Abstract
Nucleocytoplasmic coupling is mediated by outer nuclear membrane (ONM) nesprin proteins and inner nuclear membrane Sun proteins. Interactions spanning the perinuclear space create nesprin–Sun complexes connecting the cytoskeleton to nuclear components. A search for proteins displaying a conserved C-terminal sequence present in nesprins 1–3 identified nesprin 4 (Nesp4), a new member of this family. Nesp4 is a kinesin-1-binding protein that displays Sun-dependent localization to the ONM. Expression of Nesp4 is associated with dramatic changes in cellular organization involving relocation of the centrosome and Golgi apparatus relative to the nucleus. These effects can be accounted for entirely by Nesp4's kinesin-binding function. The implication is that Nesp4 may contribute to microtubule-dependent nuclear positioning.
Keywords: centrosome, LINC complex, nuclear envelope
The nuclear envelope (NE) forms the interface between the nucleus and cytoplasm acting as a selective barrier that regulates nucleo-cytoplasmic traffic (1). In addition to this partition function, accumulating evidence reveals an essential role for the NE as a determinant of higher-order nuclear and chromatin organization (2). More surprising are findings that the NE directly impacts cytoskeletal architecture and in this way may help define the mechanical properties of the cell as a whole (3). The molecular bases for these effects are now emerging, uniting aspects of cellular physiology from mechanotransduction to nuclear positioning during differentiation and development (4).
The NE is assembled from several elements, the most prominent being inner nuclear membranes (INMs) and outer nuclear membranes (ONMs) separated by a ≈40-nm gap or perinuclear space (PNS). The INM and ONM are joined where they are spanned by nuclear pore complexes (NPCs), the mediators of trafficking across the NE. The ONM also displays connections to the peripheral endoplasmic reticulum (ER). Accordingly, the INM, ONM, and ER represent a single membrane system with the PNS forming an extension of the ER lumen.
The final feature of the NE is the nuclear lamina, a protein meshwork composed primarily of A- and B-type lamins that lines the nuclear face of the INM (2). The lamina is required for NE integrity and provides chromatin-anchoring sites at the nuclear periphery. Remarkably, aberrant A-type lamin expression, which is linked to several human diseases (5), is associated with altered cytoskeletal mechanics (3). The mechanisms underlying this phenomenon represent an intriguing biological problem.
At least 60 NE membrane proteins are known (6). Although most likely reside in the INM, several ONM proteins have been identified (7). These include members of the mammalian nesprin (or syne) family (8, 9), Klarsicht (10) and Msp-300 (11, 12) in Drosophila melanogaster, Anc-1 (13), Zyg-12 (14) and Unc-83 (15, 16) in Caenorhabditis elegans, and Kms2 in the fission yeast Schizosaccharomyces pombe (17). A property that each of these has in common is that they interact with cytoskeletal components. They are also united in possessing a conserved ≈50- to 60-residue C-terminal KASH domain (Klarsicht, Anc-1, Syne homology) featuring a single transmembrane segment followed by a short luminal sequence. Localization of ONM KASH proteins depends on tethering by SUN domain proteins of the INM (18). This tethering, involving interactions spanning the PNS, was originally suggested based on findings that localization of Anc-1 depends on an INM protein, Unc-84 (19, 20), a prototype member of the SUN family.
Three mammalian nesprin genes are known [nesprins 1–3 (7)]. For nesprins 1 and 2, the primary transcripts encode a plethora of alternatively-spliced isoforms (9). The largest of these, nesprin 1 Giant (Nesp1G; ≈1,000 kDa) and nesprin 2 Giant (Nesp2G; ≈800 kDa), reside in the ONM. Smaller isoforms may be found in the INM (21, 22) and elsewhere. The large flexible cytoplasmic domains of Nesp1G and Nesp2G each feature an N-terminal actin binding domain (ABD) followed by multiple spectrin repeats. The third mammalian nesprin (Nesp3) contains an N-terminal binding site for plectin, a cytolinker molecule that may provide a bridge to the intermediate filament system (23).
The nesprins are tethered in the ONM by a pair of Unc-84-related INM proteins, Sun1 and Sun2 (24–27). Because these also interact with nuclear components, including A-type lamins (24, 25, 28), and a histone acetyl transferase [in the case of Sun1 (29)], Sun-nesprin pairs represent links in a molecular chain connecting the cytoskeleton to nuclear structures. We refer to these as LINC complexes (linker of the cytoskeleton and nucleoskeleton). The emerging theme is that all KASH proteins have one or more complementary SUN proteins that function as tethers. In this way, multiple LINC isoforms may exist in and between species.
Are nesprins 1–3 the only mammalian KASH proteins that contribute to the LINC repertoire? We sought to answer this question by searching for proteins containing KASH-like sequences. We report here the characterization of an epithelial-specific KASH protein that represents a fourth branch of the nesprin family and is noteworthy in its ability to bind kinesin-1. Furthermore, its expression is associated with a dramatic separation of the nucleus and centrosome, indicating a possible role in microtubule-dependent nuclear positioning.
