Abstract
Regulation of RNA polymerase I (Pol I) transcription is critical for controlling ribosome synthesis. Most previous investigations into Pol I transcription regulation have focused on transcription initiation. To date, the factors involved in the control of Pol I transcription elongation are poorly understood. The Paf1 complex (Paf1C) is a well-defined factor that influences polymerase II (Pol II) transcription elongation. We found that Paf1C associates with rDNA. Deletion of genes for Paf1C subunits (CDC73, CTR9, or PAF1) reduces the rRNA synthesis rate; however, there is no significant alteration of rDNA copy number or Pol I occupancy of the rDNA. Furthermore, EM analysis revealed a substantial increase in the frequency of large gaps between transcribing polymerases in ctr9Δ mutant cells compared with WT. Together, these data indicate that Paf1C promotes Pol I transcription through the rDNA by increasing the net rate of elongation. Thus, the multifunctional, conserved transcription factor Paf1C plays an important role in transcription elongation by Pol I in vivo.
Keywords: gene expression, ribosome
Synthesis of eukaryotic ribosomes is an important and complex process that involves 78 ribosomal proteins, 4 rRNA species, and ≈200 processing factors (1, 2). rRNA transcription accounts for ≈60% of total transcriptional activity in growing yeast cells, and ≈50% of polymerase II (Pol II)-dependent transcription is devoted to ribosomal protein genes (3). Given the energetic commitment that cells make to ribosome synthesis, proper control of this process is critical.
A major target for the regulation of ribosome synthesis is transcription of rDNA by polymerase I (Pol I). Although most previous investigations into the control of Pol I activity have focused on the transcription initiation step (e.g., refs. 4 and 5), recent data reveal that the elongation step of transcription by Pol I is also an important regulatory target (6). Furthermore, recent data demonstrate that the efficiency of transcription elongation by Pol I is important for optimal processing of rRNA (7). Thus, transcription elongation by Pol I is important for both regulation of rRNA synthesis and its processing.
Despite the importance of transcription elongation by Pol I, little is known about the factors involved. To date, only 5 factors [UBF (6); TFIIS (8); Fcp1p (9); Ctk1p (10); Spt4p/Spt5p (11)] have been implicated in Pol I transcription elongation. By comparison, dozens of factors that influence Pol II elongation have been identified (for a review see ref. 12). It is likely that roles for several additional factors in Pol I transcription elongation remain to be determined.
Paf1 complex (Paf1C) is a well-characterized complex that influences transcription elongation by Pol II. Paf1C contains 5 core subunits: Cdc73p, Ctr9p, Leo1p, Paf1p, and Rtf1p (13, 14). Based on Paf1C's association with transcribed genes (13, 15), genetic interactions with other elongation factors (13, 16, 17), and biochemical activity in vitro (18), it seems clear that Paf1C directly affects Pol II elongation, but the mechanism of this effect has not been determined.
Paf1C is a multifunctional complex. In addition to its direct role in Pol II transcription elongation, Paf1C affects chromatin structure, transcription initiation, and RNA processing. In yeast, Paf1C induces ubiquitination of histone H2B [by activation of Rad6p and Bre1p (19)] and methylation of histone H3 [by recruiting COMPASS (19, 20)]. Recent data suggest similar potential roles for Paf1C in Drosophila (21). Furthermore, the Rtf1p subunit of Paf1C has been shown to influence TATA box-binding protein association with the TATA box of a Pol II promoter to modulate the efficiency of transcription initiation (22). Finally, Paf1C subunits physically interact with factors involved in mRNA processing (23). Consistent with these interactions, deletion of PAF1 leads to shortened poly(A) tails on many mRNAs (15), defective 3′ processing of small nucleolar RNAs (24), and possibly impaired mRNA export from the nucleus (25). Almost all aspects of mRNA metabolism are affected by Paf1C.
In this study, we identify an additional role for Paf1C in Pol I transcription elongation. Paf1C associates with rDNA in vivo, and the rate of Pol I transcription is reduced in paf1Δ, cdc73Δ, and ctr9Δ mutants. However, this inefficient transcription is not caused by alteration in Pol I occupancy of the rDNA or changes in rDNA copy number, suggesting a role for Paf1C in Pol I transcription elongation. EM analysis of ctr9Δ cells supports a role for Paf1C in modulating the net Pol I transcription elongation rate and suggests a specific role for Paf1C in clearing transient pause sites during transcription elongation.
Results
Subunits of Paf1C Associate With rDNA.
