Abstract
We explore how inherent flexibility of a protein molecule influences the mechanism controlling allosteric transitions by using a variational model inspired from work in protein folding. The striking differences in the predicted transition mechanism for the opening of the two domains of calmodulin (CaM) emphasize that inherent flexibility is key to understanding the complex conformational changes that occur in proteins. In particular, the C-terminal domain of CaM (cCaM), which is inherently less flexible than its N-terminal domain (nCaM), reveals “cracking” or local partial unfolding during the open/closed transition. This result is in harmony with the picture that cracking relieves local stresses caused by conformational deformations of a sufficiently rigid protein. We also compare the conformational transition in a recently studied even–odd paired fragment of CaM. Our results rationalize the different relative binding affinities of the EF-hands in the engineered fragment compared with the intact odd–even paired EF-hands (nCaM and cCaM) in terms of changes in flexibility along the transition route. Aside from elucidating general theoretical ideas about the cracking mechanism, these studies also emphasize how the remarkable intrinsic plasticity of CaM underlies conformational dynamics essential for its diverse functions.
Keywords: allostery, conformational, cracking, protein, topology
To understand protein function, it is often essential to characterize large conformational changes that occur upon ligand binding. Within the population shift mechanism of allosteric conformational change (1), the bound complex is formed when a ligand selects and stabilizes a weakly populated conformational ensemble from the kinetically accessible states of the unbound folded protein. Such conformational dynamics imply an inherent flexibility or “intrinsic plasticity” of the folded state. In this article, we focus on how this inherent flexibility of a protein molecule influences the mechanism controlling the kinetics of allosteric transitions. There are a couple of possible scenarios for the transition mechanism in terms of changes in conformational flexibility. One possibility is that the flexibility adjusts smoothly to conformational deformations connecting two specific metastable states in the free-energy surface. In this case, the inherent flexibility of the protein remains relatively constant if the metastable states have similar flexibilities or else changes smoothly between the flexibilities of the metastable states if the flexibilities are different. An alternative mechanism, called “cracking” (2), has recently been proposed. In terms of conformational flexibility, cracking involves nonmonotonic changes in flexibility along the transitions route. In particular, the flexibility of specific regions of the protein may transiently increase through local unfolding. As explained in ref. 2, local unfolding can relieve specific areas of high stress during conformational deformations that would result in high free-energy barriers if the protein remained uniformly folded throughout the transition.
In the model developed to explore this idea, cracking is introduced directly into the formalism as means to incorporate nonlinear elasticity in an otherwise harmonic description of conformational fluctuations (2, 3). This work clearly shows that local unfolding can dramatically lower predicted free-energy barriers between local free-energy minima and hence facilitates faster kinetics. In contrast, the variational model developed in the present work does not assume cracking from the outset. The evidence of cracking here arises entirely from the analysis of the local changes in flexibility along transition routes predicted by an inherently nonlinear model of conformational transitions. Our results are thus an independent verification of the ideas and formalism developed in the model presented in refs. 2 and 3.
Cracking does not occur in all conformational transitions, but even when it does, the average conformations of the local minima in the free energy may show little signs that local unfolding is involved (2). Instead, cracking is a subtle consequence of the nature of the conformational deformation required to connect the two states. Because cracking is akin to folding of the whole protein, albeit on a constrained and local scale, it is reasonable to ask whether insight appropriated from the energy landscape theory of protein folding (4) can help to anticipate when cracking is likely to be important in conformational transitions between two distinct folded conformations.
One motivation for the work in this article is to investigate whether the cracking mechanism of conformational transitions, like the mechanism for folding of two-state proteins (5), is determined by the structural topology of the two metastable states. To address this question, we study the conformational transitions of the open and closed conformations of the two homologous domains of calmodulin (CaM) and a fragment of CaM involving parts of both domains. Aside from general theoretical implications, the results presented in this article are interesting from the point of view of understanding the well-known and intensely studied thermodynamic differences between the two domains of CaM (6, 7).
