710. doi: 10.1038/341705a0. [DOI] [PubMed] [Google Scholar]
- Gilbert S. D., Rambo R. P., Van Tyne D., Batey R. T. Nat. Struct. Mol. Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- Knowles T. J., PhD Thesis, University of Leeds, 2005 [Google Scholar]
- Desiderio C., Cavallaro R. A., De Rossi A., D'Anselmi F., Fuso A., Scarpa S. J. Pharm. Biomed. Anal. 2005;38:449–456. doi: 10.1016/j.jpba.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Thompson M. J., Mekhalfia A., Jakeman D. L., Phillips S. E. V., Phillips K., Porter J., Blackburn G. M. Chem. Commun. 1996:791–792. [Google Scholar]
- Couture J.-F., Hauk G., Thompson M. J., Blackburn G. M., Trievel R. C. J. Biol. Chem. 2006;281:19280–19287. doi: 10.1074/jbc.M602257200. [DOI] [PubMed] [Google Scholar]
- Parsons I. D., Persson B., Mekhalfia A., Blackburn G. M., Stockley P. G. Nucleic Acids Res. 1995;23:211–216. doi: 10.1093/nar/23.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marincs F., Manfield I. W., Stead J. A., McDowall K. J., Stockley P. G. Biochem. J. 2006;396:227–234. doi: 10.1042/BJ20060021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A., McAlpine A., Stockley P. G. FEBS Lett. 1994;348:41–45. doi: 10.1016/0014-5793(94)00579-6. [DOI] [PubMed] [Google Scholar]
- Fukuyama T., Jow C.-K., Cheung M. Tetrahedron Lett. 1995;36:6373–6374. [Google Scholar]
- Thompson M. J., Mekhalfia A., Hornby D. P., Blackburn G. M. J. Org. Chem. 1999;64:7467–7473. [Google Scholar]
- Jastorff B., Murayama A., Cramer F., Hettler H. J. Org. Chem. 1971;36:3029–3033. doi: 10.1021/jo00819a027. [DOI] [PubMed] [Google Scholar]
- Clark V. M., Todd A. R., Zussman J. J. Chem. Soc. 1951:2952–2958. [Google Scholar]
- Minnick A. A., Kenyon G. L. J. Org. Chem. 1988;53:4952–4961. [Google Scholar]
- Broadrup R. L., Wang B., Malachowski W. P. Tetrahedron. 2005;61:10277–10284. [Google Scholar]
- Jameson D. M., Sawyer W. H. Methods Enzymol. 1995;246:283–300. doi: 10.1016/0076-6879(95)46014-4. [DOI] [PubMed] [Google Scholar]
- Belfaiza J., Parsot C., Martel A., de la Tour C. Bouthier, Margarita D., Cohen G. N., Saint-Girons I. Proc. Natl. Acad. Sci. U. S. A. 1986;83:867–871. doi: 10.1073/pnas.83.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S. E. V., Manfield I., Parsons I., Davidson B. E., Rafferty J. B., Somers W. S., Margarita D., Cohen G. N., Saint-Girons I., Stockley P. G. Nature. 1989;341:711–715. doi: 10.1038/341711a0. [DOI] [PubMed] [Google Scholar]
- Phillips K., Phillips S. E. V. Structure. 1994;2:309–316. doi: 10.1016/s0969-2126(00)00032-0. [DOI] [PubMed] [Google Scholar]
- Agard N. J., Prescher J. A., Bertozzi C. R. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.