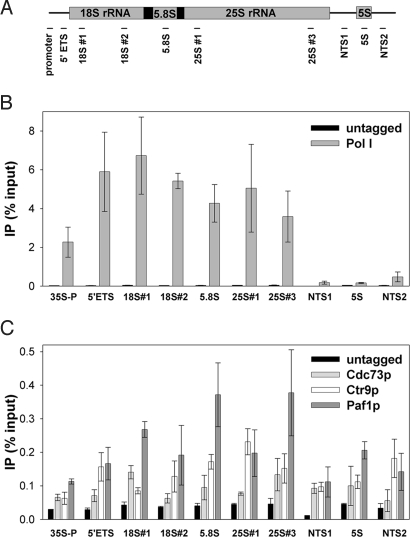
To determine whether Paf1C associates with rDNA in vivo, we constructed strains expressing his7-(HA)3-tagged Cdc73p, Ctr9p, Paf1p, and A135 (the second largest subunit of Pol I; as a positive control; Table 1). ChIP assays were performed on these strains to measure each protein's association with the rDNA, using 10 primer pairs located throughout the entire rDNA repeat (Fig. 1A).
Table 1.
Strains or plasmids used in this work
| Name | Description |
|---|---|
| Strains | |
| NOY396 | MATα ade2–1 ura3–1 trp1–1 leu2–3, 112 his3–11,15 can1–100 |
| NOY1051 | MATα ade2–1 ura3–1 trp1–1 leu2–3, 112 his3–11,15 can1–100 fob1Δ::HIS3 rpa135Δ::LEU2; pNOY117 rDNA copy number ≈143 (34) |
| NOY1064 | MATa ade2–1 ura3–1 trp1–1 leu2–3, 112 his3–11,15 can1–100 fob1Δ::HIS3; rDNA copy number ≈190 (42) |
| NOY1071 | Same as NOY1064, but rDNA copy number ≈25 (42) |
| NOY886 | Same as NOY1051, but rDNA copy number ≈42 (34) |
| DAS317 | Same as NOY396, but PAF1–his7-(HA)3-::HIS3 |
| DAS477 | Same as NOY396, but MATa RPA135–his7-(HA)3::TRP1 |
| DAS507 | Same as NOY396, but CDC73–his7-(HA)3::HIS3 |
| DAS509 | Same as NOY396, but CTR9–his7-(HA)3::HIS3 |
| DAS511 | Same as NOY396, but cdc73Δ::HIS3 |
| DAS513 | Same as NOY396, but ctr9Δ::HIS3 |
| DAS516 | Same as NOY396, but paf1Δ::HIS3 |
| Plasmids | |
| PNOY117 | pRS314 (pBluescript, CEN6, ARSH4, TRP1) derivative carrying RPA135 |
| pRS316 | pBluescript, CEN6, ARSH4, URA3; see ref. 43 |
Fig. 1.
Paf1C associates with rDNA. (A) Positions of 10 pairs of primers used in real-time PCR analyses are indicated by horizontal bars. (B) ChIP signal of Pol I (RPA135-his7-HA3; DAS477) across rDNA repeats is compared with untagged control (NOY396). Anti-HA antibody 12CA5 was used for IP of tagged proteins. IP data are expressed as a percentage of input DNA. (C) His7-(HA)3-tagged Cdc73p, Ctr9p, and Paf1p (DAS507, DAS509, DAS317) associate with the rDNA repeat. Data were quantified from at least 2 10-fold dilutions per sample from duplicate cultures. Error = ± 1 SD.
As expected, our ChIP data clearly detected strong binding of Pol I to the transcribed regions of the rDNA (Fig. 1B). The large decrease in ChIP signal observed in the nontranscribed sequence (NTS) and 5S regions demonstrates the specificity and sensitivity of the assay. Using the same primer pairs and same antibody for immunoprecipitation (IP), we also detected significant association of Cdc73p, Ctr9p, and Paf1p with the rDNA (Fig. 1C). The association of Cdc73p, Ctr9p, or Paf1p with the rDNA in the 35S rRNA coding region was ≈10-fold less than that of Pol I, but several-fold higher than background signal from the untagged negative control samples. This reduced signal relative to Pol I is expected for an elongation factor that may not bind DNA directly and is consistent with previous observations (11, 26).
Unlike Pol I, Paf1C proteins also associated with the NTS regions and the 5S rDNA. This finding is consistent with previous observations for Chd1p and Isw1p (26) and the Spt4/Spt5 complex (D.A.S., unpublished results). The reason for positive ChIP signal in the regions of rDNA that are devoid of Pol I remains unknown. However, it is known that Pol II transcripts originate from the NTS regions of the rDNA, and Pol III is active at the 5S rRNA genes (27, 28). Thus, this signal may result from association of these factors with the other polymerases that are clearly active in the spacers between 35S rRNA genes. Overall, these data confirm that Paf1C associates with the rDNA in vivo. Thus, these factors are poised to affect Pol I transcription elongation.
Deletion of CDC73, CTR9, or PAF1 Decreases rDNA Transcription in Vivo.
