Abstract
Although the transcriptome, proteome, and interactome of several eukaryotic model organisms have been described in detail, lipidomes remain relatively uncharacterized. Using Saccharomyces cerevisiae as an example, we demonstrate that automated shotgun lipidomics analysis enabled lipidome-wide absolute quantification of individual molecular lipid species by streamlined processing of a single sample of only 2 million yeast cells. By comparative lipidomics, we achieved the absolute quantification of 250 molecular lipid species covering 21 major lipid classes. This analysis provided ≈95% coverage of the yeast lipidome achieved with 125-fold improvement in sensitivity compared with previous approaches. Comparative lipidomics demonstrated that growth temperature and defects in lipid biosynthesis induce ripple effects throughout the molecular composition of the yeast lipidome. This work serves as a resource for molecular characterization of eukaryotic lipidomes, and establishes shotgun lipidomics as a powerful platform for complementing biochemical studies and other systems-level approaches.
Keywords: fatty acid elongation, S. cerevisiae, shotgun lipidomics
The lipidome of eukaryotic cells consists of hundreds to thousands of individual lipid species that constitute membranes, store metabolic energy and function as bioactive molecules (1–3). Despite the extensive characterization of proteins, their association into complexes and activities (4–6), it is still difficult to assess how perturbations within the lipid metabolic network affect the full lipidome of cells. This work shows that lipidome-wide quantification of individual molecular lipid species (molecules with defined chemical structure) by absolute quantification (expressed in mol or mol%) provides a new approach to relate lipidomics and functional genomics studies.
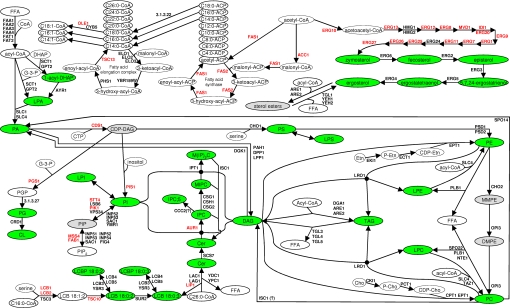
The yeast Saccharomyces cerevisiae serves as a prime model organism for studying the molecular organization and regulatory circuitry of eukaryotic lipidomes (7–9). It uses a relatively simple and conserved network of lipid metabolic pathways (Fig. 1) that synthesize a few hundred molecular lipid species constituting its full lipidome (3). The lipidome diversity is primarily determined by the fatty acid synthase (10), the Δ-9 desaturase (11) and the fatty acid elongation complex (12) that produce only saturated or mono-unsaturated fatty acids having 10 to 26 carbon atoms for the biosynthesis of glycerolipids, glycerophospholipids, and sphingolipids. Importantly, several metabolic conversions interlink sphingolipid, glycerophospholipid, and glycerolipid metabolism such that any perturbation within the metabolic network is prone to induce lipidome-wide ripple effects. Remarkably, numerous genes involved in lipid metabolism and trafficking can be mutated or deleted without apparent physiological consequences (Fig. 1).
Fig. 1.
The lipid metabolic network of S. cerevisiae. Enzymes are annotated by gene name (essential genes are indicated in red). The lipid metabolic network was compiled using the Saccharomyces Genome Database (www.yeastgenome.org) and references therein. Lipids monitored by absolute quantification are indicated by green circles. Lipids that were only identified are shown by gray circles. CL, cardiolipin; Cer, ceramide; CoA, coenzyme A; CDP-DAG, cytidine diacylglycerol; DAG, diacylglycerol; DMPE, dimethyl-phosphatidylethanolamine; FFA, free fatty acid; IPC, inositolphosphoceramide; LCB, long-chain base; LCBP, long-chain base phosphate; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPI, lysophosphatidylinositol; LPS, lysophosphatidylserine; M(IP)2C, mannosyl-diinositolphosphoceramide; MIPC, mannosyl-inositolphosphoceramide; MMPE, monomethyl-phosphatidylethanolamine; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PGP, phosphatidylglycerolphosphate; PI, phosphatidylinositol; PIP, phosphatidylinositol-phosphate; PIP2, phosphatidylinositol-bisphosphate; PS, phosphatidylserine; TAG, triacylglycerol.
