The end of the 20th century was marked by the genomics revolution, and one might say that the beginning of the 21st century is marked by efforts to bring our knowledge of a cell's proteins, known as proteomics, to a par with our growing knowledge of a cell's transcripts, known as transcriptomics. Many of us believe that the next evolution of the omics revolution will be to map all of the metabolites of a cell, known as metabolomics. Leading the way in these efforts is the work in many laboratories on one subset of the metabolome, the lipidome, aimed at mapping all of the lipids of a cell, known as lipidomics (1–4). The ultimate goal is to evolve an integrated omics picture (the “interactome”) of the genes, transcripts, proteins, and metabolites to fully describe cellular functioning. One leading effort in the lipidomics evolution is described in this issue of PNAS (5).
Ejsing et al. (5) have used advanced mass spectrometry (MS) techniques combined with state-of-the-art data analysis software to identify 342 lipid molecular species in the yeast lipidome and to quantify them; they estimate that their findings constitutes 95% of the lipid molecules present putting its coverage on a par with the early efforts at gene sequencing. This effort allowed these authors to describe the major lipid molecular species comprising the yeast Saccharomyces cerevisiae membranes. These include glycerophospholipids, sphingolipids, and sterols as well as the neutral glycerolipids. The authors have described the detailed metabolic pathways for the interacting of these diverse lipid species.
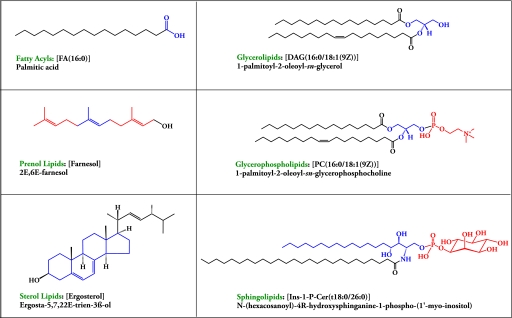
The LIPID MAPS Initiative in lipidomics (1, 4) in conjunction with the International Committee for the Classification and Nomenclature of Lipids (ICCNL) have defined 8 categories of lipids and numerous classes and subclasses (6, 7) to allow one to describe lipid molecular species. Fig. 1 illustrates examples of molecular species of lipids in each of 6 of these categories that exist in yeast, although the free fatty acids and prenols were not reported in the present study. (See www.lipidmaps.org.) The goal of lipidomics is to define and quantitate all of the lipid molecular species in a cell, but this is complicated by the extraordinary number of combinations possible with the large number of known fatty acids that can occupy in various combinations the 3 positions on the glycerol backbone of monoacylglycerols (MAGs), diacylglycerols (DAGs), and triacylglycerols (TAGs). Similarly, in monoacylglycerolphosphates (lysophospholipids) and diacylglycerolphosphates (phospholipids), fatty acyl groups can occupy 1 or 2 positions on the glycerol backbone, respectively, and the phosphate can be esterified to a large variety of polar head groups. Thus, the possible number of molecular species for a given set of fatty acids (and polar head groups) is very large and so far it has not been straightforward to define all of the molecular species and their stereochemistry for a given lipid. Even the sterol esters that sometimes occur with a single fatty acid chain, the sphingolipids with a defined fatty acid-derived hydrocarbon chain, and an amide-linked fatty acid have numerous possibilities.
Fig. 1.
Examples of lipid categories present in yeast. Six of the 8 categories of lipids in the LIPID MAPS comprehensive classification system for lipids (6, 7) are illustrated with an example for each drawn by the Structure Drawing Program (www.lipidmaps.org).
Genes and transcripts do not always predict the precise levels of active proteins/enzymes.
Ejsing et al. (5) in choosing yeast have simplified the challenge because these cells can be grown on a minimal medium lacking lipids and this organism is capable of only synthesizing a limited number of fatty acids, such as palmitic acid (16:0), stearic acid (18:0), etc., and can only desaturate in the Δ9 position to give a limited number of unsaturated fatty acids, such as palmitoleic acid (16:1 cis-9) and oleic acid (18:1 cis-9). This has greatly simplified the task involved in studying the lipidome of yeast compared with the challenges of mammalian cells, where there are tens of thousands of distinct molecular species of lipids.
There are 2 fundamentally different approaches to lipidomics analysis. (i) The LIPID MAPS Consortium has chosen to develop extraction protocols optimized for each lipid category, and in some cases specific classes of lipids, and then to use liquid chromatography (LC) to optimally separate the specific molecular species of lipids present (8–13). The LC eluate is then coupled directly to a mass spectrometer for further online analysis such as molecular fragmentation (MSMS), multiple reaction monitoring (MRM), and multiple precursor ion scanning (MPIS). In short, a kind of divide-and-conquer strategy. (ii) An alternative approach, sometimes referred to as “shotgun lipidomics” as used by Ejsing et al. (5), begins with an off-line lipid extraction, but then subjects these extracts directly to MS analysis without LC separation. In the specific study reported herein, Ejsing et al. actually subjected their yeast cells to 2 different extraction protocols, one to separate the more hydrophobic lipids and a second extraction with a more polar solvent. In some runs, they first acetylated the sample or included methylamine in the procedure to optimize the analysis. Additionally, their analysis used 2 different types of mass spectrometers, a quadrupole time-of-flight instrument in which they used MRM and MPIS and a linear ion trap-orbitrap instrument that is capable of providing extremely accurate mass determinations. With each instrument, they analyzed samples in both negative and positive ionization modes. The extremely complicated data output was then deciphered by using sophisticated analysis software. Their shotgun lipidomics approach allowed them to identify in yeast some 21 different lipids that were quantitated as 342 unique molecular species.