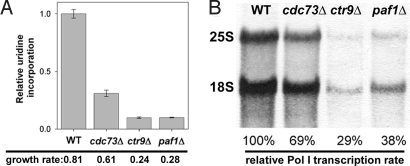
To characterize the role that Paf1C plays in Pol I transcription, we constructed 3 strains carrying cdc73Δ, ctr9Δ, or paf1Δ mutations (Table 1). The 3 mutants grew more slowly than the parental WT strain (Fig. 2A) and exhibited temperature sensitivity at 37 °C. To estimate Pol I transcription rates in vivo, the total RNA synthesis rate was measured by pulse-labeling yeast cells for 5 min with 3H-uridine. Because most RNA synthesis during exponential growth is Pol I-derived, this is a reliable method for approximating Pol I activity. Uridine incorporation into total RNA was reduced ≈3-fold in the cdc73Δ strain and ≈10-fold in both the ctr9Δ and paf1Δ stains relative to WT (Fig. 2A). This trend is consistent with the observed decreases in growth rate and suggests that Paf1C may play a significant role in Pol I transcription in vivo.
Fig. 2.
Pol I transcription is reduced in strains defective in Paf1C function. (A) Relative RNA synthesis rates were measured by using 3H-uridine incorporation in WT (NOY396), cdc73Δ (DAS511), ctr9Δ (DAS513), and paf1Δ (DAS516) strains. Growth rates are expressed as doublings per h. WT and Paf1C deletion mutants, carrying the pRS316 plasmid, were grown in SD-Ura media to A600 ≈ 0.3. Duplicate samples from duplicate cultures were quantified, averaged, and expressed relative to WT. Error = ± 1 SD. (B) RNA was purified from cells pulse-labeled with 3H-methylmethionine. After electrophoresis, RNA was transferred to a membrane and detected by autoradiography. 25S and 18S rRNA were cut from the membrane, quantified with a scintillation counter and expressed relative to WT. Data shown are from 1 of 3 independent experiments. Error was <3%.
To quantify the Pol I transcription rate, we pulse-labeled cells with 3H-methylmethionine for 5 min, chased them with excess cold methionine for 5 min (to permit completion of rRNA processing), isolated RNA, and measured the 3H incorporation into 25S and 18S rRNA (Fig. 2B). Because rRNA is methylated cotranscriptionally in yeast and the cellular pool of methylmethionine is very low, this method is a reliable way to quantify Pol I transcription in vivo (29). We observed a small reduction in the Pol I transcription rate in the cdc73Δ mutant compared with WT (69% of WT activity) and a larger reduction in ctr9Δ and paf1Δ strains (29% and 38% of WT activity, respectively). These values for inhibition of Pol I activity in the mutant strains are more reasonable for the detected defects in growth, and the greater level of inhibition of total RNA synthesis likely reflects an additional defect in uridine uptake in the mutant strains. Nevertheless, these data confirm that Pol I transcription is inhibited in the Paf1C mutant strains, consistent with their reduced growth rates and lower total RNA synthesis rates (Fig. 2A).
Deletion of CDC73, CTR9, or PAF1 Does Not Affect Pol I Occupancy of the rDNA.
Deletion of Paf1C subunits severely reduced Pol I transcription in vivo (Fig. 2); however, ChIP analysis detected no significant change in Pol I occupancy of the rDNA in Paf1C mutants relative to WT (Fig. 3). Additionally, no ChIP signal for Pol I was observed in the NTS1 region, indicating that termination of Pol I transcription was not significantly impaired in Paf1C mutants.
Fig. 3.
ChIP analysis of Pol I occupancy of the rDNA in WT (NOY396), cdc73Δ (DAS511), ctr9Δ (DAS513), and paf1Δ (DAS516). The primer pairs used are diagramed in Fig. 1A. Polyclonal anti-A190 antibody was used to immunoprecipitate Pol I. At least 2 10-fold dilutions of each sample were averaged from duplicate cultures. Error = ± 1 SD.
Processivity of Pol I was also unchanged in Paf1C mutants. Processivity describes the ability of a polymerase to elongate throughout the length of a gene. Relative processivity is estimated by comparing polymerase densities near the 5′ end of the gene with the 3′ end. Previous investigations have identified defects in Pol II processivity upon deletion of candidate elongation factors (30). Because there is no apparent decrease in the Pol I ChIP signal at the 3′ end of the rDNA, it is clear that there is no significant decrease in the relative processivity in Paf1C mutants compared with WT.
Taken together, these data indicate that deletion of Paf1C subunits does not lead to a detectable reduction in Pol I loading or processivity, despite large decreases in rRNA synthesis rates. Thus, the simplest interpretation of these data are that deletion of CDC73, CTR9, or PAF1 reduces the Pol I transcription elongation rate.
Effects of Deletion of CDC73, CTR9, or PAF1 on rDNA Copy Number.