Despite remarkable methodological advances, lipidomics seldom complements functional genomics efforts owing to three major factors. First, analysis of glycerophospholipids and sphingolipids requires two separate lipid extractions (14), which use vast amounts of sample (typically 250 million cells), numerous sample processing steps and harsh chemical treatments for recovery of sphingolipids (15). Second, the lack of appropriate internal lipid standards limits the quantification scope to profiling analysis (16) or relative quantification (14, 17), which provide no information on the stoichiometric relationship between lipid species of different classes. Third, the present methods are not designed for full characterization of molecular lipid species (e.g., PC 16:1–18:1), but instead identify 35 to 70 lipids with different sum compositions (e.g., PC 34:2) (16, 17).
Here, we present a mass spectrometric approach that enabled absolute quantification of 250 molecular lipid species constituting 21 lipid classes, including the yeast inositol-containing sphingolipids, intermediate LPA and bioactive LCBP species. Altogether, this is currently the most comprehensive description of the S. cerevisiae lipidome achieved with ≈125-fold better sensitivity and >3-fold increased lipidome coverage compared with previous approaches (14, 16, 17). To demonstrate its efficacy for functional genomics studies, we monitored in molecular detail how the full lipidome adapted to growth temperature and genomic deletion of 3-ketoacyl-CoA synthases. This works demonstrates the ability to perform global analysis of an eukaryotic lipidome, and provides a resource for further systems-level studies of lipid metabolism and membrane trafficking in yeast.
Results
Quantitative Shotgun Lipidomics Approach for S. cerevisiae.
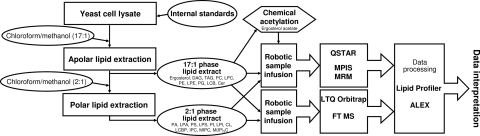
Several advances in sample processing and mass spectrometry enabled lipidome-wide absolute quantification of molecular lipid species (Fig. 2). First, we used a 2-step lipid extraction procedure that separated relatively apolar and polar lipids. Second, we improved the analytical sensitivity and specificity by optimizing the solvent systems used for infusion of lipid extracts for mass spectrometric analysis. Third, we implemented absolute quantification of molecular lipid species herein covering 21 lipid classes.
Fig. 2.
Overview of the quantitative shotgun lipidomics approach. Yeast cell lysates were spiked with internal lipid standards. Samples were processed by 2-step lipid extraction for fractionation of apolar and polar lipids. The lipid extracts were analyzed by automated shotgun lipidomics analysis in negative and positive ion mode. Lipid species were detected by MPIS or MRM analysis on a QSTAR instrument, or by FT MS analysis on a LTQ Orbitrap machine. Quantification of ergosterol was achieved by chemical acetylation followed by MRM analysis. Identification and quantification of detected lipid species were performed by Lipid Profiler and ALEX.
Yeast cell lysates spiked with internal lipid standards were first extracted with chloroform/methanol (17:1, V/V) that recovered 80–99% of relatively apolar lipids, including ergosterol, TAG, DAG, PC, LPC, PE, PG, LCB, and Cer. Subsequent extraction of the remaining aqueous phase, using chloroform/methanol (2:1, V/V) recovered 74–95% of polar lipids, including PA, LPA, PS, LPS, PI, LPI, CL, LCBP, IPC, MIPC and M(IP)2C (Table S1). This 2-step lipid extraction procedure achieved the same or better lipid recovery compared with the conventional Bligh and Dyer protocol (18), whereas increasing the recovery of the important M(IP)2C by 4-fold. Comparing mass spectrometric profiles of lipid extracts showed that partitioning into the 17:1 vs. the 2:1 extract was lipid class-dependent, rather than lipid species-dependent. Furthermore, fractionation of apolar and polar lipids increased the analytical sensitivity compared with direct analysis of unfractionated lipid extracts (data not shown).