Results and Discussion
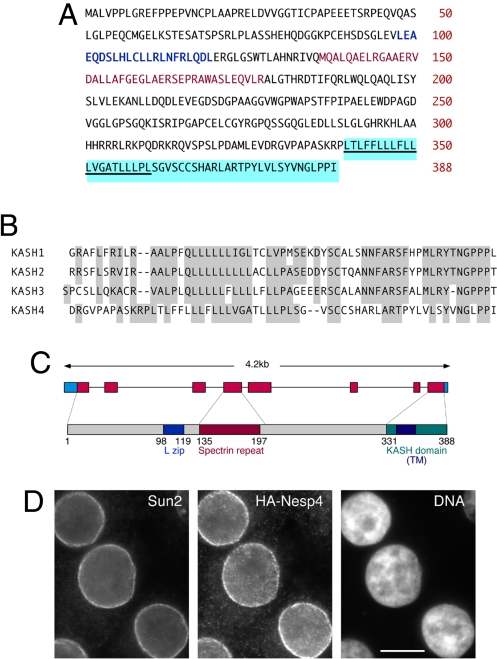
To identify new nesprin family members we performed a BLASTP search using the human Nesp2 KASH domain (KASH2) as probe. This search identified an unknown mouse protein (NP_705805) of 388 amino acid residues and predicted molecular mass of 42 kDa (Fig. 1A). Comparison of cDNA (AI428936) and genomic sequences (NC_000073.5) and analysis using FAST-DB (www.fast-db.com) suggest that NP_705805 represents the full-length protein (Fig. 1B). The region of homology with KASH2 resides at the C terminus, as expected for a KASH protein. Overall it displays 38% identity with KASH2. For comparison the KASH domains of nesprins 1–3 display pairwise identities of 64–79%. Thus, NP_705805 seems to contain a C-terminal KASH domain whose sequence has diverged from those of nesprins 1–3 (Fig. 1C).
Fig. 1.
Identification of nesprin 4. (A) Amino acid sequence of mouse protein NP_705805, now designated nesprin 4 (Nesp4). Nesp4 contains a leucine zipper (L Zip; blue), a spectrin repeat (red), and a KASH domain (KASH4; blue highlight). The putative transmembrane sequence (TM) is underlined. (B) Alignment of the KASH domains (KASH1–4) from nesprins 1–4. (C) Nesp4 gene (eight exons, 4. 2 kb) and protein domain organization. (D) Immunofluorescence microscopy of HSG cells stably expressing HA–Nesp4. The cells were labeled with antibodies against HA and Sun2. DNA is visualized with Hoechst dye. (Bar: 10 μm.)
NP_705805, and its human homologue (NP_001034965), displays little similarity with other proteins, although it does contain a partial spectrin repeat and a short leucine zipper that might promote dimerization (Fig. 1B). We expressed a full-length NP_705805 cDNA (IMAGE Clone ID 5036575), tagged with an N-terminal HA-epitope or GFP, in both human salivary gland (HSG) cells (Fig. 1D and Fig. S1) and HeLa cells (Fig. 2A and Fig. S2). Immunofluorescence microscopy, using antibodies against both Sun2 (Fig. 1D) and A-type lamins (Fig. S1), revealed that NP_705805 is targeted to the NE. Given this localization, combined with the presence of both the spectrin repeat and a C-terminal KASH domain, we propose that NP_705805 represents an additional member of the mammalian nesprin family, nesprin 4 (Nesp4). Differential permeabilization of HSG cells stably expressing HA–Nesp4 (HSG-HAN4), using digitonin versus Triton X-100 (30), revealed that the N terminus of HA–Nesp4 must be exposed on the ONM. Thus, Nesp4 must have the same topology as Nesp1–3 (Fig. S1). We cannot, however, exclude the possibility that a population of Nesp4 may also be present in the INM.
Fig. 2.
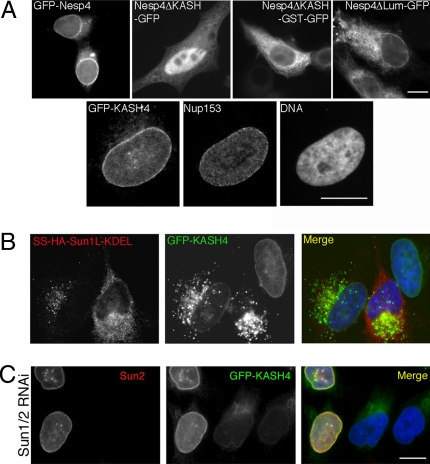
SUN- and KASH-dependent localization of Nesp4. (A) HeLa cells transiently transfected with either GFP-tagged full-length Nesp4 (GFP–Nesp4) or a variety of GFP-tagged deletion mutants (Nesp4ΔKASH–GFP, Nesp4ΔKASH–GST—GFP, and Nesp4ΔLum–GFP). Deletion of the KASH domain or the luminal portion of the KASH domain prevents localization to the NE. Conversely, GFP fused to the Nesp4 KASH domain (GFP–KASH4) localizes to the NE as revealed by colabeling with an antibody against the NPC protein Nup153. (B) Transfection of SS–HA–Sun1L–KDEL into HeLa cells stably expressing GFP–KASH4 displaces GFP–KASH4 from the NE. (C) Depletion of Sun1 and Sun2 by RNAi leads to loss of NE-associated GFP-KASH4. In the merged images, DNA (blue) was revealed by costaining with Hoechst dye. (Bars: 10 μm.)