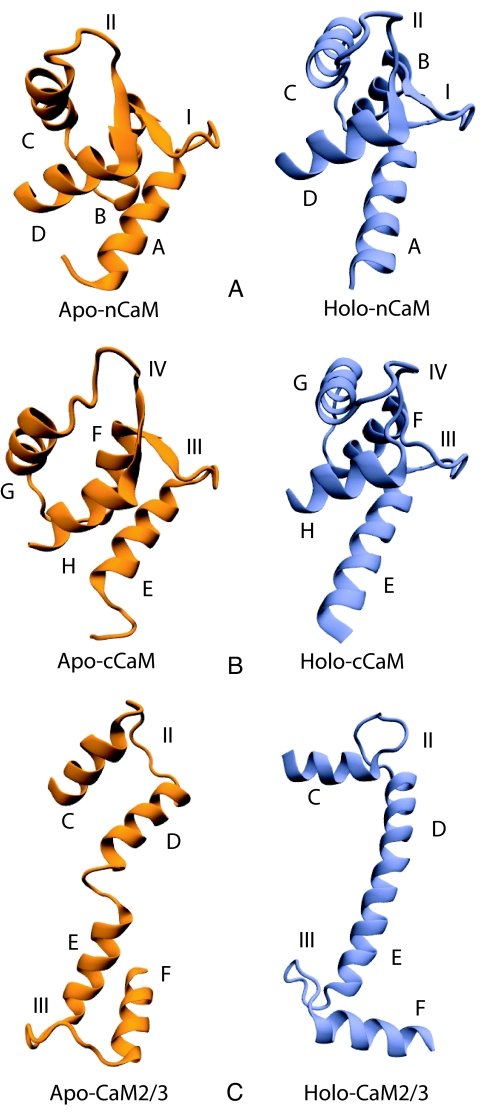
Our approach is based on a coarse-grained variational model developed to characterize protein folding (8). This model is in harmony with several recent coarse-grained simulations based on a folded-state biased interaction potential that interpolates between the contact maps of the metastable states (9–15). Interestingly, some of these recently proposed coarse-grained models can also capture the cracking or local partial unfolding during transition (10, 12, 15). In this article we compare the detailed predicted transition routes for the open/closed conformational change of the C-terminal domain of CaM (cCaM) with that of the N-terminal domain of CaM (nCaM). The open and closed state of each domain is shown in Fig. 1 A and B. Each domain of CaM contains a pair of helix–loop–helix Ca2+-binding motifs called EF-hands. In nCaM, EF-hands 1 and 2 consist of helices A/B and C/D (Fig. 1A), respectively, whereas helices E/F and G/H in cCaM (Fig. 1B) form EF-hands 3 and 4, respectively. Hence, the nCaM and cCaM are known as “odd–even” paired EF-hands because of their odd–even sequential numbering of EF-hands.
Fig. 1.
Three-dimensional structures of CaM domains and EF-hands 2 and 3 fragment. The apo-CaM and holo-CaM structures shown here are correspond to human CaM with PDB ID codes 1cfd and 1cll, respectively. (A) The closed, apo and open, holo conformations of nCaM consist of helices A/B and C/D with binding loops I and II, respectively. (B) The closed, apo and open, holo conformations of cCaM consist of helices E/F and G/H with binding loops III and IV, respectively. (C) The apo and holo conformations of CaM2/3 consist of helices C/D and E/F with binding loops II and III, respectively. These three-dimensional illustrations were made by using Visual Molecular Dynamics (VMD) (16).
Although nCaM and cCaM have similar structures, the calculated transition routes predict that the cracking is essential only in cCaM. It is well established that the two domains differ in flexibility and binding affinity for Ca2+ (6, 7). These properties are not determined by topology (because they are the same) but ultimately by more subtle structural differences encoded in the sequence of the two domains. Our results suggest that nCaM is flexible enough that cracking is not necessary to relieve stress during the conformational change, whereas the cCaM is relatively rigid so that cracking is involved in essentially the same conformational change. This is the central result of our article.
As an additional illustration of modeling conformation transitions in CaM, we investigate the conflicting Ca2+-binding mechanism in the odd–even paired EF-hands (nCaM and cCaM) and “even–odd” paired EF-hands (CaM2/3) of CaM. The fragment CaM2/3 contains EF-hands 2 and 3 with helices C/D and E/F (Fig. 1C), respectively. CaM2/3 is known as even–odd paired EF-hands because of its even–odd sequential numbering of EF-hands. In CaM2/3, an EF-hand from each domain is connected by a flexible helical linker. The extended helical linker separating the two domains shown in the X-ray structure of holo-CaM is known to be flexible in solution (17, 18), making holo-CaM structurally more flexible overall than apo-CaM (19, 20).
In contrast to cCaM and nCaM (20), CaM2/3 has a molten globule-like character in the absence of Ca2+ and folds into a structure similar to an intact domain of CaM in the presence of Ca2+ (21). Accordingly, a realistic model of Ca2+ binding to CaM2/3 would have to treat the ion binding and folding on equal footing as was done for folding azurin in the presence of zinc ions in ref. 22. At the risk overly simplifying the binding mechanism of CaM2/3, we keep the focus of this article on conformational transition routes between two specified conformations instead of binding-induced folding. In particular, we take the two states of CaM2/3 from the X-ray structure of intact CaM as shown Fig. 1C. Although this is a convenient way to define the conformation endpoint structures (and has a certain consistency with our studies of cCaM and nCaM), both the apo-CaM2/3 and the holo-CaM2/3 structures are expected to be low-probability conformations of CaM2/3 in solution: the apo-CaM structure assumed here is unstable to a molten globule state, and the holo-CaM structure inferred from the X-ray structure of CaM is unstable to a structurally compact domain like holo-cCaM or holo-nCaM (21).
Our study of the transition between the conformations of CaM2/3 defined by the structures of the intact CaM suggests that the highly flexible central linker region would locally unfold before gaining helical structure. Even though this model of the binding dynamics of CaM2/3 is based on low-probability conformations in solution (17, 18), our predicted transition route can rationalize the sequential Ca2+ binding in CaM2/3 (21) in terms of the differences in conformational flexibility of Ca2+-binding loops in CaM2/3. Finally, we address how our comparative study of conformational transitions in odd–even and even–odd paired EF-hands reveals the interplay between intrinsic plasticity, target binding affinity, and function of CaM.
Results
Conformational Flexibility of the CaM Domains.