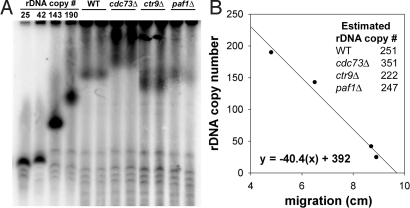
Because the rDNA in eukaryotic cells exists as a tandemly-repeated array, recombination events can lead to variation in the rDNA copy number (11, 28, 31). A potential decrease in the rDNA copy number could reduce rRNA synthesis rate. To control for this possibility, we measured the rDNA copy number in Paf1C mutants compared with reference strains with known rDNA copy numbers.
We observed no decrease in the rDNA copy numbers in Paf1C mutants. The rDNA copy numbers were estimated by using contour-clamped homogenous field electrophoresis (CHEF) followed by Southern blot hybridization. Because ≈60% of chromosome XII in yeast is rDNA sequences (32), the migration distance of chromosome XII through a gel is heavily influenced by changes in the rDNA array. To quantify the rDNA copy number in the Paf1C mutants and the WT control, we compared the mobility of chromosome XII with that from strains with known rDNA copy numbers (Fig. 4). Because the Paf1C mutants are Fob1+, recombination between rDNA repeats is efficient, and there is significant variation in rDNA copy number within a culture (33). Migration of chromosome XII was measured from Southern blot hybridizations by using an rDNA probe to clearly detect the location of rDNA (Fig. 4A). From a linear regression of chromosome migration distance versus known rDNA copy number in reference strains, we estimated that ctr9Δ, and paf1Δ mutants carry approximately an equal number of rDNA copies as WT, and the rDNA copy number was increased ≈40% in the cdc73Δ mutant (Fig. 4B). This increase in rDNA copy number may contribute to the reduced severity of the cdc73Δ mutation on Pol I transcription (Fig. 2).
Fig. 4.
rDNA copy number was measured by CHEF and Southern blot hybridization. (A) To visualize chromosome XII, we used a 32P-labeled rDNA probe in Southern blot hybridization. Reference strains with known rDNA copy numbers (NOY1071, 25 copies; NOY886, 42 copies; NOY1051, 143 copies; and NOY1064, 190 copies) were included. DNA was isolated from 2 independent cultures of WT (NOY396), cdc73Δ (DAS511), ctr9Δ (DAS513), and paf1Δ (DAS516) and analyzed as described (see Materials and Methods). (B) A linear regression was generated by plotting copy number of reference strains versus the migration distance from the wells to the middle of the chromosome XII signal. From this regression, rDNA copy number was estimated for WT, cdc73Δ, ctr9Δ, and paf1Δ.
Taken together, the effects of Paf1C on Pol I transcription are independent of changes in processivity and number of engaged polymerases or rDNA copy number, which is consistent with Paf1C affecting the Pol I transcription elongation rate.
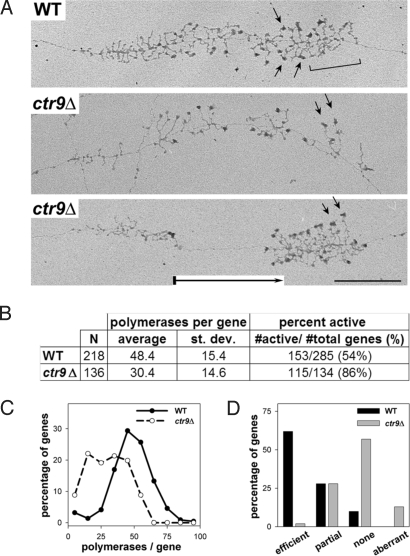
EM Analysis Revealed Impaired Transcription Elongation in ctr9Δ Cells.
To further test our model for Paf1C's role in Pol I transcription, we analyzed Pol I transcription in vivo by using EM analysis of Miller chromatin spreads of the ctr9Δ strain. EM analysis detected a relatively small decrease in the Pol I density per gene relative to WT (representative raw data in Fig. 5A). Quantification of these data showed that the average number of polymerases per gene was reduced 37% in the ctr9Δ strain compared with WT (Fig. 5 B and C). Thus, the initiation rate per gene is lower in the mutant strain than in WT and the overall defect in transcription initiation is slightly greater than the net defect in transcription elongation (see Discussion).
Fig. 5.
EM analysis of Miller chromatin spreads reveals defects in Pol I transcription elongation and rRNA processing. (A) Representative rRNA genes from Miller spreads of WT (NOY396) and ctr9Δ (DAS513) cells. WT rRNA genes showed cleavage of nascent transcripts, indicated by a bracket (Top). Genes from ctr9Δ often exhibited polymerase-free gaps, indicated by a boxed arrow in the gene in Bottom. Arrows indicate uncleaved transcripts. (Scale bar: 0.5 μM.) (B) Quantification of Pol I densities and percentage of active rDNA genes in WT and ctr9Δ strains. Error and size of dataset are indicated. (C) Percentage of genes with different polymerase occupancies is shown for WT and ctr9Δ strains. (D) Cotranscriptional cleavage efficiency was classified into 4 categories (efficient, partial, none, and aberrant) and plotted versus the frequency of detection.