Localization of Nesp4 to the NE is KASH-dependent. Replacement of the KASH domain with GFP (Nesp4ΔKASH-GFP) leads to a cytoplasmic and nuclear distribution (Fig. 2A). Insertion of a GST module between the Nesp4 and GFP sequences (Nesp4ΔKASH–GST–GFP) results in an exclusive cytoplasmic localization. Presumably its greater bulk limits nuclear entry. Replacement of the luminal portion of the KASH domain with GFP (Nesp4ΔLum–GFP) also results in loss of NE association. However, because the predicted transmembrane domain remains intact, the mutant protein becomes distributed throughout what appears to be the peripheral ER. Conversely, GFP–KASH4, generated by fusion of the Nesp4 KASH domain (KASH4) to the C terminus of GFP, localizes to the NE (Fig. 2A). These data indicate that the KASH domain is both necessary and sufficient for the ONM localization of Nesp4. This view is reinforced by findings that GFP–KASH4 or GFP–Nesp4 will displace HA–Nesp4, endogenous Nesp2G (Fig. S2), and Nesp3 (data not shown) from the ONM. Similarly, overexpression of GFP–KASH2 (Fig. S2) will displace ONM-associated HA–Nesp4. Evidently KASH4 competes with other nesprins for NE-associated tethering molecules.
Localization of GFP-KASH4 or GFP-Nesp4 to the ONM is SUN-dependent and can be abolished by expression of SS–HASun1LKDEL (Fig. 2B), a SUN protein dominant negative mutant (24). Similarly, depletion of Sun1/2 by RNA interference results in failure to retain GFP–KASH4 at the NE (Fig. 2C). Our conclusion is that Nesp4 is tethered in the ONM by SUN–KASH interactions and that it defines additional mammalian LINC complex isoforms.
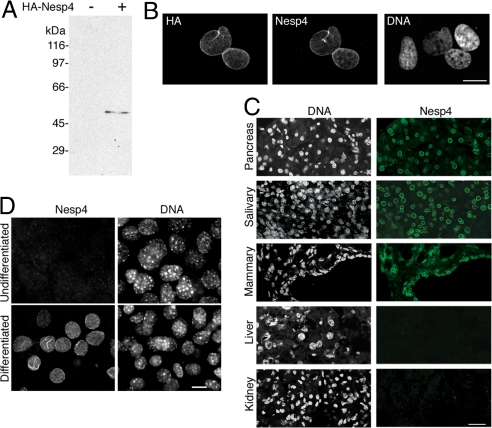
Antibodies raised against recombinant Nesp4 detect mouse HA–Nesp4, expressed in HeLa or HSG cells, by both Western blot analysis and immunofluorescence microscopy (Fig. 3 A and B). However, endogenous Nesp4 was not detected in any common cell line. We therefore surveyed multiple mouse tissue cryosections by immunoflourescence microscopy. Surprisingly, most tissues displayed no evidence of Nesp4 expression. However, Nesp4 was present in the NEs of salivary gland, exocrine pancreas, bulbourethral gland, and mammary tissue (Fig. 3C). The occasional kidney cell was also Nesp4-positive. These data imply that Nesp4 is restricted mainly to secretory epithelia. Northern blot analysis of pregnant mouse mammary RNA revealed a 1.4- to 1.7-kb transcript, consistent in size with Nesp4 cDNA sequences (Fig. S3). Quantitative RT-PCR using multiple primers gave no evidence for alternative splicing (data not shown).
Fig. 3.
Nesp4 is expressed in secretory epithelial cells. (A) Western blot analysis of HeLa cells using rabbit anti-Nesp4. The cells were either nontransfected (−) or expressed HA-Nesp4 (+). A single band (≈54 kDa) corresponding to HA–Nesp4 is evident only in the transfected sample. The relatively slow migration of HA–Nesp4 (its predicted molecular mass is 42 kDa) is likely caused by its high proline content (9%). (B) HeLa cells expressing HA–Nesp4 and labeled with rabbit anti-Nesp4 and mouse anti-HA. The rabbit anti-Nesp4 only decorates the NE of transfected cells (C) Immunofluorescence microscopy of mouse tissue cryosections using the rabbit anti-Nesp4 antibody. (D) Immunofluorescence microscopy of both undifferentiated and differentiated HC11 mouse mammary cells using the anti-Nesp4 antibody. In all positive cells Nesp4 is localized at the NE. All specimens were stained with Hoechst dye to reveal the DNA. (Bars: B and D, 10 μm; C, 40 μm.)
