Abstract
Viruses are a paradigm of the economy of genome resources, reflected in their multiplication strategy and for their own structure. Although there is enormous structural diversity, the viral genome is always enclosed within a proteinaceous coat, and most virus species are haploid; the only exception to this rule are the highly pleomorphic enveloped viruses. We performed an in-depth characterization of infectious bursal disease virus (IBDV), a non-enveloped icosahedral dsRNA virus with a bisegmented genome. Up to 6 natural populations can be purified, which share a similar protein composition but show higher sedimentation coefficients as particle density increases. Stoichiometry analysis of their genome indicated that these biophysical differences correlate with the copy number of dsRNA segments inside the viral capsid. This is a demonstration of a functional polyploid icosahedral dsRNA virus. We show that IBDV particles with greater genome copy number have higher infectivity rates. Our results show an unprecedented replicative strategy for dsRNA viruses and suggest that birnaviruses are living viral entities encompassing numerous functional and structural characteristics of positive and negative ssRNA viruses.
Keywords: cargo volume, IBDV, icosahedral dsRNA viruses, life cycle, viral polyploidy
Viruses are entities with limited coding capacity, but with extensive diversity in their genetic material [single-stranded (ss) RNA, ssDNA, double-stranded (ds) RNA or dsDNA]. An important limitation to the size of the viral genome is its container, the protein capsid. Virus particles have evolved to incorporate only a single copy of their genome within the protein shell, a widespread but not universal strategy.
Capsid arrangement of 1 or more protein subunits defines the volume available for the viral genome (1). Helical viruses are considered open structures, since a genome of any size can be enclosed by varying helix length. Icosahedral viral shells form a container of limited internal volume for nucleic acid packaging. Retroviruses represent a structural variation with a fullerene-based capsid and are exceptional, since each virion particle incorporates 2 copies of the positive-sense ssRNA (2). Enveloped pleomorphic viruses with helical nucleocapsids (encompassing negative ssRNA) have relaxed constraints on packaged genome size (3), leading to the incorporation of additional RNA segments, as in the case of influenza virus, an orthomyxovirus with a segmented genome (4). Furthermore, paramyxoviruses (with non-segmented genomes) are extremely pleomorphic and can package several genomes efficiently; paramyxoviruses might therefore be considered polyploid (5).
Most dsDNA, ssRNA, and dsRNA viruses are spherical and build a defined cargo space, with sizes ranging from 20 nm to > 220 nm (6). One genome copy is often packed with a high degree of compaction, compatible with the inner particle dimensions (7). Although variable nucleic acid densities are reported, it is clear that there are upper (and lower) limits for the size of the nucleic acid to be encapsidated.
Here, we analyzed the biophysical and biochemical properties of a non-enveloped icosahedral dsRNA virus, the infectious bursal disease virus (IBDV), an avian virus of the family Birnaviridae (8). IBDV has a bipartite dsRNA genome (segments A and B) that is packaged into a single virus particle, ≈70 nm in diameter, exhibiting T = 13 levo symmetry (9, 10). Considering that the IBDV capsid average internal radius is 26.5 nm (9, 11), which implies a cargo space of ≈77,900 nm3, and that the dsRNA average size is 6 kpb, a genomic packing density of ≈10 bp/100 nm3 is obtained. This value contrasts with those previously determined for icosahedral viruses with densities ≈40 bp/100 nm3.
IBDV Segment A (3.2 kpb) has 2 partially overlapping open reading frames (ORFs). The smaller of the two encodes VP5, a nonstructural host membrane-associated protein; the larger encodes a polyprotein that is cotranslationally processed by the viral protease VP4 (12), yielding the capsid precursor protein pVP2, as well as VP3 and VP4. pVP2 is further processed at its C-terminal region to give the mature VP2. Segment B (2.8 kpb) is monocistronic and encodes VP1, the RNA-dependent RNA polymerase [RdRp (13)], which is packaged inside the virion.
VP2 and a variable amount of pVP2 assemble into 260 trimers to form the capsid structural units (accounting for ≈38 MDa); VP3 is a multifunctional protein that interacts with itself (14), pVP2 (11), VP1 (15, 16), and with the dsRNA to make ribonucleoprotein complexes (17), a unique feature among dsRNA viruses.