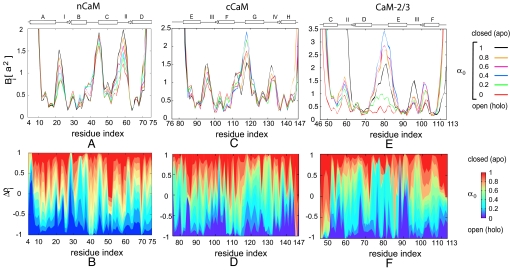
We first compare the inherent flexibility of the two domains of CaM by calculating the mean square fluctuations Bi for each residue in the nCaM and cCaM for the open/closed conformational transitions of each domain. The Bi (related to the temperature factors from X-ray crystallography) contains information about the degree of structural order and conformational flexibility of each residue. Fig. 2A shows Bi along the open/closed conformational transition for each residue of nCaM (23). The magnitude of the fluctuations for each residue of cCaM for the open/closed transition route is shown in Fig. 2C. Comparison of Fig. 2 A and C shows that although the binding loops and helix linker in both domains are very flexible, the apo (closed)-nCaM is inherently more flexible than apo (closed)-cCaM. For nCaM, the flexibility of the binding loops I and II and the B/C helix linker between EF-hands 1 and 2 decreases upon domain opening. Similarly, the flexibility of the Ca2+-binding loops III and IV decreases considerably during the opening of cCaM. The fluctuations of the F/G helix linker between EF-hands 3 and 4 has different behavior along the transition route. The flexibility of this region of the protein increases to a relatively high flexibility before reducing to the flexibility of the folded open conformation. (Fig. 2C). We also note that the F/G helix linker in the open structure is more flexible than in the closed structure. The calculated conformational flexibility suggests that unlike the B/C helix linker in the apo (closed)-nCaM the F/G helix linker in the apo (closed)-cCaM is relatively less flexible. In contrast to the B/C helix linker of nCaM, the increase and then decrease in flexibility of the F/G helix linker during the domain opening of cCaM indicates cracking or local partial unfolding in this less flexible F/G helix linker (See the change in fluctuations of the F/G helix linker during domain opening in Fig. 2C.) The model also predicts that binding loop II in nCaM has the highest flexibility compared with other binding loops. This result agrees with a recent molecular dynamics simulation study of CaM D129N mutant, where the binding loop II consistently exhibited higher mobility than the other three Ca2+-binding loops (24).
Fig. 2.
Structural characterization of transition routes. (A, C, E) The fluctuations Bi vs. residue index for selected values of the interpolation parameter α0 of nCaM, cCam, and CaM2/3, respectively. Here, a = 3.8 Å is the distance between successive monomers in our model. The secondary structures for nCaM, cCaM, and CaM2/3 are indicated above each plot. Helices are represented by the rectangular boxes, binding loops and helix linker are indicated by lines, and small β-sheets are shown by arrows. (B, D, and F) Difference between the normalized native density Δρi (a measure of structural similarity) vs. residue index for different α0 of nCaM, cCaM, and CaM2/3, respectively. In each plot the change in color from red to blue indicates the progress along the transitin from the apo to holo conformations. This is normalized to be −1 at the holo state minimum(α0 = 0; blue) and 1 at the apo state minimum (α0 = 1; red). (Parts A and B reproduced with permission from S Tripathi and JJ Portman, Journal of Chemical Physics, Vol. 128, page 205104, 2008. Copyright 2008, American Institute of Physics.)
Cracking in the Conformational Transition of cCaM.
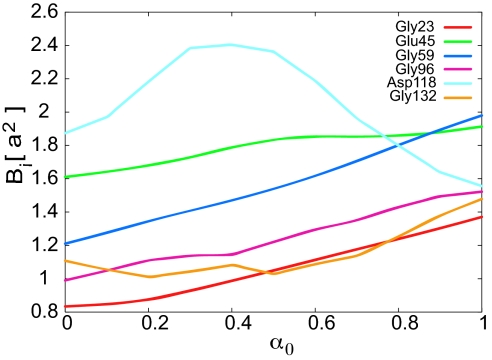
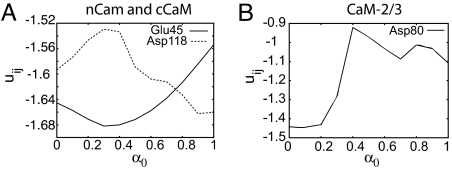
To compare the conformational flexibility of the two CaM domains in more detail we have plotted the change in fluctuations Bi for some specific residues in Fig. 3. Residue Asp-118 of the F/G helix linker from cCaM shows very different behavior relative to the rest of the residues in Fig. 3. Residue Asp-118 shows the highest Bi near the transition state at α0 = 0.4, whereas Bi of the other residues shown decrease monotonically during closed to open transition. The increase in flexibility of the helix linker region in cCaM near the transition state is caused by local transient unfolding or cracking. For further analysis of cracking in the cCaM we also compare the contact pair potential energy uij for residues Glu-45 and Asp-118 from the helix linker of nCaM and cCaM, respectively. Fig. 4A shows that for residue Asp-118, uij increases in the transition state region, whereas uij for residue Glu-45 from the helix linker of nCaM decreases in the transition state region. This striking difference in uij implies that the increase in contact energy of the F/G helix linker during opening of cCaM leads to some transient cracking in this region near the transition state, enhancing the inherent flexibility of this linker during the open/closed conformational change.