Based on these findings, we examined Nesp4 expression in HC11 mouse mammary cells (31). Upon hormonal induction, HC11 cells differentiate to yield a morphologically heterogeneous population secreting milk proteins. Undifferentiated HC11 cultures contain few Nesp4-positive cells (Fig. 3D). However, during the 1- to 2-week differentiation process Nesp4-positive cells accumulate until they represent 40–50% of the population. In all of these cells, Nesp4 is localized exclusively at the NE (Fig. 3D). Additional observations of HC11 cells reveal many that lack Nesp4 but still acquire an epithelioid appearance (delineated by the junctional protein ZO1; Fig. S4A) and at the same time express milk components such a whey acidic protein (Fig. S4B). Clearly Nesp4 expression is not a prerequisite for HC11 differentiation, at least with respect to milk protein production.
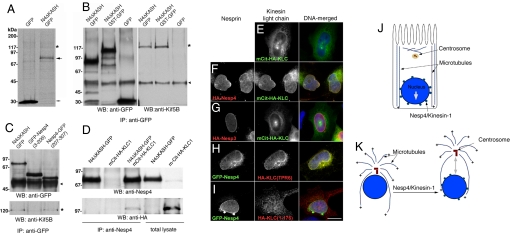
We used anti-GFP immunoprecipitation (IP) of Nesp4ΔKASH–GFP (Fig. 2) expressed in HEK293 cells to identify Nesp4-interacting proteins. In pilot IPs using 35S-labeled cells, in addition to Nesp4ΔKASH–GFP itself, a faint band of ≈120 kDa was consistently observed (Fig. 4A). This band was absent from IPs containing GFP alone. Scale-up of the IPs and analysis by mass spectrometry revealed that the 120-kDa band contained Kif5B, the heavy-chain subunit of kinesin-1 [also known as conventional kinesin or KIF5 (32)]. Parallel yeast two-hybrid analysis (33), using a membrane proximal region of Nesp4 (residues 129–339) as bait, was performed by Myriad Genetics. This analysis revealed kinesin light chains (KLCs) 1, 2, 3, and 4 as candidate Nesp4 binding partners. Together, these data imply that Nesp4 is a kinesin-binding protein. To verify that we immunoprecipitated Nesp4ΔKASH–GFP, Nesp4ΔKASH–GST–GFP, and GFP alone from HeLa cells. IPs were analyzed by Western blot using anti-Kif5B. Fig. 4B shows that Kif5B is found in a complex with the Nesp4 cytoplasmic domain. In all likelihood, given the two-hybrid data, formation of this complex is mediated by the KLCs. This notion is supported by the finding that a Nesp4 deletion mutant [Nesp4-GFP(207–307)], encompassing 200 residues contained within the two-hybrid bait construct, will also coimmunoprecipitate with Kinesin-1 (Fig. 4C). A mutant lacking this region [GFP-Nesp4(3–206)] shows no such interaction.
Fig. 4.
Nesp4 interacts with kinesin-I. (A) Anti-GFP IP analysis of 35S-Met/Cys-labeled HEK293 cells expressing either Nesp4ΔKASH–GFP (large arrow) or GFP alone (small arrow). A faint ≈120-kDa band (*) present only in the Nesp4ΔKASH–GFP IPs was found, by mass spectrometry, to contain Kif5B, the kinesin-1 heavy chain. (B) Unlabeled HeLa cells expressing Nesp4ΔKASH–GFP, Nesp4ΔKASH–GST–GFP, or GFP were processed for IP using anti-GFP. IPs were analyzed by Western blot using either anti-GFP or anti-Kif5B. Kif5B (*) was clearly evident in IPs containing either Nesp4ΔKASH–GFP or Nesp4ΔKASH–GST–GFP. Ig heavy chains are indicated (arrowhead). (C) Similar analysis of HeLa cells expressing Nesp4ΔKASH–GFP, GFP–Nesp4(3–206), and Nesp4–GFP(207–307). Kinesin-1 coimmunoprecipitates only with Nesp4ΔKASH–GFP and Nesp4–GFP(207–307). (D) HeLa cells were transfected with various combinations of Nesp4ΔKASH–GFP and with KLC1 fused to a fluorescent protein and HA-tagged mCit-HA-KLC1. IP of Nesp4 followed by Western blot analysis using both anti-Nesp4 and anti-HA antibodies reveals association between Nesp4 and mCit–HA–KLC1. Immunofluorescence microscopy of HeLa cells expressing various combinations of nesprins and KLC1. (E) mCit–HA–KLC1 alone localizes to the cytoplasm. (F) HA–Nesp4 recruits mCit–HA–KLC1 to the NE. (G) Nesp3 has no effect on mCit–HA–KLC1. (H) An HA-tagged version of the KLC1 TPR domain [HA-KLC(TPR6)] is recruited to the NE by GFP–Nesp4. (I) The distribution of HA–KLC(1–176), which lacks the TPR domain, is unaffected by GFP–Nesp4. In the merged images, DNA, revealed by staining with Hoechst dye, is shown in blue. (Bar: 10 μm.) (J) In polarized epithelial cells microtubules are noncentrosomal. The bulk are arranged in lateral bundles with their plus (+) ends oriented toward the basal membrane. Kinesin-1 on the NE should drive the nucleus toward the base of the cell. (K) Nesp4-mediated recruitment of kinesin-1 to the NE in fibroblasts should induce separation of the nucleus and centrosome.