IBDV is an icosahedral virus that can package more than 1 complete genome copy. Moreover, multiploid IBDV particles propagate with higher efficiency than haploid virions. These features suggest that IBDV and, indeed, all birnaviruses have a number of structural and functional features (18, 19) that represent evolutionary links to unrelated viruses.
Results
Purification of IBDV Natural Populations.
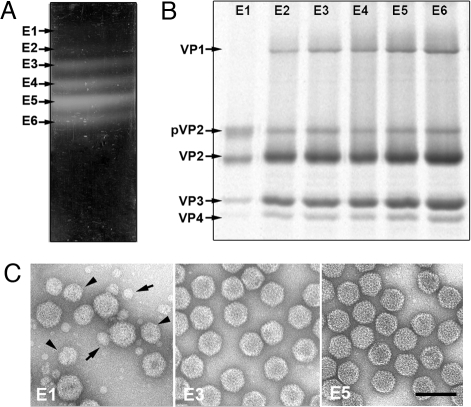
Despite the considerable detail available about the high resolution structure of most IBDV proteins, biochemical quantitation of its components is limited to sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) analysis and Coomassie blue staining; these calculations assume that polypeptides bind the dye in linear proportion to their molecular weight (20). Several bands are visible when IBDV virions, isolated from infected cultured cells or from the bursa of Fabricius from infected birds, are purified in CsCl gradients (21, 22). To determine the number of copies of IBDV structural proteins with accuracy, we purified IBDV virions by ultracentrifugation in CsCl gradients. At least 6 major bands were visible and termed E1 to E6 from top to bottom (Fig. 1A). The E5 fraction made up 48% of the total population. When cells were inoculated using E3–E6 fractions individually, the same 6 major populations were recapitulated (data not shown). These fractions were analyzed by SDS/PAGE (Fig. 1B) and electron microscopy (Fig. 1C). By visual inspection of the stained gel, it could be deduced that whereas E2–E6 fractions had similar protein profiles for structural proteins VP1, pVP2 and VP2, VP3, and VP4, the E1 fraction had a higher relative amount of pVP2 and contained no visible VP1. Electron microscopy analysis of the E2–E6 fractions showed homogeneous populations of 65- to 70-nm diameter icosahedral particles that correspond to T = 13 capsids (Fig. 1C, E3 and E5). In the E1 fraction, however, we observed complete virion-like particles, partially broken particles, and isometric assemblies (11) whose size corresponded to T = 1 (≈23 nm) and T = 7 (≈53 nm) particles (Fig. 1C, E1). The colocalization of these structures indicated that they have the same protein:RNA ratio. Moreover, considering that T = 1 capsids are built only of VP2 trimers (10), E1 fraction isometric assemblies must be devoid of nucleic acids. Due to its structural heterogeneity, the E1 fraction was excluded from further analyses.
Fig. 1.
Purification of IBDV natural populations. (A) A typical CsCl linear gradient for IBDV purification illuminated from the bottom after centrifugation to equilibrium, containing at least 6 IBDV fractions (denoted E1 to E6, from top to bottom). E1–E6 bands represent 3, 5, 12, 16, 48, and 16% of the total virion particles. (B) E1–E6 bands were collected by side puncture, analyzed by SDS/PAGE and developed by Coomassie staining. IBDV structural proteins are indicated. (C) Electron microscopy of purified E1–E6 populations negatively stained with 2% uranyl acetate. T = 7 capsid-like (arrows) and T = 1 capsid-like particles (arrowheads) are indicated for the E1 population. (Scale bar, 100 nm.)
Stoichiometry of IBDV Structural Components.
To determine whether E2–E6 capsids are biochemically identical, we used autoradiograms of SDS/PAGE gels to quantitate the structural proteins in [35S]Met-labeled preparations of all 5 types of particles (Table 1; supporting information (SI) Fig. S1).
Table 1.