Next, we optimized the solvent system for sample infusion and shotgun lipidomic analysis. The use of 0.2 mM methylamine increased the ionization efficiency in negative ion mode and enhanced the detection sensitivity 3- to 25-fold for the anionic lipid classes PA, PG, PI, PS, IPC, MIPC, and M(IP)2C compared with commonly used 7.5 mM ammonium acetate (19, 20). Comparative analysis of the ionization efficiency in positive and negative ion mode showed that only sterols, PC, DAG, TAG, and Cer should be analyzed in positive ion mode (Fig. S1).
Until now, no mass spectrometric method has allowed absolute quantification of the yeast sphingolipids IPC, MIPC and M(IP)2C. In wild-type S. cerevisiae, the most abundant sphingolipid species contain a C18 phytosphingosine (LCB 18:0;3) with an amide-linked fatty acid moiety having 26 carbon atoms, no double bonds and a single hydroxyl group (e.g., M(IP)2C 18:0;3/26:0;1) (9). To enable absolute quantification of yeast sphingolipids we isolated IPC, MIPC and M(IP)2C standards from the double hydroxylase mutant sur2Δscs7Δ, which synthesizes sphingolipids with LCB 18:0;2 (C18 dihydrosphingosine) and a 26:0;0 fatty acid moiety (e.g., M(IP)2C 18:0;2/26:0;0) (21, 22). For the absolute quantification of glycerolipids, glycerophospholipids, and sterols we used commercially available lipid standards (Table S2).
In summary, the lipidome-wide quantification of endogenous lipid species was achieved by spiking cell lysates with defined amounts of 21 internal lipid class-specific standards. Lipid species recovered in the 17:1 and 2:1 phase lipid extracts were quantified by 6 successive automated MS and MS/MS experiments (Table S2) followed by data processing using dedicated software (19). The lipidomics platform featured a broad dynamic quantification range covering 3 to 4 orders of magnitude and a detection limit in the low pmol range (Fig. S2). We estimated that the quantitative analysis covered ≈95% of the S. cerevisiae lipidome by monitoring lipid species constituting the 21 major lipid classes of the 30 lipid classes in yeast (Fig. 1). We also detected several low abundant biosynthetic intermediates, phosphoinositides and ergosterol esters (7). However, their quantification was omitted because of the lack of applicable lipid standards. Importantly, the analysis required only 2 million yeast cells (≈0.2 OD units), which is 125-fold less sample compared with previous methods (14, 17).
Molecular Composition of the S. cerevisiae Lipidome.
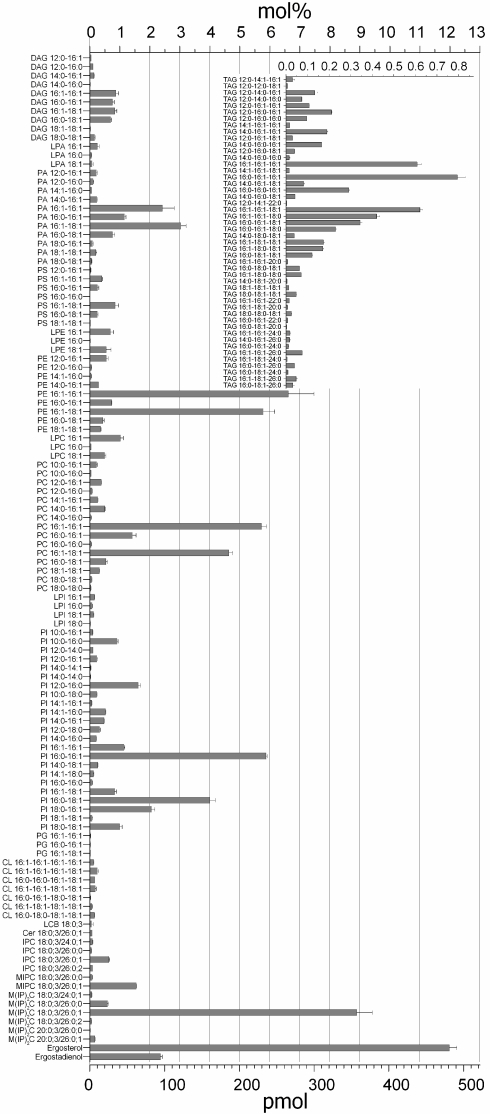