If Nesp4 binds kinesin via the light chains it should recruit KLCs to the NE. We therefore introduced a fluorescent protein-tagged version of KLC1 [mCit–HA–KLC1 (34)] into HeLa cells. mCit–HA–KLC1 alone adopts a generalized cytoplasmic distribution (Fig. 4E). However, coexpression with HA–Nesp4, but not HA–Nesp3 (or Nesp1α; data not shown), causes accumulation of mCit–HA–KLC1 at the NE (Fig. 4 F and G). Similar results were obtained with a KLC mutant [Fig. 4H; HA-KLC(TPR6)] containing the presumptive cargo-binding tetratricopeptide repeat (TPR) domain but lacking the heptad repeat region that interacts with the kinesin heavy chain (35). In contrast, KLC lacking the TPR domain [Fig. 4I; HA-KLC(1–176)] is unaffected by Nesp4 expression. Evidently Nesp4 interacts specifically with the KLC TPR domain. Additional evidence for this interaction between Nesp4 and KLC-1 is provided by the finding that Nesp4ΔKASH–GFP will coimmunoprecipitate with mCit–HA–KLC1 when both recombinant proteins are coexpressed in HeLa cells (Fig. 4D). Based on these findings we would predict that if Nesp4 does indeed function as a binding partner for kinesin-1, then endogenous kinesin should be detectable at the NE of differentiated HC11 cells. Immunofluorescence microscopy of such cells using antibodies against both Nesp4 and Kif5b suggests that this is indeed the case (Fig. S5).
What significance can be attached to the Nesp4–kinesin-1 association? We speculate that Nesp4 could contribute to microtubule-dependent nuclear positioning in secretory epithelial cells. A feature of such cells is that they contain lateral bundles of microtubules that extend from the apical surface with their plus ends oriented toward the basal membrane (36). Nesp4-mediated recruitment to the NE of kinesin-1, a plus-end-directed motor protein, would be predicted to promote nuclear migration toward the base of the cell (Fig. 4J).
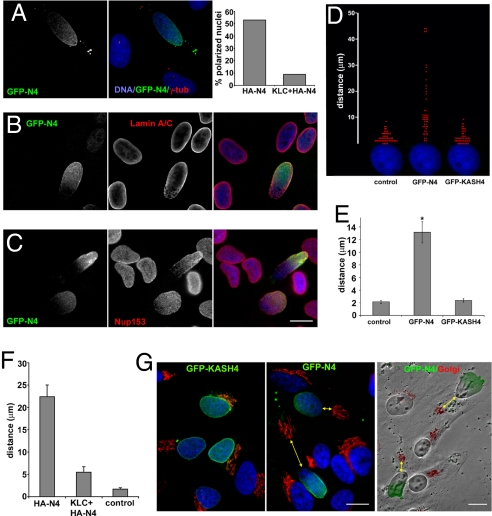
At present we have no epithelial cell system in which to examine this proposed function for Nesp4. Although differentiated HC11 cells do express Nesp4, their heterogeneity makes them unsuitable for any study of epithelial morphogenesis (Fig. S4). Nonetheless, we can use nonpolarized cells to determine how Nesp4 might affect the interaction of the nucleus with the cytoskeleton and whether it might induce changes in cytoarchitecture. When expressed in HSG cells, both HA–Nesp4 and GFP–Nesp4 usually display a uniform distribution about the nuclear surface. However, in a minority of cells HA–Nesp4 and GFP–Nesp4, but not GFP–KASH4, accumulate at a pole of the nucleus that is invariably distal to the centrosome. The effect is most prevalent in HeLa cells (≈50% of cells are affected; Fig. 5 A–C and Fig. S6) where expression of Nesp4 leads to the polarization of other NE components, including lamins and NPCs (Fig. 5 B and C). This effect may be prevented by treatment of the cells with nocodazole (data not shown) or overexpression of mCit–HA–KLC1 (Fig. 5A). The latter should behave in a dominant negative fashion by saturating both kinesin heavy chains and Nesp4. Because microtubules are anchored at the centrosome by their minus ends, the polarization of Nesp4 may be explained by its kinesin-1-mediated anterograde movement across the nuclear surface. In other words, Nesp4 behaves as a kinesin cargo, providing a functional binding site for kinesin-1 at the NE.
Fig. 5.
Nesp4 expression perturbs centrosome and Golgi positioning. (A) In ≈50% of HeLa cells, exogenous Nesp4 (GFP–N4) concentrates at a pole of the nucleus that is always distal to the centrosome (γ-tubulin). (B and C) Both A-type lamins (B) and NPCs (Nup153; C), copolarize with Nesp4. Nesp4 polarization is abolished by overexpression of KLC1 (A). Expression of Nesp4 causes separation of the nucleus from the centrosome. (D) In the scatter plot, the position of individual centrosomes (red dots) in multiple cells is displayed relative to the nucleus (in blue). (E) Centrosomes in nontransfected control cells or cells expressing GFP–KASH4 lie within 2 μm of the nucleus. Those in cells expressing GFP–Nesp4 (GFP–N4) are on average ≈13 μm from the nucleus (n = > 47; *, P = 0.0001). (F) This effect of Nesp4 can be abrogated by overexpression of KLC1. (G) Nesp4 has similar effects on the Golgi apparatus, causing it to depart from its normal perinuclear location (yellow arrows). In merged images, DNA is revealed by staining with Hoechst dye. In the right image in G, GFP–Nesp4 and the Golgi apparatus (in red) are overlaid on the same field visualized by using phase-contrast optics. (Bars: 10 μm.)