Stoichiometry of IBDV polypeptides and dsRNA based on 35S labeling
| Protein, kDa | Residues (Met) | E2 | E3 | E4 | E5 | E6 | IBDV 35S-Avg* | IBDV blue-Avg† |
|---|---|---|---|---|---|---|---|---|
| VP1 (97) | 879 (21) | 7 ± 0.7 | 9 ± 1.4 | 13 ± 0.8 | 16 ± 1 | 15 ± 1.6 | 13 ± 3 | 41 ± 6 |
| pVP2 (54) | 512 (9) | 191 ± 13 | 75 ± 13 | 63 ± 5 | 37 ± 3 | 43 ± 3 | 55 ± 17 | 96 ± 12 |
| VP2 (48) | 441 (9) | 589 ± 79 | 705 ± 54 | 717 ± 59 | 743 ± 23 | 737 ± 42 | 725 ± 41 | 684 ± 28 |
| VP3 (28) | 256 (9) | 415 ± 30 | 487 ± 42 | 470 ± 55 | 468 ± 24 | 401 ± 50 | 457 ± 50 | 601 ± 60 |
| VP4 (25) | 244 (6) | 27 ± 2 | 33 ± 4 | 60 ± 3 | 61 ± 11 | 24 ± 3 | 45 ± 18 | 50 ± 20 |
*These values indicate the corresponding normalized average copy number of IBDV structural proteins/capsid for each population (E3-E6).
†Average copy number estimated from Coomassie blue-stained gels. These values are similar to previous analysis (20).
The biochemical quantitation data were comparable for IBDV E3–E6, supporting a similar protein composition for these populations independently of their buoyant density. Our calculations identified 12 copies of VP1/virion, an unexpectedly large copy number for the RdRp if we considerer that the IBDV genome complement is formed by 2 dsRNA molecules. Members of the family Reoviridae, whose genome consists of 10–12 segments of dsRNA packaged within the same particle, thus contain 12 RNA-polymerase complexes (Table s1). VP3 is found at a relatively constant amount, although it is not icosahedrally ordered. Finally, the viral protease VP4 can be considered as another structural component, although it is found in highly variable amounts.
For comparative purposes, we also performed stoichiometric analysis based on Coomassie blue-stained gels, as described in ref. 20. We obtained a systematic overestimation of copy ratios for proteins VP1 and VP3, indicating preferential staining of these polypeptides (Fig. S1).
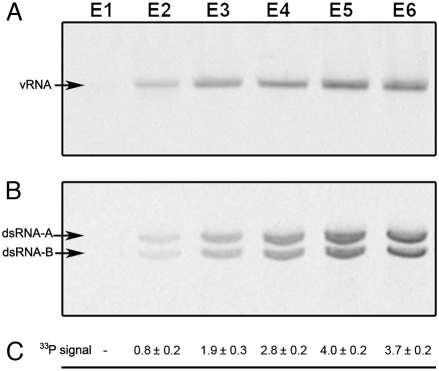
We also analyzed the nucleic acid component of the distinct IBDV populations from [33P]HPO4−2-metabolically labeled virions to determine whether the differences in buoyant density correlated with the dsRNA segment copy number inside the viral capsid. E3–E6 IBDV populations, which are infectious (see below) and have a similar protein composition, and the E2 population were analyzed by agarose gel electrophoresis and autoradiographed at the same protein concentration using intact (Fig. 2A) or SDS- and proteinase K-treated virions (Fig. 2B). The existence of a single band of defined mobility demonstrated the structural integrity of the virion particles E2–E6 (Fig. 2A, labeled as vRNA). Purified dsRNA segments A and B, obtained after SDS and proteinase K treatments, were observed as 2 well-defined bands (Fig. 2B, labeled as dsRNA-A and dsRNA-B). At equal particle concentrations, the amount of viral genomic dsRNA incorporated into the virion capsid increased from lower-density (E2) to higher-density (E6) IBDV populations, with equimolar amounts of segments A and B (Fig. 2B). These differences in radioactive content corresponded to different amounts of packaged genomic segments, since intermediate size segments were not detected; consequently, we normalized incorporated radioactivity relative to that of the E2 fraction (Fig. 2C). The data showed that the respective genome content of E3, E4, E5, and E6 populations are, respectively, 2-, 3-, 4-, and 4-fold higher than that of the E2 population. In this model, the number of packaged dsRNA segments is a natural integer of those ratios, and the IBDV particle is therefore able to package at least 4 dsRNA segments.
Fig. 2.
Genome stoichiometry of IBDV populations. Agarose gel electrophoresis and autoradiography of intact (A) or SDS- and proteinase K–treated (B) [33P]HPO4−2-labeled E1–E6 IBDV populations. The same amount of total protein was analyzed for each IBDV population. vRNA corresponds to viral RNA packaged inside intact virion particles; dsRNA-A and dsRNA-B correspond to purified dsRNA segments A and B, obtained after SDS and proteinase K treatments. (C) Relative IBDV dsRNA content based on 33P labeling for E2–E6 populations. Counts in the bands were measured and normalized with respect to the E2 population value. Average and standard deviation values were calculated from triplicate experiments.