Fig. 3.
Change in fluctuations Bi of the residues from binding loops and helix linker of CaM domains along the open/closed conformational transition route for different α0. Here, a = 3.8 Å is the distance between successive monomers in our model. Residues Gly-23, Gly-59, and Glu-45 are from binding loop I, loop II, and the helix linker of the nCaM, respectively. Residues Gly-96, Gly-132, and Asp-118 are from binding loop III, loop IV, and the helix linker of the cCaM, respectively.
Fig. 4.
Average pair potentials uij (in kilocalories per mole) of selected residues along the open/closed conformational transition route for different α0. (A) Residues Glu-45 and Asp-118 are from the nCaM and cCaM, respectively. (B) Residue Asp-80 is from the central linker of CaM.
Open/Closed Transition Mechanism of the CaM Domains.
To elucidate further the predicted conformational transition mechanisms of domain opening in CaM, we consider a structural order parameter that measures the similarity to the open (holo) or closed (apo) state conformations, Δρi, described in Methods. This order parameter is defined such that Δρi = 1 corresponds to the closed conformation and Δρi = −1 corresponds to the open conformation of nCaM or cCaM. The transition routes illustrated by Δρi for each residue of nCaM and cCam are shown in Fig. 2 B and D, respectively. As shown in Fig. 2D, the model predicts that the structural change in binding loop IV occurs earlier than binding loop III during the domain opening of cCaM. (See the sharp change in color near residue number 130 in Fig. 2D.) Also, helix G has an earlier conformational transition than helix F (Fig. 2D). This sequence is quite different from nCaM as shown in Fig. 2B. For nCaM, the structural change in helix B is predicted to be earlier than the structural change in helix C for the closed to open conformational transition. Finally, our results show that the F/G helix linker in the cCaM has abrupt transition near the open state. (See the transition near residue number 115 in Fig. 2D). In contrast, the transition in the B/C helix linker in nCaM is more gradual (see Fig. 2B).
Conformational Flexibility and Cracking of the CaM2/3 Fragment.
The engineered even–odd EF-hands paired CaM2/3 fragment (composed of EF-hands 2 and 3) has been shown recently to have distinct transition characteristics from the odd–even EF-hands paired nCaM and cCaM (21). NMR spectroscopy has shown that Ca2+-free (apo) CaM2/3 does not have a stable folded structure but rather shows characteristics of a molten globule state. Ca2+ binding induces the folding of this even–odd paired EF-hand motifs CaM2/3, and the Ca2+-bound (holo) CaM2/3 adopts a similar structure as holo-nCaM or cCaM [see Protein Data Bank (PDB) ID code 2hf5] (21). In this article, we study the transition between the two metastable conformations, apo-CaM2/3 and holo-CaM2/3 taken directly from the folded apo-CaM (PDB ID code 1cfd) and holo-CaM (PDB ID code 1cll) as another example of flexibility influenced transitions in CaM. Here, we focus primarily on the conformational flexibility and transition of the central linker to an α-helix (even though this conformation has low probability in solution). The central linker of CaM was also studied by molecular dynamics (MD) simulation (18). Although our model does not accommodate the calcium-induced folding of CaM2/3 described in ref. 21, the model does capture certain aspects of the apo/holo conformational transition of this CaM2/3 fragment. In particular, the model predicts the apo-CaM2/3 to holo-CaM2/3 conformational transition mechanism that agrees well with the sequential Ca2+-binding mechanism of CaM2/3 suggested by the NMR measurements.
The magnitude of the fluctuations of each residue in CaM2/3 (shown in Fig. 2E) suggests that the changes in conformational flexibility of binding loops II and III of CaM2/3 are very different. In particular, helix C and binding loop II are highly flexible in the apo conformation of CaM2/3. The high intrinsic flexibility of apo-CaM2/3 may account for the molten globule state characteristics described in ref. 21. Fig. 2E also shows that binding loop II is more flexible than loop III. This result is in harmony with the NMR measurements of Ca2+ binding in CaM2/3 (21) that show that Ca2+ binds to loop III with higher affinity (lower Ca2+ concentration) than loop II. Our results also indicate that the flexibility of binding loop III decreases monotonically along the transition route from the apo-CaM2/3 to the holo-CaM2/3 structure. In contrast, the flexibility of helix F increases along the transition route from apo to holo transition of CaM2/3 fragment. In particular, the N-terminal part (close to binding loop III of helix F) partially unfolds when adopting the holo-CaM2/3 conformation. We also notice in Fig. 2E that the domain linker between EF-hands 2 and 3 (residue numbers 74–81) during the apo to holo transition has a relatively large change in conformational flexibility. The fluctuation amplitude Bi of this linker increases in the intermediate states and then decreases abruptly as the helix forms near the holo state. The local unfolding of the domain linker enhances the flexibility of this linker region dramatically, first relaxing in structure and then stabilizing in the holo-CaM2/3 structure. This local unfolding signaled by increase and decrease in flexibility is similar to the cracking exhibited in cCaM.