If the nucleus is engaged with centrosomal microtubules via Nesp4 and kinesin-1, we would predict that this interaction could drive the separation of the nucleus from the centrosome (Fig. 4K). In fact, this is exactly what occurs. In nontransfected HeLa cells or HeLa cells expressing GFP–KASH4, the centrosome is located within 2 μm of the nucleus (Fig. 5 A, D, and E and Fig. S6). Expression of either HA-Nesp4 or GFP-Nesp4 increases this distance to ≈13 μm (Fig. 5 A and D–F and Fig. S4). Indeed, values of 40–50 μm are not unusual. To put these numbers in perspective, loss of emerin or A-type lamins leads to a 3- to 4-μm separation between the centrosome and nucleus (37, 38). Our findings suggest that Nesp4 expression does not simply disrupt centrosome tethering, but actively drives the separation of these structures. Furthermore it can be repressed by coexpression of mCit–HA–KLC1 (Fig. 5F). The only reasonable conclusion is that this effect of Nesp4 is mediated by recruitment of kinesin-1 to the NE (Fig. 4K). Whether the nucleus moves relative to the centrosome or vice versa is unknown. However, studies on centrosome reorientation in migrating fibroblasts suggests that the centrosome itself might remain relatively immobile (39).
In nonepithelial cells, the Golgi apparatus displays a peri-nuclear localization in association with the centrosome (40). Given its effect on centrosome/nuclear positioning, the prediction is that introduction of Nesp4 would cause similar dislocation of the Golgi apparatus. Again, this is precisely what happens. Cells expressing Nesp4, but not GFP–KASH4, display obvious disengagement of the Golgi apparatus from the nucleus (Fig. 5G). Indeed we have observed Nesp4-expressing cells in which the Golgi apparatus lies adjacent to the plasma membrane at the cell margins. This raises the question of whether it might lead to perturbations in membrane trafficking, particularly with respect to the distribution and targeting of exocytic vesicles and the delivery of membrane and secretory proteins to the plasma membrane. Clearly expression of a single ONM protein can induce dramatic changes in cytoarchitecture, effects that are unprecedented for a NE protein. These observations raise the possibility that Nesp4 might indeed contribute to the establishment of secretory epithelial morphology, by promoting kinesin-dependent apical migration of the centrosome and Golgi apparatus and basal localization of the nucleus. We should be in a better position to explore this possibility as mice rendered deficient in Nesp4 become available.
In conclusion, we have identified Nesp4 as a kinesin-1-binding protein of the NE and have been able to demonstrate clear consequences of this binding in vivo. The identification of Nesp4 now completes the association between various mammalian LINC complex isoforms and all major branches of the cytoskeleton. However, Nesp4 is not the only member of the nesprin family to interact with a kinesin. A Nesp1 isoform binds kinesin-2, mediating the transport of membrane to the midbody of cells undergoing cytokinesis (41). This finding raises the possibility that other nesprins at the NE might also bind kinesins. Consequently, Nesp4-expressing cells may not be unique in recruiting kinesin to the NE. Both our findings and those from other laboratories make it increasingly evident that NE represents a nexus of cytoskeletal interactions (7). The implication, of course, is that perturbations in the structure and composition of the NE may have far-reaching effects on cell and tissue organization.
Materials and Methods
Plasmids.
A mouse nesprin 4 (NP_705805) cDNA (clone ID 5036575) was obtained from Invitrogen. HA–Nesp4 was inserted into pcDNA 3.1(−), GFP–Nesp4 and GFP–KASH4 were inserted into pEGFP-C1 (Clontech), Nesp4ΔKASH–GFP, Nesp4ΔKASH—GST–GFP, and Nesp4ΔLum-GFP were inserted into pEGFP-N1 (Clontech), and GFP and Nesp4ΔKASH–GFP were inserted into pLNCX2 (Clontech), all by standard PCR cloning. GFP–KASH1 and GFP–KASH2 were a gift from Catherine Shanahan (Kings College, London). The HA–Nesprin 3 plasmid (23, 26) was a gift from Arnoud Sonnenberg (Netherlands Cancer Institute, Amsterdam). HA–KLC1(TPR6), HA–KLC1(1–176), and mCit–HA–KLC1 plasmids (34) were a gift from Kristen Verhey (University of Michigan Medical School, Ann Arbor).
Antibodies.