Biophysical Analysis of IBDV Populations.
To obtain an independent estimate of the molecular weight of the E2–E6 populations, and to help interpret the quantitative biochemical results, we used analytical ultracentrifugation to study equivalent samples of the IBDV populations (Fig. S2). Most E2–E6 IBDV particles sedimented as a defined species with standard sedimentation coefficient values (s20,w) of 290, 336, 376, 418, and 416 S, respectively (Table 2). Dynamic light scattering experiments with the same viral particles gave similar standard translational diffusion coefficients D20,w (Table 2), compatible with a main scattering species for each IBDV population. From the combined s20,w and D20,w coefficients, the Svedberg equation allows determination of the molar mass of the E2–E6 viral particles in a shape-independent manner (23). We established that the main species of E2–E6 particles have molar masses of 52, 54, 56, 59, and 58 MDa, respectively. Considering the stoichiometry and molar masses of their structural proteins, the molecular mass of the protein component of the IBDV virion is 52 ± 2 MDa. In view of the dsRNA segment sizes (3.2 and 2.8 kbp), and assuming a molar mass of 682 Da/bp, the molar mass of the IBDV unit genome component is ≈4 MDa.
Table 2.
Biophysical properties of IBDV populations.
| IBDV | S20,w* | D20,w, cm2/s | Molecular mass, MDa | dsRNA molecules† |
|---|---|---|---|---|
| E2 | 290 ± 20 | 5.05 ± 0.25 | 52 ± 4 | 1 (54) |
| E3 | 336 ± 12 | 5.47 ± 0.20 | 54 ± 2 | 2 (56) |
| E4 | 370 ± 8 | 5.59 ± 0.15 | 56 ± 3 | 3 (58) |
| E5 | 418 ± 10 | 5.72 ± 0.12 | 59 ± 2 | 4 (60) |
| E6 | 416 ± 15 | 5.75 ± 0.10 | 58 ± 2 | 4 (60) |
*Standard sedimentation coefficient averaged over four independent experiments.
†dsRNA segment copy numbers compatible with experimental MW. MW of viron particle is indicated in parenthesis, assuming 52 ± 2 MDa for the protein component and 2 MDa for packaged dsRNA molecule.
These analyses support the argument that molar masses of E2–E6 virions are compatible with the incorporation of an increasing number of dsRNA molecules, with an average increment of 1 dsRNA molecule (average molecular mass of ≈2 MDa). Our data permit correlation of the relative amount of nucleic acid for IBDV populations with the number of dsRNA copies packaged in the virion, and suggest that IBDV populations correspond to empty particles (E1 fraction) and particles that package 1 (E2), 2 (E3), 3 (E4), and 4 (E5 and E6) dsRNA molecules.
Functional Analysis of IBDV Populations.
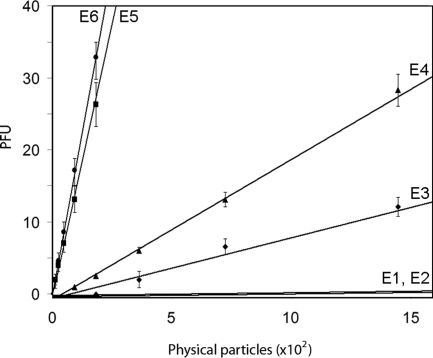
To determine whether differences in the packaged genome are related to the specific infectivity of viral particles [the ratio of physical particles (pp) to plaque-forming units (pfu)], we titrated the IBDV populations by plaque assay (Fig. 3). E1 particles were used as a negative control. The coincident infectivity profiles of E1 and E2 populations showed residual intrinsic infectivity limited by the purification of IBDV populations in the same CsCl gradient.
Fig. 3.
Titration of IBDV populations on QM7 monolayers. Number of particles was estimated from protein concentration measurements of purified E1–E6 populations. Error bars indicate the standard deviations of titrations of 3 independent experiments.