Local intermediate unfolding (i.e., cracking) of the domain linker during the apo to holo transition of CaM2/3 can also be seen through a change in the pair potential energy uij for the Asp residue from the linker at position 80 along the transition route as shown in Fig. 4B. During the apo to holo transition of CaM2/3 the energy of residue Asp-80 increases from the apo-CaM2/3 structure (α0 = 1) and decreases abruptly at the transition state (α0 = 0.4) to the minimum at holo-CaM2/3 structure (α0 = 0) as the helix forms. This increase and then sharp decrease in the contact energy of residue Asp-80 signal cracking (local unfolding and refolding) of the linker in CaM2/3 to a stable α-helix. We note also that the energy relative to the holo-CaM2/3 energy of Asp-80 is higher than that of Asp-118 from the F/G helix linker of cCaM. This emphasizes the importance of cracking in the apo-CaM2/3 to holo-CaM2/3 transition.
Conformational Transition Mechanism of CaM2/3 Fragment.
For CaM2/3, the order parameter Δρi = 1 corresponds to the apo state, and Δρi = −1 corresponds to the holo state of CaM2/3. Fig. 2F indicates that the structural change in binding loop III is initiated earlier than the structural change in binding loop II. Also, the conformational change in binding loop II is much more gradual than that of binding loop III. Similar to the identification of the Ca2+-binding affinity with the changes in inherent flexibility, the early structural change in binding loop III may imply the stepwise binding of Ca2+ in CaM2/3 with loop III having a higher binding affinity than loop II.
Discussion
CaM, a small (148-residue) Ca2+-binding protein with very high plasticity, may be an ideal system to demonstrate flexibility-influenced conformational transitions. The protein consists of structurally similar N- and C-terminal globular domains connected by a flexible tether also known as central or interdomain linker (17). The Ca2+-induced structural rearrangement in CaM results in the solvent exposure of a large hydrophobic surface responsible for molecular recognition of various cellular targets (25).
Although similar in structure and fold, the two CaM domains are quite different in terms of their flexibility, melting temperatures, and Ca2+-binding affinities (6, 7). In the two homologous (46% sequence identity) domains of CaM, Ca2+ binding occurs sequentially, first, in the binding sites of cCaM and then in the binding sites of nCaM (7). Despite large cooperativity in the Ca2+-binding process within each domain, the two domains reflect different Ca2+ and target affinities. The N-terminal pair of EF-hands binds to Ca2+ ions with much lower affinity than the C-terminal EF-hands pair (7). NMR (26, 27), and MD simulation (28) studies have shown that the Ca2+-bound nCaM is considerably less open than the cCaM. This was not observed in the X-ray crystal structure of the protein. Experimentally, it has been shown that the more-conserved cCaM has a greater affinity for Ca2+ and some CaM targets, whereas the nCaM is less specific in its choice of target motif (29, 30). Heat denaturation studies have shown that cCaM of Ca2+-free (apo) CaM starts to denature slightly above the physiological temperature (6). The denaturation of the apo-cCaM was observed at a lower concentration of denaturant than denaturation of the nCaM, whereas the order was reversed for Ca2+-CaM (31). From temperature-jump fluorescence spectroscopy by Rabl et al. (32), the instability of the cCaM was also observed from the study of unfolding of apo-CaM. They suggested that the cCaM was partially unfolded at native conditions. Recent NMR experiments done by Lundstrom and Akke (33) monitoring relaxation rates involving 13Cα spins in adjacent residues of E140Q mutant of cCaM revealed transient partial unfolding of helix F. This was interpreted as a global exchange process involves a partially unfolded minor state that was not detected (27, 34). A very recent conformational dynamics simulation of the cCaM by Chen et al. (35) has shown the presence of an unfolded apo state. These observations further suggest that the conformational exchange may be more complex than a simple two-state process.
Differences in inherent flexibility of the two domains can account for their distinct physical characteristics and the complexity in the conformational transition mechanism (26). Our results are consistent with a MD simulation study (36), which has shown that the nCaM is inherently more flexible with lower binding affinity than the cCaM. The more open conformation and lower intrinsic flexibility of the cCaM is also probably the key to understanding initial binding between this domain and CaM target enzymes (36, 37).
The variational model presented in this article predicts that the different inherent flexibilities of the two domains of CaM also lead to distinct transition mechanisms, even though the folded state topology of the two domains is the same. The mechanism controlling the open/closed transition does not involve cracking for the relatively flexible nCaM, whereas the mechanism controlling the conformation transition of the more rigid cCaM exhibits transient partial local unfolding in its helix linker between the binding loops. This partial local unfolding or cracking in the cCaM shows the complexity of the open/closed conformational transition mechanism of cCaM.