The following antibodies were used in this study: the monoclonal anti-lamin A/C (XB10) (42), anti-Nup153 (SA1) (43), anti-HA (12CA5; Covance), anti-γ-tubulin (GTU-88, Sigma), anti-golgi (58K-9; Sigma); polyclonal rabbit anti-HA (ab9110; AbCam), anti-Sun2 (44), anti-GFP (ab290; AbCam), and polyclonal goat anti-kinesin 1 (ab15075; AbCam). Polyclonal rabbit anti-Nesp4 was raised against a GST–Nesp4(1–90) fusion protein by Rockland Immunochemicals. GST-fusion protein purification and affinity purification of antisera was performed as described (24). Secondary antibodies, conjugated with AlexaFluor dyes or peroxidase, were from Biosource International/Invitrogen.
Cell Culture and Transfections.
HeLa, HSG, and HEK293 cells were maintained in 6.0% CO2 at 37 °C in DMEM supplemented with 10% FBS, 10% penicillin/streptomycin, and 2 mM l-glutamine. Transfections were performed by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. HC11 cells (provided by Kermit Caraway, University of Miami, Miami) were maintained and differentiated as described (31).
Generation of Stable Cell Lines.
HSG cells stably expressing HA–Nesp4 and HeLa cells stably expressing GFP–KASH4 were generated by G418 (600 μg/mL) selection and subcloning. HEK293 cells stably expressing GFP or Nesp4ΔKASH–GFP were generated by retroviral transduction. Briefly, the appropriate pLNCX2 plasmid DNA was transiently transfected into the amphotropic retroviral packaging cell line Phoenix A (obtained from Garry Nolan, Stanford University, Stanford, CA). Infective supernatant was recovered 48 h after transfection. Retroviral transduction of HEK293 cells was performed in the presence of 4 μg/mL polybrene. Infected cells were selected with G418.
Immunofluorescence Microscopy.
Cells grown on glass coverslips were fixed in 3% formaldehyde (prepared in PBS from paraformaldehyde, PFA/PBS) and immunolabeled as appropriate (24). To quantify separation of the centrosome/Golgi apparatus from the nucleus the distance from the centrosome to the nearest point on the nuclear periphery was measured by using IPLab Spectrum software (BD Biosciences) calibrated with a stage micrometer.
Tissue Imaging.
Tissues removed from mice after CO2 asphyxiation were frozen in liquid nitrogen-cooled 2-methyl butane, and sections (≈10 μm) were cut by cryostat and mounted on glass slides. Sections were fixed with 3% PFA/PBS for 10 min, permeabilized with 0.2% Triton X-100, and processed as described (45). Images were collected by using a Leica TCS SP5 confocal microscope system running Leica Application Suite 1.8.2 software.
Immunoblots and IPs.
For immunoblots, proteins were fractionated by SDS/PAGE and analyzed by Western blot (46). For radiolabeled IPs, 3.5-cm plates of HEK293 cells transiently expressing NespΔKASH–GFP or GFP were labeled with 50 μCi of 35S-translabel (MP Biomedicals) for 16 h before lysis in 0.5 mL of buffer [150 mM NaCl, 50 mM Tris (pH 7.4), 2.5 mM MgCl2, 0.5% Triton X-100, 1 mM DTT, 10 μg/ml each of chymostatin, leupeptin, antipain, and pepstatin, and 1 mM PMSF]. Lysates were passed through a 21-gauge needle (10×) and centrifuged 16,000 × g for 10 min at 4 °C. The supernatants were rotated for 4 h at 4 °C with protein A Sepharose beads (Sigma) coupled to rabbit anti-GFP. Samples were analyzed by SDS/PAGE and fluorography (46). For the larger-scale IPs, 10-cm plates of HEK293 cells were lysed in 1 mL of buffer and processed as described above. After SDS/PAGE, radiolabeled samples were processed as described (46). A gel with nonradiolabeled samples was silver-stained, and bands unique to the NespΔKASH–GPF IP were excised and submitted to the University of Florida's Interdisciplinary Center for Biotechnology Research proteomics laboratory for tryptic digest before mass spectrometry analysis (QTRAP 4000; Applied Biosystems). Tandem mass spectrophotometric data were searched by using Mascot version 2.2 (Matrix Science) and compiled by using Scaffold 1.7 (Proteome Software).
Northern Blot Analysis.
Twenty micrograms of total RNA samples isolated from mouse mammary tissue were analyzed by Northern blot using a 32P-labeled full-length Nesp4 cDNA probe using established protocols (47).
Yeast-Two-Hybrid Analysis.
A fragment of the mouse Nesp4 cytoplasmic domain (residues 129–339) was used in a yeast two-hybrid analysis to screen 3 separate cDNA libraries: pooled 7- to 19-day total mouse embryo, mouse uterus/mammary gland mix, and mouse ovary. The analysis was carried out by Myriad Genetics using published procedures (33).
Supplementary Material
Acknowledgments.
We thank Kristen Verhey for kinesin cDNA constructs and very helpful advice and Arnoud Sonnenberg and Catherine Shanahan for antibodies and cDNAs. This work was supported by a grant from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808602106/DCSupplemental.