Plaque assays showed linear relationships with different slopes between pfu and pp number for the IBDV populations (Fig. 3). As a measure of fitness of the virus populations, we calculated pp/pfu ratios. The values obtained were ≈7 × 103 for E1 and E2, ≈100 for E3, ≈50 for E4, and ≈6 for E5 and E6. These results indicated the molecular basis of the observed phenotype, i.e., IBDV populations with a larger number of packed dsRNA segments were not only functional, but also correlated with higher infectivity ratios. The data obtained in our experimental settings suggest that infectivity is a function of nucleic acid content, but other factors (e.g., the pVP2/VP2 ratio) cannot strictly be excluded. Accordingly, it appears that the IBDV genetic program allocates 96% of the total infectivity within 64% of the viral progeny, corresponding to the polyploid virus population (E5 and E6) (Table s2). These findings suggest that IBDV particles composed of standard structural proteins in similar ratios can package multiple genomes to achieve higher specific infectivity.
Discussion
We characterized an icosahedral dsRNA virus, IBDV, and found that throughout its evolution, this virus has maintained a much larger capsid size than is necessary for enclosing a single genome copy. This finding is indeed surprising, since no other icosahedral viruses have been described for which selection pressure has acted to yield similar consequences.
Viral polyploidy is uncommon among viruses. The only reported polyploid viruses are those with a lipid bilayer, such as retroviruses, which have a diploid genome encased in a fullerene-based capsid, and several negative-strand RNA viruses, such as para- and orthomyxoviruses, which have a helical nucleocapsid arrangement with total or partial polyploidy, respectively. The volume enclosed by an icosahedral shell is nonetheless determined by capsid geometry, and defines genome size as well as the copy number of the encapsidated proteins.
Calculations based on structural analyses indicate a high degree of compaction for dsRNA and dsDNA viruses (1); this is reflected in an average genome density of ≈40 bp/100 nm3 (Table s1). To date, the exception to this rule is represented by fungal dsRNA virus cores that consistently exhibit a lower density (≈20 bp/100 nm3). These spacious capsids were explained on the basis of facilitating dsRNA mobility in the transcriptional and replicative active cores (24). For IBDV, given the volume occupied by the ≈12 copies of RdRp and the ≈450 copies of VP3 (assuming a protein density of 1.3 g/cm3), there is a clear discrepancy between the available space provided by the T = 13 capsid and that occupied by a single copy of the bipartite genome (Table s1). Packaging of 2 complete genomes or in general terms, 4 dsRNA molecules (≈12 kpb), as determined experimentally in this study, involves a dsRNA density value similar to that for dsRNA fungal virus. Even in capsids with a smaller triangulation number, such as the T = 7 capsid observed by heterologous expression of selected IBDV proteins (11), space would be sufficient to accommodate a single genome copy. Furthermore, fungal viruses with multipartite dsRNA genomes, such as chrysovirus (25) and partitivirus (26), package each dsRNA segment in separate particles rather than in a single capsid. This feature is also described for comovirus, a group of plant viruses with a genome consisting of 2 molecules of positive-strand ssRNA (27).
IBDV segments probably bear specific cis-acting packaging signals to discriminate between viral and cellular RNA. Random segment incorporation by highly multipartite genome viruses (such as reo- and orthomyxoviruses) would lead to a vanishingly small proportion of fully infectious particles. Thus, whereas multiple gene segments of influenza virus (28) and bacteriophage φ6 (29) are incorporated in a specific order, our data for IBDV (with only a bipartite genome) provide no evidence of a mechanism responsible for incorporating these segments in a given order. This random packaging is supported by the equimolar amounts of segments A and B observed in E2–E6 populations, and the fact that E2 particles, which package only 1 dsRNA molecule, have equal representation of segments A and B.
A random packaging mechanism would involve a pfu/pp ratio of 0.33 if only 2 dsRNA molecules were incorporated, whereas the incorporation of 4 molecules would increase 3-fold the probability of generating an infectious IBDV particle (Fig. S3). In this context, IBDV polyploidy results in a mechanism that notably increases the probability of the assembly of virion particles with a complete functional genome. E4–E6 populations, which might contain more than 1 complete genome, account for ≈80% of total particles (Table s2); functional analysis showed a direct correlation between the number of packaged segments and particle infectivity. Polyploidy also reduces sensitivity to genetic and environmental perturbation, i.e., increases robustness.
The molecular differences remain to be established between E5 and E6 particles, whose infectivity and sedimentation behavior differ slightly. Differences in small single-stranded oligonucleotide content, which forms up to 25% of the RNA in purified reovirus, may account for a higher sedimentation coefficient for the E6 population (30). Alternatively, E5 and E6 particles may have distinct properties, as reported for nodavirus infectious virions and virus-like particles with cellular RNA, which are crystallographically identical but have different sensitivities to proteases (31).