Not all of the rDNA repeats are transcriptionally active in growing yeast cells (34). The fraction of active versus inactive rDNA genes was also quantified by EM. We found a 32% increase in the fraction of active rDNA genes in ctr9Δ strain relative to WT (Fig. 5B), thus offsetting the decrease in initiation rate per gene. From these data, we can calculate the average number of polymerases engaged in transcription in the WT and mutant cells [(rDNA copy number)(% active)(no. pols per gene)]. Based on this equation, there are ≈6,500 engaged polymerases per WT cell and ≈5,800 per ctr9Δ cell. Because the rRNA synthesis rate in the ctr9Δ cells is only 29% of the WT level, the net elongation rate of Pol I in ctr9Δ cells is 3.1-fold slower than in WT cells [fold defect in rRNA synthesis (100% divided by 29%) multiplied by the ratio of number of polymerases engaged in transcription in the mutant versus WT (5,800 divided by 6,500) = 3.1].
Processing of rRNA Is Impaired in ctr9Δ Cells.
Previous studies have suggested that transcription elongation by Pol I is functionally linked to rRNA processing (7, 11). Our EM analysis revealed defects in rRNA processing in ctr9Δ cells. The majority of pre-rRNAs in WT cells are initially processed at sites A0, A1, and A2 to release the 20S precursor to 18S RNA. In all control yeast strains examined by EM, the separating cleavage at A2 can be seen to occur cotranscriptionally as transcripts reach the 3′ end of the gene (35). As shown in Fig. 5A, WT cells displayed this cotranscriptional cleavage event (bracketed within Fig. 5A Top). However, deletion of CTR9 caused a reduction in the efficiency of this cleavage event compared with WT (Fig. 5 A and D), with ≈60% of active rDNA genes lacking this normal cotranscriptional cleavage. This observation is consistent with published data from the Jaehning laboratory [showing accumulation of rRNA precursors in paf1Δ cells (25)]. Thus, these data are consistent with the model that the efficiency of transcription elongation by Pol I affects rRNA processing.
Polymerase-Free Gaps Are More Frequently Observed in ctr9Δ Cells Compared with WT.
EM analysis also revealed large gaps between transcribing Pol I complexes in ctr9Δ mutant (Fig. 5A Bottom, indicated by boxed arrow). The percentage of genes with large polymerase-free gaps (≥ ¼ of the gene length) was 36% in ctr9Δ (n = 143) compared with 12% in WT (n = 302). These gaps likely reflect sites of paused complexes. Increased susceptibility to pausing/arresting has been shown to correlate with slower elongation rates (36).
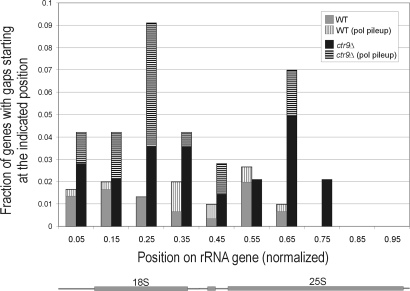
The positions of the 5′ ends of these gaps were mapped to determine whether there was any evidence for specific sites that Pol I was less capable of clearing in Paf1C mutants (Fig. 6). The presence or absence of a polymerase pileup at the 5′ end of the gap, perhaps caused by a roadblock effect, was also noted. This analysis showed that gap start sites were distributed across the gene in both WT and mutant strains. However, in ctr9Δ, there were 2 regions, 1 within the 18S RNA sequence and 1 within the 25S RNA sequence that had relatively more gap start sites. Because these gap sites were significantly more frequent in ctr9Δ cells, and a large portion of the gaps clearly involved polymerase blocks or pileups, we propose that Paf1C might play a specific role in transcribing through these regions of the gene.
Fig. 6.
Gaps between transcribing Pol I complexes are more frequently observed in ctr9Δ cells than in WT. Position of 5′ ends of gaps (gap start sites) is shown on an rRNA gene normalized in length. Also indicated is the portion of gap start sites with polymerase pileups (see Materials and Methods). The absence of gap start sites in the 3′ quarter of the gene is caused by the minimum length requirement (¼ gene length) for gaps to be included in the analysis.