By shotgun lipidomics analysis we quantified 162 molecular lipid species in wild-type S. cerevisiae BY4741 (Fig. 3). The most abundant lipid species was ergosterol comprising 12.0 mol% of the lipidome (equivalent to 481 pmol/0.2 OD units), followed by 8.9 mol% M(IP)2C 18:0;3/26:0;1. The corresponding sphingolipid intermediates Cer 18:0;3/26:0;1, IPC 18:0;3/26:0;1 and MIPC 18:0;3/26:0;1 were 128-, 14-, and 6-fold less abundant compared with M(IP)2C 18:0;3/26:0;1, respectively. This result shows efficient substrate shunting under steady state conditions by the IPC synthetase (Aur1), the MIPC synthetases (Csg1-Csh1-Csg2), and the M(IP)2C synthetase (Ipt1). The most abundant glycerophospholipid species contained 16:1, 18:1, and 16:0 fatty acids moieties corroborating previous findings (16, 23).
Fig. 3.
Molecular composition of the wild-type S. cerevisiae lipidome. 162 molecular lipid species were quantified. S. cerevisiae BY4741 was cultured in synthetic defined medium at 24 °C. The insert shows TAG species. The abundance of lipid species is depicted in mol% and absolute amount (pmol per 0.2 OD units). Error bars indicate ±SD. (n = 4, 2 independent analyses of lipid extracts from 2 independent cultures).
By calculating the lipid class composition (sum of all lipid species constituting the same lipid class), we found that the BY4741 lipidome contained 20.3% PI, 14.9% PE, 14.3% PC, 8.3% PA and 1.8% PS species (Fig. S3B). Although it is commonly believed that PC dominate the yeast glycerophospholipidome (23), our analysis of wild-type strains BY4742, BY4743, NY13 and CTY182 consistently identified PI as a major lipid class constituting 17% to 30% of the lipidome.
We also observed that S. cerevisiae modulates its lipid composition at elevated growth temperature (37 °C) in three major ways (Fig. S3): by (i) synthesizing more PI species, and less PE and TAG species; (ii) preferential incorporation of 16:0 and 18:1 fatty acid moieties into glycerophospholipids; and (iii) synthesizing sphingolipids with a LCB 20:0;3 backbone; detected as a 18-fold increase in M(IP)2C 20:0;3/26:0;1 compared with growth at 24 °C.
Lipidome-Wide Consequences of Defective Fatty Acid Elongation.
Elo1, Elo2 and Elo3 are 3-ketoacyl-CoA synthases embedded in the ER where they form a complex with Ybr159w, Phs1 and Tsc13 (12). This enzyme complex catalyzes the 4-step cycle that successively elongates precursor acyl-CoA by 2 methylene groups. Although the substrate specificities of the 3-ketoacyl-CoA synthases have been characterized (12, 24–27), it is still unknown how their genomic deletion affects the molecular composition of the S. cerevisiae lipidome. Whereas elo1Δ shows no apparent phenotype, elo2Δ and elo3Δ accumulate aberrant sphingolipid species with shortened fatty acid moieties (25) together with compromised vacuolar organization and protein trafficking to the cell surface (28, 29).