The viral replicative cycle of dsRNA viruses is marked by the ubiquitous presence of a T = 2 core in which individual mRNA is transcribed and negative-strand RNA is synthesized from genome segments that remain enclosed (32). This common, although not universal, capsid isolates the viral genome from suppression of gene expression by host responses. Birnaviruses are a well-established exception and have a single-shelled T = 13 capsid. Moreover, ribonucleoprotein complexes of birnaviruses (17) might also represent a functional evolutionary link with negative-strand ssRNA viruses, in which ribonucleoprotein filaments are common. Furthermore, IBDV RdRP activity is regulated by the VP3 C-terminal region, which acts as a transcriptional activator; in a similar manner, the polymerase complex of negative-strand RNA viruses can only copy the RNA sequence in the ribonucleoprotein complex, not naked RNA. It is currently impossible to identify conserved structural motifs, since the available VP3 structure lacks the RNA binding region, precisely where the N proteins from 3 negative-strand ssRNA virus families show a common topology (33). All these features together must involve profound differences with regard to the classical view of the dsRNA virus life cycle. In this scenario, where there is an excess of packaging capacity, reassortment of segments A and B—either after coinfection with other viral strains or, more probably, from naturally occurring variants during cell infection—might have been a essential mechanism in birnavirus evolution.
Materials and Methods
Purification of Virions.
QM7 quail muscle cells were infected with IBDV Soroa strain at a multiplicity of infection (MOI) of 1–2 pfu per cell. When cytopathic effect was complete (2 days postinfection), the cell medium was supplemented with polyethylene glycol 8000 and NaCl to a final concentration of 3.5% and 0.5 M, respectively. After an incubation (12 h, 4 °C), the virus was pelleted at 1,000 × g for 30 min and the resulting pellet was resuspended in PES buffer [25 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.2), 150 mM NaCl, and 20 mM CaCl2]. The virus was purified by pelleting through a 25% (wt/wt) sucrose cushion at 170,000 × g for 150 min, followed by a CsCl equilibrium gradient centrifugation for 14 h at 130,000 × g at 4 °C, adjusting the initial density of the solution to 1.33 g/ml by addition of CsCl. At least 6 virus-containing opalescent bands were visible when the tube was illuminated with white light from the bottom, and they were separately collected by side puncturing. Fractions (≈200 μl) were named E1 to E6 from top to bottom, dialyzed against PES buffer, and identified by SDS/PAGE as described in ref. 11.
Radiolabeling of Infected Cells.
QM7 cells were infected with IBDV at an MOI of 2 pfu/ml. After 1 h of adsorption, the inoculum was replaced with DMEM supplemented with 2% FCS containing 100 μCi/ml [35S]methionine or [33P]HPO4−2, respectively. Cell medium of IBDV-labeled virions was mixed with media of unlabeled, IBDV-producing cells and further purified by the standard protocol described above. [35S]Met- or [33P]HPO4−2-labeled virions were analyzed by SDS/PAGE, or in 0.7% agarose gels containing 90 mM Tris·HCl (pH 8), 90 mM boric acid, and 20 mM EDTA. To analyze [33P]HPO4−2-labeled dsRNA content, E1–E6 samples were incubated with 1% SDS (3 min, 100 °C) and treated with proteinase K (2 mg/ml; 1 h, 37 °C). dsRNA was then extracted with TriZol (Invitrogen) and purified using the silica-based minispin column (Quiagen). Gels were dried on 3MM paper and visualized by phosphorimaging (Storm 869, Molecular Dynamics). Data were quantitated by using Quantity One software (BioRad). To calculate protein stoichiometry of the viral particle, the counts in the bands were measured and normalized considering the number of Met residues (21, 9, 9, and 6 for VP1, (p)VP2, VP3, and VP4, respectively; the C-terminal region of pVP2 that is processed during virus maturation has no Met residues). The copy numbers of VP1, VP3, and VP4 molecules were normalized to the internal standard given by the 780 copies of VP2 and pVP2, as unequivocally established by X-ray crystallography and the 3D cryoelectron microscopy of virions (10, 11). The ratio of pVP2 to VP2 for E2–E6 populations was determined in each 35S-labeling experiment. Data are averages from triplicate experiments.
Electron Microscopy of IBDV Particles.