The use of some specialized MS scan modes (e.g., MRM) allows for high sensitivity; however, a drawback is that one only finds what one is looking for. Combined with the use of internal standards, these techniques allow for the identification and quantification of the lipids known to be present, but do not, by themselves, lead to the finding of novel or unexpected molecular species. Ejsing et al. (5) made critical use of multiple lipid standards obtained from Avanti Polar Lipids which had been developed under LIPID MAPS auspices (14), but they also had to develop their own standards for some specific yeast phytosphingolipids. The yeast lipidome is characterized by the usual TAG, DAG, and phospholipid species found in mammalian cells, but in addition has unusual phytosphingolipid species in which the long-chain base and the fatty acyl amide moieties are characterized by hydroxylated species (e.g., phytoceramides) and an abundance of the sterol ergosterol rather than the mammalian cholesterol.
Much is known about the yeast lipidome from prior traditional studies on the enzymes of lipid metabolism and enhanced considerably by gene sequencing and genetic manipulation (15, 16). However, genes and transcripts do not always predict the precise levels of active proteins/enzymes, and knowledge of the actual lipid metabolite levels is more predictive of metabolic implications. The article of Ejsing et al. (5) provides quantitative information on the levels of the lipid metabolites and their changes with specific gene deletions that enhances our understanding of metabolism enormously. Furthermore, they have been able to demonstrate in quantitative terms the subtle changes in lipid molecular species when yeast is grown at different temperatures and the “ripple” effects through multiple classes of membrane lipids.
Acknowledgments.
Dr. Eoin Fahy aided in the preparation of Fig. 1. This work was supported by National Institutes of Health Large Scale Collaborative “Glue” Grant U54 GM069338 for the LIPID MAPS initiative and National Institutes of Health Grants R01 GM 20,501 and R01 GM 64,611.
Footnotes
The author declares no conflict of interest.
See companion article on page 2136.
References
- 1.Dennis EA, et al. The LIPID MAPS approach to lipidomics. In: Feng L, Prestwich G, editors. Functional Lipidomics. London: CRC Press/Taylor & Francis Group; 2005. pp. 1–15. [Google Scholar]
- 2.van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 4.Schmelzer K, Fahy E, Subramaniam S, Dennis EA. The LIPID MAPS initiative in lipidomics. Methods Enzymol. 2007;432:169–181. doi: 10.1016/S0076-6879(07)32007-7. [DOI] [PubMed] [Google Scholar]
- 5.Ejsing CS, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fahy E, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Fahy E, et al. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2008 doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krank J, Murphy RC, Barkley RM, Duchoslav E, McAnoy A. Qualitative analysis and quantitative assessment of changes in neutral glycerol lipid molecular species within cells. Methods Enzymol. 2007;432:1–20. doi: 10.1016/S0076-6879(07)32001-6. [DOI] [PubMed] [Google Scholar]
- 9.Ivanova P, Milne S, Byrne MO, Xiang Y, Brown HA. Glycerophospholipid identification and quantitation by electrospray mass spectrometry. Methods Enzymol. 2007;432:21–57. doi: 10.1016/S0076-6879(07)32002-8. [DOI] [PubMed] [Google Scholar]
- 10.Deems RA, Buczynski MW, Bowers-Gentry RC, Harkewicz R, Dennis EA. Detection and quantitation of eicosanoids via high performance liquid chromatography/electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:59–82. doi: 10.1016/S0076-6879(07)32003-X. [DOI] [PubMed] [Google Scholar]
- 11.Sullards MC, et al. Structure-specific, quantitative methods for analysis of sphingolipids by liquid-chromatography tandem mass spectrometry: “Inside-out” sphingolipidomics. Methods Enzymol. 2007;432:83–115. doi: 10.1016/S0076-6879(07)32004-1. [DOI] [PubMed] [Google Scholar]
- 12.Garrett TA, Guan Z, Raetz CR. Analysis of ubiquinones, dolichols and dolichol diphosphate-oligosaccharides by liquid chromatography-electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:117–142. doi: 10.1016/S0076-6879(07)32005-3. [DOI] [PubMed] [Google Scholar]
- 13.McDonald JG, Thompson B, McCrum EC, Russell DW. Extraction and analysis of sterols in biological matrices by high-performance liquid chromatography electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:143–168. doi: 10.1016/S0076-6879(07)32006-5. [DOI] [PubMed] [Google Scholar]
- 14.Moore JD, Caufield WV, Shaw W. Quantitation and standardization of lipid internal standards for mass spectroscopy. Methods Enzymol. 2007;432:351–367. doi: 10.1016/S0076-6879(07)32014-4. [DOI] [PubMed] [Google Scholar]
- 15.Carman GM, Henry SA. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 2007;282:37293–37297. doi: 10.1074/jbc.R700038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannun YA, Obeid LM. Principles of bioactive lipid signaling: Lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]