A recent NMR experiment studied the EF-hand association effects on the structure, Ca2+ affinity, and cooperativity of CaM (21). The EF-hands in CaM domains are odd–even paired with EF-hands 1 and 2 in nCaM and EF-hands 3 and 4 in cCaM. This arrangement of EF-hands in CaM is thought to be a consequence of its evolution from a biologically related ancestor EF-hand by gene duplication (38). The even–odd pairing of EF-hands 2 and 3 (CaM2/3) (Fig. 1C) has been characterized by NMR in ref. 21. In this fragment, EF-hands 2 and 3 are connected by the central linker between nCaM and cCaM. Although from the crystal structure of holo Ca2+-CaM the interdomain linker region appears to be a long rigid α-helix (39) several NMR relaxation experiments have demonstrated that this central linker is flexible in solution near its midpoint, and the two domains do not interact (17, 18). In contrast to the high-affinity and positive cooperativity for Ca2+ binding in the two odd–even paired EF-hand domains of CaM, the CaM2/3 binds Ca2+ in a sequence. First in the high-affinity EF-hand 3 and then in the EF-hand 2 with much lower affinity (21). Although not a direct focus of our article, we also note that a peptide binding to CaM2/3 has also been characterized recently by McIntosh and co-workers (40). It was found that CaM2/3 adopts Ca2+-bound structure with peptide binding very much similar to those of nCaM or cCaM. These observations reflect the very high plasticity of the EF-hand association and mediate Ca2+-dependent recognition of target proteins.
In this article we study the transition between the two states of the fragment CaM2/3 inferred by the structures of intact CaM. Our results reveal that the central linker in CaM2/3 is highly flexible and dominates the mechanism for this transition. The folding of the linker to an α-helix is preceded by a further increase in its flexibility and local unfolding. This cracking leads to a very sharp transition of the helix linker from apo-CaM2/3 to holo-CaM2/3 conformation. Although our model did not predict partial local unfolding or cracking of helix F from the open/closed conformational transition of cCaM, the apo/holo conformational transition of CaM2/3 reveals cracking in some region of helix F as we have discussed already in comparison with the NMR study (33). The predicted change in conformational flexibility of CaM2/3 reveals the stepwise binding of Ca2+ ions with binding loop III having higher affinity than binding loop II. Nevertheless, the relevance of this transition to the binding dynamics of CaM2/3 is somewhat limited because the model is based on low-probability structures in solution.
Methods
Preparation of Structures.
The structures of the two conformations of the transition are rotated to have the same center of mass and minimum root mean square deviation of the Cα positions. The conformational transition in cCaM is modeled from the residues 76–147 of apo-CaM (PDB ID code 1cfd) (19) and holo-CaM (PDB ID code 1cll) (41) structures, whereas the conformational transition in nCaM is modeled from the residues 4–75 of the same PDB structures. The apo to holo conformational transitions of the even–odd paired EF-hand motifs CaM2/3 are modeled from EF-hands 2 and 3, residues 46–113 of apo-CaM (PDB ID code 1cfd) and holo-CaM (PDB ID code 1cll) structures.
Variational Model of Conformational Transitions.
A conformation of a protein in our model (described in more detail in ref. 23) is described by the N position vectors of the α-carbons of the polypeptide backbone, {ri}. Partially ordered ensembles of polymer configurations are described by a coarse-grained reference Hamiltonian
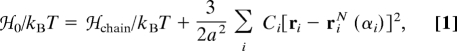 |
where T is the temperature and kB is the Boltzmann constant. Here, Hchain is a harmonic potential that enforces chain connectivity of a freely rotating polymer with mean bond length a = 3.8 Å (42) and mean valance angle cos θ = 0.8 (43). The second term includes N variational parameters, {Ci}, that control the magnitude of the fluctuations about α-carbon position vectors
Here, we have introduced another set of N variational parameters, {αi} (ranging between 0 and 1), that specify the backbone positions as an interpolation between the apo-CaM and holo-CaM conformations, {riNapo} and {riNholo}, respectively. The probability for a particular configurational ensemble specified by the variational parameters {Ci,αi} at temperature T is given by the variational free-energy F({C},{α}) = E({C},{α}) − TS({C},{α}). Here, S({C},{α}) is the entropy loss caused by the localization of the residues around the mean positions ri(αi)}. The energy is determined by the two-body interactions between distant residues E({C},{α}) = Σ[i,j] εij uij, where uij is the average of the pair potential u(rij) over H0 (42) and εij is the strength of a fully formed contact between residues i and j, given by Miyazawa–Jernigan interaction parameters (44). The sum is restricted to a set of contacts [i,j] determined by pairs of residues in the proximity in each of the metastable conformations (23). Each metastable conformation has a distinct but overlapping set of contacts [i,j]apo, [i,j]holo, and [i,j] is the union of these sets of contacts. In this model, the interaction energy for contacts that occur exclusively in only one metastable structure is given by exp(−uij/kBT) = 1 + exp(−〈uij〉0/kBT). This two-state model provides an interpolation at the individual contact level of the interaction energy determined by the two metastable conformations and is similar to some other native state-biased potentials describing conformational transitions. (10–12). Analysis of the free-energy surface parameterized by {C,α} follows the program developed to describe folding (42): the mechanism controlling the kinetics of the transitions is determined by the ensemble of structures characterized by the monomer density at the saddlepoints of the free energy. At this point, we simplify our model and restrict the interpolation parameter αi to be the same for all residues, αi = α0 (45). With this simplification, the numerical problem of finding saddlepoints with respect to {C,α} simplifies to minimizing the free-energy F({C},α0) with respect to {C} for a fixed α0.
Conformational Flexibility.