Samples (≈5 μl) were applied to glow-discharged carbon-coated grids for 2 min. Samples were stained with 2% aqueous uranyl acetate. Micrographs were recorded with a JEOL 1200 EXII electron microscope operating at 100 kV at a nominal magnification of ×40,000.
Biophysical Analysis of E2–E6 IBDV Population Particles.
Sedimentation velocity.
Experiments were done using IBDV particles E2–E6 (at 0.1–0.5 mg/ml) equilibrated in PES buffer. Sedimentation velocity runs were carried out at 10,000 rpm and 20 °C in an XLA-analytical ultracentrifuge (Beckman-Coulter) using an An50Ti rotor and 12-mm double-sector centerpieces, and measuring absorbance at the appropriate wavelength. Sedimentation coefficient distribution was calculated by least-squares boundary modeling of sedimentation velocity data using the c(s) method (34). These s-values were corrected to standard conditions [water, 20 °C, and infinite dilution (23)] to obtain the corresponding standard s-values (s20,w).
Dynamic light scattering (DLS).
DLS experiments were conducted with a Protein Solutions DynaPro-MS/X instrument. Buffers were filtered by a 100-nm filter with a syringe, and protein solutions were centrifuged at 10,000 × g to remove dust particles. Samples (20 μl) were inserted in the 90° light scattering cuvette at 20 °C. Diffusion coefficients of the scattering solute particles (assuming a globular shape of the protein species) were calculated using the SEDFIT program adapted for DLS by replacing the single species Lamm equation solutions by the intensity correlation function (35). These experimental values were corrected for buffer and concentration (23) to get the standard D0 coefficients. Molecular masses of E2–E6 IBDV particles were calculated by the Svedberg equation.
Virus Plaque Titration.
Plaque assays were determined on QM7 cell monolayers (106 cells per assay). Briefly, virus stocks were serially diluted with serum-free DMEM, incubated (1 h) to allow the virus to adsorb to the cells, and 0.5% agar in DMEM supplemented with 2% FCS was added to each plate. Cells were inoculated in triplicate with each virus dilution. After 48 h, cells were fixed in 10% formaldehyde and stained with 2% crystal violet. For gradient-purified E1–E6 band IBDV populations, the amount of total protein was determined using the BCA protein assay kit (Pierce), and infections were performed at the same protein concentration for each IBDV population.
Supplementary Material
Acknowledgments.
We thank C. Mark for editorial help. This work was supported by grants BFU 2005–06487 and BIO 2006–09407 from the Spanish Dirección General de Investigación (MEC).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808498106/DCSupplemental.
References
- 1.Casjens S. In: Structural Biology of Viruses. Chiu W, Burnett RM, Garcea RL, editors. New York: Oxford Univ Press; 1997. pp. 3–37. [Google Scholar]
- 2.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: An inseparable pair. Nat Rev Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 3.Noda T, et al. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 4.Enami M, Sharma G, Benham C, Palese P. An influenza virus containing nine different RNA segments. Virology. 1991;185:291–298. doi: 10.1016/0042-6822(91)90776-8. [DOI] [PubMed] [Google Scholar]
- 5.Rager M, Vongpunsawad S, Duprex WP, Cattaneo R. Polyploid measles virus with hexameric genome length. EMBO J. 2002;21:2364–2372. doi: 10.1093/emboj/21.10.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison SC. In: Fields Virology. 5th Ed. Knipe DM, et al., editors. Vol 1. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 59–98. [Google Scholar]
- 7.Cerritelli ME, et al. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 8.Delmas B, et al. In: Virus Taxonomy. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Amsterdam: Elsevier Academic; 2005. pp. 561–569. [Google Scholar]
- 9.Castón JR, et al. C terminus of infectious bursal disease virus major capsid protein VP2 is involved in definition of the T number for capsid assembly. J Virol. 2001;75:10815–10828. doi: 10.1128/JVI.75.22.10815-10828.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulibaly F, et al. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell. 2005;120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Saugar I, et al. Structural polymorphism of the major capsid protein of a double-stranded RNA virus: An amphipathic α helix as a molecular switch. Structure. 2005;13:1007–1017. doi: 10.1016/j.str.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Feldman AR, Lee J, Delmas B, Paetzel M. Crystal structure of a novel viral protease with a serine/lysine catalytic dyad mechanism. J Mol Biol. 2006;358:1378–1389. doi: 10.1016/j.jmb.