Discussion
The data presented here demonstrate that subunits of Paf1C (Cdc73p, Ctr9p, and Paf1p) associate with the rDNA and contribute to the mechanism of Pol I transcription elongation in vivo. Deletion of CDC73, CTR9, or PAF1 leads to reduced rRNA synthesis rates (Fig. 2), but Pol I occupancy of the rDNA is not reduced as severely (as observed using ChIP and EM; Figs. 3 and 5). The simplest interpretation of these data is that Paf1C plays an important, positive role in Pol I transcription elongation under normal growth conditions. Thus, we have identified an additional role for Paf1C in Pol I transcription in yeast, and this role may be conserved in higher eukaryotes as well.
In addition to defects in Pol I transcription rates in Paf1C mutant strains, we observed defects in rRNA processing. Cotranscriptional cleavage of the 35S pre-rRNA was significantly reduced (as observed with EM; Fig. 5), consistent with previous studies that detected accumulation of rRNA precursors in paf1Δ mutants (25). The observation of rRNA processing defects when Pol I transcription elongation is compromised supports the previously proposed model that transcription elongation by Pol I is coupled to rRNA processing (7).
Mechanism by Which Paf1C Influences Pol I Transcription.
Genetic and biochemical studies have implicated Paf1C in Pol II transcription elongation, but the mechanism by which Paf1C affects Pol II remains controversial (15). Our ChIP and EM analyses showed that there was little (or no) change in Pol I occupancy of the rDNA or processivity in cdc73Δ, ctr9Δ, and paf1Δ strains compared with WT, despite large decreases in rRNA synthesis rates in the mutant strains. These data suggest that Paf1C positively influences Pol I transcription elongation in vivo.
Our data also indicate that transcription initiation by Pol I is directly or indirectly affected by Paf1C. Because the >3-fold reduction in Pol I elongation rate was not accompanied by a large increase in the polymerase density per gene, the initiation rate was also reduced. Indeed, in the ctr9Δ strain, polymerase density per gene was slightly lower than WT (Fig. 5), thus the initiation rate was affected slightly more than the elongation rate. It is not clear whether Paf1C directly affects the transcription initiation rate, or whether promoter clearance and early elongation events indirectly affect the assembly of the initiation complex.
From our EM data, we observed a 3-fold increase in the frequency of detection of gaps between Pol I complexes in the ctr9Δ cells compared with WT (Fig. 6). We propose that a major role for Paf1C in vivo is to directly increase the transcription elongation rate of Pol I through the rDNA, making the enzyme less prone to pause. Whether disruption of Paf1C affects the pausing of the transcription elongation complex at specific sites or whether a general decrease in the elongation rate of Pol I renders the complex more prone to pausing remains to be determined, although analysis of gap positions suggests contributions from both.
Immunofluorescence studies have shown that subunits of Paf1C can be detected in the nucleolus (25). However, those analyses suggested that the complex was largely absent from the nucleolus in WT cells, but enriched when the genes for Paf1 subunits (PAF1 or RTF1) were deleted. Our ChIP analyses (Fig. 1) clearly show that Paf1C is in close proximity to the rDNA, even in growing WT cells.
The simplest mechanism we can propose accounting for all of the known data are that Paf1C directly associates with transcribing Pol I complexes and modulates its elongation efficiency. Although this is the simplest model, we cannot exclude more complicated indirect models by which Paf1C might affect Pol I transcription. For example, Paf1C recruits complexes that modify histones in transcribed regions of Pol II genes (19, 20). Thus, it is possible that Paf1C cooperates with other modulators of chromatin to facilitate Pol I transcription. Furthermore, because Paf1C mutations influence Pol II gene expression (37), we cannot exclude potential indirect effects of changes in the expression of other potential transcription elongation factors on Pol I transcription. Detailed in vitro analyses are required to clearly identify direct effects of Paf1C on Pol I transcription elongation.
Paf1C Affects the Chromatin Structure of Inactive rDNA.
We observed an increase in the percentage of active rDNA repeats in the ctr9Δ strain (Fig. 5B) relative to WT. This observation may reflect a role for Ctr9p in maintenance of silent chromatin structure within the rDNA. Previous studies have shown that PAF1 is required for efficient rDNA silencing (38). Both of these observations are consistent with the model that Paf1C directly or indirectly affects the maintenance of silent chromatin within the rDNA. The mechanism by which this occurs is a topic for future investigation.
Does Paf1C Regulate Pol I Transcription?
Recent evidence suggests that transcription elongation by Pol I is regulated to control rRNA synthesis, but the mechanisms responsible for this regulation are largely unknown. The most detailed study of Pol I elongation as a regulatory target showed that the transcription factor UBF affects transcription elongation by Pol I in mammalian cells and that this effect on elongation was regulated in response to growth factor stimulation (6). The data presented here do not address whether Paf1C serves as a regulatory target for the control of rRNA synthesis. However, preliminary data suggest that deletion of CTR9 or PAF1 also leads to inappropriate regulation of Pol I transcription in yeast cells.