Main-chain conformational flexibility is characterized by the mean square fluctuations {Bi(α0)} of each α-carbon of the polypeptide chain from its average positions, δri along the transition route as α0 goes from 0 to 1 (23). These natural-order parameters for the reference Hamiltonian H0, Bi = 〈δri2〉0, contains information about the degree of structural order of each residue (8, 42).
Conformational Transition Mechanism.
The main-chain dynamics responsible for the detailed mechanism of the conformational transition in our analysis is based on the order parameters introduced to describe the folding mechanism ρiapo/holo = 〈exp(−3αN(ri − riNapo/holo)2/2a2〉0 with αN = 0.5 defining the width of a Gaussian window about the metastable structure {riNapo/holo}. In particular, it is convenient to characterize the relative similarity to the apo structure along the transition route through the normalized measure
where ρiapo(α0) is the monomer density of the ith residue with respect to the apo conformation described by {riNapo} (23). Similarly, we represent the relative structural similarity to the holo conformation as
where ρiholo(α0) is the monomer density of the ith residue with respect to the holo conformation described by riNholo}. In the holo state, ρiapo (0) = 0 and ρiholo (0) = 1, whereas in the apo state ρiapo (1) = 1 and ρiholo (1) = 0. To represent the structural changes more clearly, it is convenient to consider the difference,
for each residue. This difference shifts the relative degree of localization to be between Δρi(1) = 1 and Δρi(0) = −1 corresponding to the apo and holo conformations, respectively.
Acknowledgments.
We thank Ad Bax for insightful comments. This work was supported in part by grant from the Ohio Board of Regents Research Challenge program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Weber G. Ligand binding and internal equilibriums in proteins. Biochemistry. 1972;11:864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- 2.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscape of functional transitions in proteins. Proc Natl Acad Sci USA. 2003;100:12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyashita O, Wolynes PG, Onuchic JN. Simple energy landscape model for the kinetics of functional transitions in proteins. J Phys Chem B. 2005;109:1959–1969. doi: 10.1021/jp046736q. [DOI] [PubMed] [Google Scholar]
- 4.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways and the energy landscape of protein folding: A synthesis. Proteins Struct Funct Genet. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 5.Onuchic JN, Wolynes PG. The theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Tsalkova TN, Privalov PL. Thermodynamic study of domain organization in troponin C and calmodulin. J Mol Biol. 1985;181:533–544. doi: 10.1016/0022-2836(85)90425-5. [DOI] [PubMed] [Google Scholar]
- 7.Linse S, Helmersson A, Forsen S. Calcium binding to calmodulin and its globular domains. J Biol Chem. 1991;266:8050–8054. [PubMed] [Google Scholar]
- 8.Portman JJ, Takada S, Wolynes PG. Variational theory for site-resolved protein folding free energy surfaces. Phys Rev Lett. 1998;81:5237–5240. [Google Scholar]
- 9.Zuckerman DM. Simulation of an ensemble of conformational transitions in a united-residue model of calmodulin. J Phys Chem B. 2004;108:5127–5137. [Google Scholar]
- 10.Best RB, Chen YG, Hummer G. Slow protein conformational dynamics from multiple experimental structures: The helix/sheet transition of arc repressor. Structure. 2005;13:1755–1763. doi: 10.1016/j.str.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Maragakis P, Karplus M. Large-amplitude conformational change in proteins explored with a plastic network model: Adenylate kinase. J Mol Biol. 2005;352:807–822. doi: 10.1016/j.jmb.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Okazaki K, Koga N, Takada S, Onuchic JN, Wolynes PG. Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: Structure-based molecular dynamics simulations. Proc Natl Acad Sci USA. 2006;103:11844–11849. doi: 10.1073/pnas.0604375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu JW, Voth GA. Coarse-grained free energy functions for studying protein conformational changes: A double-well network model. Biophys J. 2007;93:3860–3871. doi: 10.1529/biophysj.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Q, Wang J. Single molecule conformational dynamics of adenylate kinase: Energy landscape, structural correlations, and transition state ensembles. J Am Chem Soc. 2008;130:4772–4783. doi: 10.1021/ja0780481. [DOI] [PubMed] [Google Scholar]