References
- 1.Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: Structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- 2.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 3.Lammerding J, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart CL, Roux KJ, Burke B. Blurring the boundary: The nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- 5.Worman HJ, Bonne G. Laminopathies: A wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 7.Wilhelmsen K, Ketema M, Truong H, Sonnenberg A. KASH-domain proteins in nuclear migration, anchorage, and other processes. J Cell Sci. 2006;119:5021–5029. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- 8.Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J Biol Chem. 2000;275:31986–31995. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Q, et al. Nesprins: A novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114:4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- 10.Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr Biol. 1999;9:1211–1220. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- 11.Rosenberg-Hasson Y, Renert-Pasca M, Volk T. A Drosophila dystrophin-related protein, MSP-300, is required for embryonic muscle morphogenesis. Mech Dev. 1996;60:83–94. doi: 10.1016/s0925-4773(96)00602-8. [DOI] [PubMed] [Google Scholar]
- 12.Volk T. A new member of the spectrin superfamily may participate in the formation of embryonic muscle attachments in Drosophila. Development. 1992;116:721–730. doi: 10.1242/dev.116.3.721. [DOI] [PubMed] [Google Scholar]
- 13.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 14.Malone CJ, et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 15.McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starr DA, et al. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- 17.King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: “Velcro” that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- 19.Lee KK, et al. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starr DA, Han M. ANChors away: An actin-based mechanism of nuclear positioning. J Cell Sci. 2003;116:211–216. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- 21.Mislow JM, et al. Nesprin-1α self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 2002;525:135–140. doi: 10.1016/s0014-5793(02)03105-8. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q, et al. Nesprin-2 is a multiisomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- 23.Wilhelmsen K, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crisp M, et al. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haque F, et al. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ketema M, et al. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J Cell Sci. 2007;120:3384–3394. doi: 10.1242/jcs.014191. [DOI] [PubMed] [Google Scholar]
- 27.Padmakumar VC, et al. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 28.Hasan S, et al. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 2006;580:1263–1268. doi: 10.1016/j.febslet.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 29.Chi YH, Haller K, Peloponese JM, Jr, Jeang KT. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in decondensation of mitotic chromosomes. J Biol Chem. 2007;282:27447–27458. doi: 10.1074/jbc.M703098200. [DOI] [PubMed] [Google Scholar]
- 30.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ball RK, Friis RR, Schoenenberger CA, Doppler W, Groner B. Prolactin regulation of β-casein gene expression and of a cytosolic 120-kDa protein in a cloned mouse mammary epithelial cell line. EMBO J. 1988;7:2089–2095. doi: 10.1002/j.1460-2075.1988.tb03048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navone F, et al. Cloning and expression of a human kinesin heavy chain gene: Interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J Cell Biol. 1992;117:1263–1275. doi: 10.1083/jcb.117.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 34.Cai D, Hoppe AD, Swanson JA, Verhey KJ. Kinesin-1 structural organization and conformational changes revealed by FRET stoichiometry in live cells. J Cell Biol. 2007;176:51–63. doi: 10.1083/jcb.200605097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adio S, Reth J, Bathe F, Woehlke G. Review: Regulation mechanisms of kinesin-1. J Muscle Res Cell Motil. 2006;27:153–160. doi: 10.1007/s10974-005-9054-1. [DOI] [PubMed] [Google Scholar]
- 36.Bacallao R, et al. The subcellular organization of Madin–Darby canine kidney cells during the formation of a polarized epithelium. J Cell Biol. 1989;109:2817–2832. doi: 10.1083/jcb.109.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JS, et al. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ, Hutchison CJ. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 40.Rogalski AA, Singer SJ. Associations of elements of the Golgi apparatus with microtubules. J Cell Biol. 1984;99:1092–1100. doi: 10.1083/jcb.99.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan J, Beck KA. A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J Cell Sci. 2004;117:619–629. doi: 10.1242/jcs.00892. [DOI] [PubMed] [Google Scholar]
- 42.Raharjo WH, Enarson P, Sullivan T, Stewart CL, Burke B. Nuclear envelope defects associated with LMNA mutations cause dilated cardiomyopathy and Emery–Dreifuss muscular dystrophy. J Cell Sci. 2001;114:4447–4457. doi: 10.1242/jcs.114.24.4447. [DOI] [PubMed] [Google Scholar]
- 43.Bodoor K, et al. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J Cell Sci. 1999;112:2253–2264. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- 44.Hodzic DM, Yeater DB, Bengtsson L, Otto H, Stahl PD. Sun2 is a novel mammalian inner nuclear membrane protein. J Biol Chem. 2004;279:25805–25812. doi: 10.1074/jbc.M313157200. [DOI] [PubMed] [Google Scholar]
- 45.Roux KJ, Amici SA, Notterpek L. The temporospatial expression of peripheral myelin protein 22 at the developing blood–nerve and blood–brain barriers. J Comp Neurol. 2004;474:578–588. doi: 10.1002/cne.20154. [DOI] [PubMed] [Google Scholar]
- 46.Liu Q, et al. Functional association of Sun1 with nuclear pore complexes. J Cell Biol. 2007;178:785–798. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ausubel FM, et al., editors. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.