In higher eukaryotes, the homologues of Paf1C have been identified (39, 40). Overexpression of hPaf1 enhances cell growth and induces tumor formation; however, transient overexpression of HRPT-2 (the human homologue of CDC73) suppresses cell proliferation (40, 41). The involvement of Paf1C in dysregulation of cell growth during cancer development suggests that Paf1C directly or indirectly contributes to regulation of ribosome synthesis in mammalian cells. Transcription elongation by Pol I as a target for the control of rRNA synthesis is a topic that is rapidly gaining interest and one that will require significant attention in future studies.
Relationship Between Spt4p and Paf1C at the rDNA.
We have shown that both Spt4p/Spt5p and Paf1C affect Pol I transcription elongation in vivo, in addition to their known roles in Pol II transcription (11). It has been reported that Spt4p recruits Paf1p to transcribing Pol II complexes (17). Is it possible that Spt4p plays a similar role in Pol I transcription? Phenotypic data suggest this may not be the case. Deletion of SPT4 had a modest effect on Pol I elongation (11). In contrast, deletion of CTR9 or PAF1 induces large decreases in the Pol I elongation rate and in overall cell growth rate compared with WT cells. Although both Spt4p/Spt5p and Paf1C contribute to Pol I transcription elongation, it seems that the two complexes function differently. However, more detailed experiments are required to determine the relative contributions of Paf1C and Spt4p/Spt5p to Pol I transcription in vivo and in vitro.
Conclusions
We have identified an important role for the conserved Paf1C in transcription elongation by Pol I. The importance of transcription elongation by Pol I to regulation of rRNA synthesis (6) and processing of rRNA (refs. 7 and 26 and this study) is becoming clear. However, by comparison to the list of factors known to influence transcription elongation by Pol II, very little is known about the control of Pol I elongation. Thus, many questions remain to be addressed regarding the mechanisms that control transcription elongation of Pol I through rDNA.
Materials and Methods
Strains and Media.
Strains and plasmids used in this study are listed in Table 1. Cells were cultured in either yeast extract/peptone/dextrose or synthetic glucose complete (SD) media and were grown at 30 °C with aeration (4). Epitope tags [his7-(HA)3] were incorporated into Cdc73p, Ctr9p, and Paf1p by using standard methods (11).
ChIP.
ChIP analysis was performed as described (11) except that cells were treated with formaldehyde for only 6 min to cross-link in vivo. ChIP signals were quantified by real-time PCR using an ABI 7900HT system (Applied Biosystems). Each reaction contained 1× GeneChoice buffer (GeneChoice), 200 μM dNTPs, 100 μg/ml BSA, 0.01× SYBR green (Invitrogen), 25 units/mL Taq polymerase (NEB), and 450 nM each primer. We note that our quantification was performed over at least a 10-fold range of input and IP DNA concentrations. Because this analysis takes into account the true precision of the experiment over a range of DNA concentrations, error bars are larger than would be expected for simple triplicate analysis.
Measurement of RNA Synthesis in Vivo.
Duplicate cultures of WT, cdc73Δ, ctr9Δ, and paf1Δ, carrying pRS316 were grown in SD-Ura medium overnight to exponential phase (OD600 ≈0.3). 3H-uridine (5 μCi) was mixed with 500 μL of each culture and incubated at 30 °C for 5 min with aeration. Then the samples were treated with 2.5 mL of 10% trichloroacetic acid (TCA) with 2.5 mg/ml of uridine. After filtration through nitrocellulose, each membrane was washed with 5% TCA, dried, and counted in a Beckman LS 6500 multipurpose scintillation counter.
To measure Pol I transcription rates, we grew cells in SD-met medium and pulse-labeled cells for 5 min with 20 μCi/mL 3H-methyl methionine (see also ref. 29). We followed that with a chase of cold methionine (500 μg/mL) for 5 min. RNA was prepared from these cells, run on a formaldehyde agarose gel, transferred to a membrane, and detected by autoradiography. Individual RNA species were excised from the blot (together with nearby regions of the blot for assessment of background) and were quantified with a scintillation counter.
rDNA Copy Number Determination.
rDNA copy number was determined by using CHEF followed by Southern blot hybridization as described (31). Four reference strains (NOY886, NOY1051, NOY1064, and NOY1071) bearing known rDNA copy number were included. For Southern hybridization, an rDNA probe was amplified by using a forward primer (5′-GGGGCACCTGTCACTTTGGAAA-3′) and a reverse primer (5′-GCTGATTTGAGAGGAGGTTAC-3′). The resulting 600-bp probe was then labeled with 32P-dCTP (Redi Prime kit; GE Health U.K. Limited) and hybridized as described (31). A linear regression was constructed by plotting the migration distance of chromosome XII in known copy number reference strains versus the rDNA repeat number. From this regression, the average rDNA copy number was calculated for Paf1C deletion mutants and the WT control.