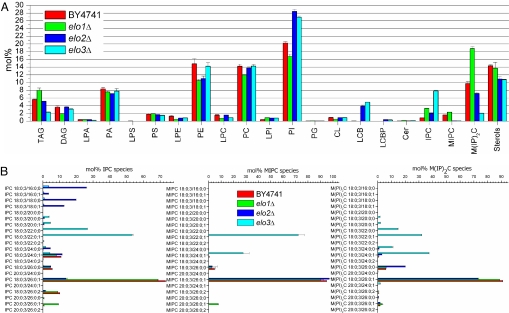
Comparative lipidome analysis of elo1Δ, elo2Δ, elo3Δ and wild type quantified in total 250 molecular species constituting 21 lipid classes (Fig. S4, Fig. 4). This is at least 3-fold better lipidome coverage than previously achieved (14, 16, 17). The content of M(IP)2C, MIPC and IPC in elo1Δ increased by 1.4- to 3.8-fold, whereas the levels of PI, PC and PE were reduced. Apparently, the deletion of Elo1 promotes elongation of shorter chain acyl-CoA to C26:0-CoA mediated by functional Elo2 and Elo3. This concomitantly enhances the production of LCB for synthesis of sphingolipids, and in turn reduces the concentration of substrate PI species. Furthermore, the elo1Δ mutant contained more glycerophospholipid species with 14:0 and 14:1 fatty acid moieties (Fig. S4), which corroborates the known substrate preference of Elo1 (24, 26).
Fig. 4.
Comparative lipidomics of wild-type BY4741, elo1Δ, elo2Δ, and elo3Δ. (A) Lipid class composition. (B) Molecular composition of sphingolipid species. Error bars indicate ±SD. (n = 4).
The lipidome of elo2Δ and elo3Δ showed specific changes in the molecular composition of sphingolipid and glycerophospholipid species. PI (≈27%), the substrate for synthesis of IPC and M(IP)2C, was the most abundant lipid class in both elo2Δ and elo3Δ (Fig. 4A). Importantly, elo2Δ and elo3Δ accumulated 3.8% and 4.9% of the sphingolipid intermediate LCB 18:0;3, respectively (Fig. S4). In addition, we observed an increased concentration of LCBP 18:0;3 (not detected in wild type). The sphingolipidome of elo2Δ and elo3Δ contained less MIPC and M(IP)2C, but accumulated 2.4- to 9.0-fold more IPC compared with wild type. elo2Δ produced a heterogeneous assortment of IPC species including shorter chain IPC 18:0;3/16:0;0 and IPC 18:0;3/18:0;0, which were not converted into corresponding MIPC and M(IP)2C species, so that MIPC 18:0;3/26:0;1 and M(IP)2C 18:0;3/26:0;1 were the major species (Fig. 4B). In comparison, elo3Δ accumulated IPC 18:0;3/22:0;1 as the major IPC species; whereas MIPC 18:0;3/24:0;1 and M(IP)2C 18:0;3/24:0;1 were the predominant MIPC and M(IP)2C species, respectively. These results confirm previous observations that elo2Δ and elo3Δ synthesize sphingolipids with shortened fatty acid moieties (25, 27). Furthermore, we detected low amounts of lysoIPC 18:0;3 (phytosphingosine linked to inositol-phosphate) in both elo2Δ and elo3Δ (data not shown). Taken together, these results demonstrated a promiscuous substrate specificity of the IPC synthase Aur1, whereas the MIPC synthases (Csg1-Csh1-Csg2) showed a restricted substrate specificity, using primarily IPC species with C26 or C24 fatty acid moieties (Fig. 4B). These results also explain the successful metabolic conversion of fluorescent Cer tracers into IPC; and the inability to convert fluorescent IPC into MIPC and M(IP)2C (30).
Although the concentration of PI and PC in elo2Δ and elo3Δ was similar, we observed that the molecular composition of PI and PC species were dramatically different (Fig. S5). elo2Δ contained less PI and PC species with 16:0, 16:1 and shorter fatty acid moieties, which was offset by increased content of species with 18:1 and 18:0 fatty acids. In contrast, the elo3Δ lipidome showed differences primarily in the composition of PC species (and to a smaller extent within PE, PS and PA) characterized by increased abundance of PC 16:1–16:1 and PC 14:1–16:1 and reduced content of PC 16:0–16:1 and PC 16:0–18:1 compared with wild type.