- 15.Whitford PC, Miyashita O, Levy Y, Onuchic JN. Conformational transitions of adenylate kinase: Switching by cracking. J Mol Biol. 2007;366:1661–1671. doi: 10.1016/j.jmb.2006.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humphrey W, Dalke A, Schulten K. Vmd: Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 17.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: The central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 18.van der Spoel D, de Groot BL, Hayward S, Berendsen HJ, Vogel HJ. Bending of the calmodulin central helix: A theoretical study. Protein Sci. 1996;5:2044–2053. doi: 10.1002/pro.5560051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuboniwa H, et al. Solution structure of calcium-free calmodulin. Nat Struct Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 20.Tjandra N, Kuboniwa H, Ren H, Bax A. Rotational dynamics of calcium-free calmodulin studied by 15N NMR relaxation measurements. Eur J Biochem. 1995;230:1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- 21.Lakowski TM, Lee GM, Okon M, Reid RE, McIntosh LP. Calcium-induced folding of a fragment of calmodulin composed of EF-hands 2 and 3. Protein Sci. 2007;16:1119–1132. doi: 10.1110/ps.072777107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zong C, et al. Establishing the entatic state in folding metallated Pseudomonas aeruginosa azurin. Proc Natl Acad Sci USA. 2007;104:3159–3164. doi: 10.1073/pnas.0611149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tripathi S, Portman JJ. Inherent flexibility and protein function: The open/closed conformational transition in the N-terminal domain of calmodulin. J Chem Phys. 2008;128:205104. doi: 10.1063/1.2928634. [DOI] [PubMed] [Google Scholar]
- 24.Likic VA, Strehler EE, Gooley PR. Dynamics of Ca2+-saturated calmodulin D129N mutant studied by multiple molecular dynamics simulations. Protein Sci. 2003;12:2215–2229. doi: 10.1110/ps.0377803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of X-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 26.Chou JJ, Li S, Klee CB, Bax A. Solution structure of Ca2+-calmodulin reveals flexible hand-like properties of its domains. Nat Struct Biol. 2001;8:990–996. doi: 10.1038/nsb1101-990. [DOI] [PubMed] [Google Scholar]
- 27.Evenas J, Forsen S, Malmendal A, Akke M. Backbone dynamics and energetics of a calmodulin domain mutant exchanging between closed and open conformations. J Mol Biol. 1999;289:603–617. doi: 10.1006/jmbi.1999.2770. [DOI] [PubMed] [Google Scholar]
- 28.Vigil D, Gallagher SC, Trewhella J, Garcia AE. Functional dynamics of the hydrophobic cleft in the N-domain of calmodulin. Biophys J. 2001;80:2082–2092. doi: 10.1016/S0006-3495(01)76182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baley PM, Findlay WA, Martin SR. Target recognition by calmodulin: Dissecting the kinetics and affinity of interaction using short peptide sequences. Protein Sci. 1996;5:1215–1228. doi: 10.1002/pro.5560050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barth A, Martin SR, Bayley PM. Specificity and symmetry in the interaction of calmodulin domains with the skeletal muscle myosin light chain kinase target sequence. J Biol Chem. 1998;273:2174–2183. doi: 10.1074/jbc.273.4.2174. [DOI] [PubMed] [Google Scholar]
- 31.Masino L, Martin SR, Bayley PM. Ligand binding and thermodynamic stability of a multidomain protein, calmodulin. Protein Sci. 2000;9:1519–1529. doi: 10.1110/ps.9.8.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabl CR, Martin SR, Neumann E, Bayley PM. Temperature jump kinetic study of the stability of apocalmodulin. Biophys Chem. 2002;101:553–564. doi: 10.1016/s0301-4622(02)00150-3. [DOI] [PubMed] [Google Scholar]
- 33.Lundstrom P, Mulder FAA, Akke M. Correlated dynamics of consecutive residues reveal transient and cooperative unfolding of secondary structure in proteins. Proc Natl Acad Sci USA. 2005;102:16984–16989. doi: 10.1073/pnas.0504361102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evenas J, Malmendal A, Akke M. Dynamics of the transition between open and closed conformations in a calmodulin C-terminal domain mutant. Structure. 2001;9:185–195. doi: 10.1016/s0969-2126(01)00575-5. [DOI] [PubMed] [Google Scholar]
- 35.Chen YG, Hummer G. Slow conformational dynamics and unfolding of the calmodulin C-terminal domain. J Am Chem Soc. 2007;129:2414–2415. doi: 10.1021/ja067791a. [DOI] [PubMed] [Google Scholar]
- 36.Barton NP, Verma CS, Caves LSD. Inherent flexibility of calmodulin domains: A normal-mode analysis study. J Phys Chem B. 2002;106:11036–11040. [Google Scholar]
- 37.Yamniuk AP, Vogel HJ. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama S, Moncrief ND, Kretsinger RH. Evolution of EF-hand calcium-modulated proteins. II. Domains of several subfamilies have diverse evolutionary histories. J Mol Evol. 1992;34:416–448. doi: 10.1007/BF00162998. [DOI] [PubMed] [Google Scholar]
- 39.Babu YS, et al. Three-dimensional structure of calmodulin. Nature. 1985;315:37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 40.Lakowski TM, et al. Peptide binding by a fragment of calmodulin composed of EF-hands 2 and 3. Biochemistry. 2007;46:8525–8536. doi: 10.1021/bi700265j. [DOI] [PubMed] [Google Scholar]
- 41.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 Å resolution. J Mol Biol. 1992;228:1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 42.Portman JJ, Takada S, Wolynes PG. Microscopic theory of protein folding rates. I. Fine structure of the free energy profile and folding routes form a variational approach. J Chem Phys. 2001;114:5069–5081. [Google Scholar]
- 43.Bixon M, Zwanzig R. Optimized Rouse–Zimm theory for stiff polymer chains. J Chem Phys. 1978;68:1896–1902. [Google Scholar]
- 44.Miyazawa S, Jernigan RL. Residue–residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J Mol Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 45.Kim MK, Jernigan RL, Chirikjian GS. Efficient generation of feasible pathways for protein conformational transitions. Biophys J. 2002;83:1620–1630. doi: 10.1016/S0006-3495(02)73931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]