EM Analysis.
EM Miller chromatin spreading analysis was performed as described (11). Polymerase-free gaps that were at least ¼ of the total gene length were analyzed by mapping their position within the gene. For each gap, it was also noted whether or not there were 3 or more very closely positioned (apparently touching) polymerases at its 5′ end, indicating a polymerase pileup. When the gaps were at the extreme 5′ (or 3′) end of the gene, the position of the gene start (or end) site was estimated by using the average length of neighboring genes.
Acknowledgments.
We thank Dr. N. P. Higgins for use of his CHEF-mapper electrophoresis system and Drs. Y. Osheim and S. French for help with EM data analysis. This work was supported by American Cancer Society Grant IRG-60-001-47 (to D.A.S.), National Cancer Institute Grant CA-13148-31 (to D.A.S.), and National Institutes of Health Grant GM63952 (to A.L.B.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Wade CH, Umbarger MA, McAlear MA. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast. 2006;23:293–306. doi: 10.1002/yea.1353. [DOI] [PubMed] [Google Scholar]
- 2.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 3.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 4.Claypool JA, et al. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol Biol Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefanovsky V, et al. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell. 2006;21:629–639. doi: 10.1016/j.molcel.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 7.Schneider DA, et al. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell. 2007;26:217–229. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnapp G, Graveley BR, Grummt I. TFIIS binds to mouse RNA polymerase I and stimulates transcript elongation and hydrolytic cleavage of nascent rRNA. Mol Gen Genet. 1996;252:412–419. doi: 10.1007/BF02173006. [DOI] [PubMed] [Google Scholar]
- 9.Fath S, et al. Dephosphorylation of RNA polymerase I by Fcp1p is required for efficient rRNA synthesis. J Biol Chem. 2004;279:25251–25259. doi: 10.1074/jbc.M401867200. [DOI] [PubMed] [Google Scholar]
- 10.Bouchoux C, et al. CTD kinase I is involved in RNA polymerase I transcription. Nucleic Acids Res. 2004;32:5851–5860. doi: 10.1093/nar/gkh927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider DA, et al. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci USA. 2006;103:12707–12712. doi: 10.1073/pnas.0605686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 13.Krogan NJ, et al. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- 16.Squazzo SL, et al. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu H, Hu C, Wong CM, Hinnebusch AG. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol Cell Biol. 2006;26:3135–3148. doi: 10.1128/MCB.26.8.3135-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rondón AG, Gallardo M, García-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood A, et al. The Paf1 complex is essential for histone monoubiquitination by the Rad6–Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 20.Krogan NJ, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 21.Adelman K, et al. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stolinski LA, Eisenmann DM, Arndt KM. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4490–4500. doi: 10.1128/mcb.17.8.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nordick KM, Hoffman MG, Betz JL, Jaehning JA. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryotic Cell. 2008;7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheldon KE, Mauger DM, Arndt KM. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porter SE, Penheiter KL, Jaehning JA. Separation of the Saccharomyces cerevisiae Paf1 complex from RNA polymerase II results in changes in its subnuclear localization. Eukaryotic Cell. 2005;4:209–220. doi: 10.1128/EC.4.1.209-220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones HS, et al. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat Struct Mol Biol. 2007;14:123–130. doi: 10.1038/nsmb1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.French SL, et al. Visual analysis of the yeast 5S rRNA gene transcriptome: Regulation and role of La protein. Mol Cell Biol. 2008;28:4576–4587. doi: 10.1128/MCB.00127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 29.Warner JR. Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:423–428. doi: 10.1016/0076-6879(91)94033-9. [DOI] [PubMed] [Google Scholar]
- 30.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Oakes M, et al. Transcription factor UAF, expansion, and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol Cell Biol. 1999;19:8559–8569. doi: 10.1128/mcb.19.12.8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y.-H., et al. Chromosome XII context is important for rDNA function in yeast. Nucleic Acids Res. 2006;34:2914–2924. doi: 10.1093/nar/gkl293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.French SL, et al. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osheim YN, et al. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 36.Powell W, Reines D. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J Biol Chem. 1996;271:6866–6873. doi: 10.1074/jbc.271.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang M, et al. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu B, et al. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodard GE, et al. Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene. 2004;24:1272–1276. doi: 10.1038/sj.onc.1208274. [DOI] [PubMed] [Google Scholar]
- 41.Rozenblatt-Rosen O, et al. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cioci F, et al. Silencing in yeast rDNA chromatin: Reciprocal relationship in gene expression between RNA polymerase I and II. Mol Cell. 2003;12:135–145. doi: 10.1016/s1097-2765(03)00262-4. [DOI] [PubMed] [Google Scholar]
- 43.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]