Discussion
In a comparative lipidomics study, we determined the absolute abundance of 250 lipid species covering 21 major lipid classes. This analysis enabled us to determine the lipidome-wide stoichiometric relationship between molecular lipid species in S. cerevisiae. Furthermore, 26 lipid species from an additional 5 lipid classes were detected, but omitted from quantification because of the lack of applicable lipid standards (data not shown). We estimate that the analysis covered ≈95% of the S. cerevisiae lipidome including M(IP)2C, LPA and bioactive LCBP species that escaped characterization by previous lipidomic strategies (14, 16). The most detailed analysis presented here required 100 min per sample. We note that the lipidomics platform can be operated with higher sample throughput as required for screening routines simply by lowering the structural specificity of the analysis (31). By monitoring the absolute abundance of individual lipid species we demonstrated that differences in growth temperature and defects in the lipid biosynthesis machinery produced alterations throughout the entire yeast lipidome. These ripple effects could not have been predicted from known activities of lipid biosynthesis enzymes (32). We also note that phenotypes seen upon deletion of genes expected to affect a specific lipid class might derive from seemingly unrelated compensatory changes within the lipidome that can only be assessed by global lipid analysis. Finally, we note that minor modifications of the methodology should enable the characterization of mammalian glycosphingolipids, bioactive LPA and LCBP species (33, 34) and phosphoinositides (35).
Our results indicate that the biosynthetic aperture for LCB synthesis (Lcb1-Lcb2-Tsc3, Tsc10, Sur2), the fatty acid elongation complex (Elo2/Elo3-Ybr159w-Phs1-Tsc13) and the ceramide synthases (Lag1/Lac1-Lip1) mediate efficient substrate shunting for de novo ceramide biosynthesis, whereas Elo1 shunts C16:0/C18:0-CoA toward glycerophospholipid incorporation by an unknown acyltransferase. The Elo1 activity counterbalances the flux of acyl-CoA toward Elo2 and Elo3, which mediate the production of C26:0-CoA for ceramide synthesis and subsequent production of IPC, MIPC and M(IP)2C. This model is corroborated by three lines of evidence:(i) elo1Δ contained high levels of IPC, MIPC and M(IP)2C 18:0;3/26:0;1 produced by the elongation of C16:0-CoA to C26:0-CoA by functional Elo2 and Elo3 and concomitant synthesis of LCB 18:0;3;(ii) the intermediate sphingolipid LCB 18:0;3 accumulated in both elo2Δ and elo3Δ; and(iii) elo2Δ (having functional Elo1) produced more glycerophospholipid species with 18:0 and 18:1 fatty acid moieties. Furthermore, the results indicated that Elo2 and Elo3 interact (in cooperation with Ybr159w, Phs1, and Tsc13) (12) to mediate efficient elongation of C16:0-CoA to C26:0-CoA, as indicated by the accumulation of IPC species with shortened amide-linked fatty acid moieties in both elo2Δ and elo3Δ.
In summary, we argue that lipidome-wide absolute quantification of molecular lipid species changes the scope of functional genomics approaches for characterizing metabolic networks. Contrary to other omics approaches, lipidomics-based mass spectrometry provides direct measurement of products and substrates within the network. Therefore, there is no need to infer the content of lipids from expression level or bulk concentration of enzymes (36). The interpretation of phenotypes, seemingly unrelated to defects in lipid metabolism, should be simplified because lipidomics reveals their direct correlation with specifically altered abundances of lipid species manifested as global ripple effects. Conversely, the methodology will also impact lipid biology in its broad sense because information on the absolute abundance of molecular lipid species under any biological condition could prompt the development of biophysical models that adequately addresses the complexity of biological membranes. Finally, lipidome-wide quantification should also improve our understanding of the molecular architecture of membrane domains and cellular organelles because the stoichiometry between molecular lipid and protein constituents can now be accurately determined (13).
Materials and Methods
Annotation of Lipid Species.