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 13.Pan J, Vakharia VN, Tao YJ. The structure of a birnavirus polymerase reveals a distinct active site topology. Proc Natl Acad Sci USA. 2007;104:7385–7390. doi: 10.1073/pnas.0611599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casañas A, et al. Structural insights into the multifunctional protein VP3 of birnaviruses. Structure. 2008;16:29–37. doi: 10.1016/j.str.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 15.Garriga D, et al. Activation mechanism of a noncanonical RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 2007;104:20540–20545. doi: 10.1073/pnas.0704447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maraver A, Clemente R, Rodríguez JF, Lombardo E. Identification and molecular characterization of the RNA polymerase-binding motif of infectious bursal disease virus inner capsid protein VP3. J Virol. 2003;77:2459–2468. doi: 10.1128/JVI.77.4.2459-2468.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hjalmarsson A, Carlemalm E, Everitt E. Infectious pancreatic necrosis virus: Identification of a VP3-containing ribonucleoprotein core structure and evidence for O-linked glycosylation of the capsid protein VP2. J Virol. 1999;73:3484–3490. doi: 10.1128/jvi.73.4.3484-3490.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahlquist P. Virus evolution: Fitting lifestyles to a T. Curr Biol. 2005;15:R465–R467. doi: 10.1016/j.cub.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat Rev Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobos P, et al. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J Virol. 1979;32:593–605. doi: 10.1128/jvi.32.2.593-605.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller H, Lange H, Becht H. Formation, characterization and interfering capacity of a small plaque mutant and of incomplete virus particles of infectious bursal disease virus. Virus Res. 1986;4:297–309. doi: 10.1016/0168-1702(86)90008-0. [DOI] [PubMed] [Google Scholar]
- 22.Muller H, Becht H. Biosynthesis of virus-specific proteins in cells infected with infectious bursal disease virus and their significance as structural elements for infectious virus and incomplete particles. J Virol. 1982;44:384–392. doi: 10.1128/jvi.44.1.384-392.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Holde K. Physical biochemistry. Englewood Cliffs, NJ: Prentice Hall; 1985. pp. 110–136. [Google Scholar]
- 24.Castón JR, et al. Structure of L-A virus: A specialized compartment for the transcription and replication of double-stranded RNA. J Cell Biol. 1997;138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castón JR, et al. Three-dimensional structure of Penicillium chrysogenum virus: A double-stranded RNA virus with a genuine T=1 capsid. J Mol Biol. 2003;331:417–431. doi: 10.1016/s0022-2836(03)00695-8. [DOI] [PubMed] [Google Scholar]
- 26.Ochoa WF, et al. Partitivirus structure reveals a 120-subunit, helix-rich capsid with distinctive surface arches formed by quasisymmetric coat-protein dimers. Structure. 2008;16:776–786. doi: 10.1016/j.str.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomonossoff GP, Johnson JE. The synthesis and structure of comovirus capsids. Prog Biophys Mol Biol. 1991;55:107–137. doi: 10.1016/0079-6107(91)90003-b. [DOI] [PubMed] [Google Scholar]
- 28.Muramoto Y, et al. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J Virol. 2006;80:2318–2325. doi: 10.1128/JVI.80.5.2318-2325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mindich L. Packaging, replication and recombination of the segmented genome of bacteriophage Phi6 and its relatives. Virus Res. 2004;101:83–92. doi: 10.1016/j.virusres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Bellamy AR, Joklik WK. Studies on the A-rich RNA of reovirus. Proc Natl Acad Sci USA. 1967;58:1389–1395. doi: 10.1073/pnas.58.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bothner B, et al. Crystallographically identical virus capsids display different properties in solution. Nat Struct Biol. 1999;6:114–116. doi: 10.1038/5799. [DOI] [PubMed] [Google Scholar]
- 32.Reinisch KM. The dsRNA viridae and their catalytic capsids. Nat Struct Biol. 2002;9:714–716. doi: 10.1038/nsb1002-714. [DOI] [PubMed] [Google Scholar]
- 33.Luo M, Green TJ, Zhang X, Tsao J, Qiu S. Structural comparisons of the nucleoprotein from three negative strand RNA virus families. Virol J. 2007;4:72. doi: 10.1186/1743-422X-4-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: Strategies and application to model systems. Biophys J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuck P, Taraporewala Z, McPhie P, Patton JT. Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes. J Biol Chem. 2001;276:9679–9687. doi: 10.1074/jbc.M009398200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.