Lipid species were annotated by molecular composition (19, 22). Lysoglycerophospholipid, glycerophospholipid, DAG, and TAG species were annotated as: 〈lipid class〉 〈no. of C in the first fatty acid moiety〉:〈no. of double bonds in the first fatty acid moiety〉-〈no. of C in the second fatty acid moiety〉:〈no. of double bonds in the second fatty acid moiety〉-〈no. of C in the third fatty acid moiety (only TAG)〉:〈no. of double bonds in the third fatty acid moiety (only TAG)〉. For example, LPC 16:1, PI 16:0–18:1, DAG 16:1–16:1, TAG 16:0–16:1–16:1. Sphingolipid species were annotated as: 〈lipid class〉 〈no. of C in the LCB moiety〉:〈no. of double bonds in the LCB moiety〉;〈no. of hydroxyl groups in the LCB moiety〉/〈no. of C in the fatty acid moiety〉:〈no. of double bonds in the fatty acid moiety〉;〈no. of hydroxyl groups in the fatty acid moiety〉. For example, IPC 18:0;3/26:0;2; an IPC species containing a C18 phytosphingosine (with no double bonds and 3 hydroxyl groups) and a C26 amide-linked fatty acid moiety with no double bonds and 2 hydroxyl groups.
Yeast Strains and Culture Conditions.
S. cerevisiae BY4741 (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0), and elo1Δ, elo2Δ and elo3Δ produced in the BY4741 background (EUROSCARF, Frankfurt am Main, Germany). Yeast were cultured in SD media with 2% raffinose. For lipidomic experiments, yeast were precultured, diluted to 0.2 OD600 units/ml and cultured to 2 OD units/ml, washed and stored at −80 °C.
Two-Step Lipid Extraction at 4 °C.
Yeast were resuspended in 150 mM NH4HCO3 (pH 8) and disrupted by zirconia beads (0.5 mm; BioSpec Products). Yeast cell lysates were diluted to 0.2 OD units (≈2·106 cells) per 200 μL, and mixed with 20 μL of internal lipid standard mixture (see SI Materials and Methods for details). Samples were extracted with 990 μL chloroform/methanol (17:1, V/V) for 120 min. The lower organic 17:1 phase lipid extract was collected. The remaining aqueous sample material was reextracted with 990 μL chloroform/methanol (2:1, V/V) for 120 min. The lower organic 2:1 phase lipid extract was collected. The lipid extracts were vacuum evaporated. Finally, the lipid extracts were dissolved in 100 μL chloroform/methanol (1:2, V/V). Quantification of lipid extraction recovery was performed as described in SI Materials and Methods.
Mass Spectrometry.
Lipid extracts were analyzed in negative and positive ion mode on a QSTAR Pulsar-i instrument (MDS Analytical Technologies) and a LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific) both equipped with the robotic nanoflow ion source TriVersa NanoMate (Advion Biosciences) as described in refs. 19, 22, and 31. Detected lipid species were identified and quantified using Lipid Profiler (MDS Analytical Technologies) and proprietary Analysis of Lipid EXperiments (ALEX) software as described in ref. 19.
Further details appear in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Christoph Thiele (Max Planck Institute of Cell Biology and Genetics) for providing synthetic PI 17:0–17:0 and constructive comments; Teresa Dunn (Uniformed Services University of the Health Sciences, Bethesda, MD) for providing the sur2Δscs7Δ double mutant; Harald Pichler (Technische Universität, Graz, Austria) for comments on sterol analysis; Igor Chernushevich for advice on QqTOF mass spectrometry; and Reinaldo Almeida and Mark Baumert for expert advice on NanoMate operation. This work was supported by Deutsche Forschungsgemeinschaft SFB/TR 13 projects A1 (K.S.) and D1 (A.S.) and EUFP6 PRISM (K.S.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 2089.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811700106/DCSupplemental.
References
- 1.van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 3.Yetukuri L, Ekroos K, Vidal-Puig A, Oresic M. Informatics and computational strategies for the study of lipids. Mol Biosyst. 2008;4:121–127. doi: 10.1039/b715468b. [DOI] [PubMed] [Google Scholar]
- 4.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 5.Breslow DK, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Natter K, et al. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol Cell Proteomics. 2005;4:662–672. doi: 10.1074/mcp.M400123-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 8.Carman GM, Henry SA. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 2007;282:37293–37297. doi: 10.1074/jbc.R700038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson RC. New insights into sphingolipid metabolism and function in budding yeast. J Lipid Res. 2008;49:909–921. doi: 10.1194/jlr.R800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tehlivets O, Scheuringer K, Kohlwein SD. Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta. 2007;1771:255–270. doi: 10.1016/j.bbalip.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Martin CE, Oh CS, Jiang YD. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim Biophys Acta. 2007;1771:271–285. doi: 10.1016/j.bbalip.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Denic V, Weissman JS. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 2007;130:663–677. doi: 10.1016/j.cell.2007.06.031. [DOI] [PubMed] [Google Scholar]
- 13.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 14.Guan XL, Wenk MR. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast. 2006;23:465–477. doi: 10.1002/yea.1362. [DOI] [PubMed] [Google Scholar]
- 15.Hanson BA, Lester RL. The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J Lipid Res. 1980;21:309–315. [PubMed] [Google Scholar]
- 16.Schneiter R, et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benghezal M, Roubaty C, Veepuri V, Knudsen J, Conzelmann A. SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J Biol Chem. 2007;282:30845–30855. doi: 10.1074/jbc.M702719200. [DOI] [PubMed] [Google Scholar]
- 18.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 19.Ejsing CS, et al. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem. 2006;78:6202–6214. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- 20.Liebisch G, et al. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS) Biochim Biophys Acta. 2006;1761:121–128. doi: 10.1016/j.bbalip.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Haak D, Gable K, Beeler T, Dunn T. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J Biol Chem. 1997;272:29704–29710. doi: 10.1074/jbc.272.47.29704. [DOI] [PubMed] [Google Scholar]
- 22.Ejsing CS, et al. Collision-induced dissociation pathways of yeast sphingolipids and their molecular profiling in total lipid extracts: A study by quadrupole TOF and linear ion trap-orbitrap mass spectrometry. J Mass Spectrom. 2006;41:372–389. doi: 10.1002/jms.997. [DOI] [PubMed] [Google Scholar]
- 23.Daum G, et al. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast. 1999;15:601–614. doi: 10.1002/(SICI)1097-0061(199905)15:7<601::AID-YEA390>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 24.Toke DA, Martin CE. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:18413–18422. doi: 10.1074/jbc.271.31.18413. [DOI] [PubMed] [Google Scholar]
- 25.Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 26.Schneiter R, Tatzer V, Gogg G, Leitner E, Kohlwein SD. Elo1p-dependent carboxy-terminal elongation of C14:1Delta(9) to C16:1Delta(11) fatty acids in Saccharomyces cerevisiae. J Bacteriol. 2000;182:3655–3660. doi: 10.1128/jb.182.13.3655-3660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul S, et al. Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae. J Biol Chem. 2006;281:9018–9029. doi: 10.1074/jbc.M507723200. [DOI] [PubMed] [Google Scholar]
- 28.Kohlwein SD, et al. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:109–125. doi: 10.1128/MCB.21.1.109-125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proszynski TJ, et al. A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proc Natl Acad Sci USA. 2005;102:17981–17986. doi: 10.1073/pnas.0509107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huitema K, van den Dikkenberg J, Brouwers JF, Holthuis JC. Identification of a family of animal sphingomyelin synthases. EMBO J. 2004;23:33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwudke D, et al. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal Chem. 2007;79:4083–4093. doi: 10.1021/ac062455y. [DOI] [PubMed] [Google Scholar]
- 32.Alvarez-Vasquez F, et al. Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature. 2005;433:425–430. doi: 10.1038/nature03232. [DOI] [PubMed] [Google Scholar]
- 33.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 34.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 35.Pettitt TR, Dove SK, Lubben A, Calaminus SDJ, Wakelam MJO. Analysis of intact phosphoinositides in biological samples. J Lipid Res. 2006;47:1588–1596. doi: 10.1194/jlr.D600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.de Godoy LM, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.