Abstract
Telomere integrity is maintained through end-protection proteins that block nuclease degradation and prevent telomeres from being recognized as DNA breaks. Although less well understood, end protection proteins may also play a role in facilitating telomere replication. Here, we show that overproduction (OP) of the yeast telomere capping protein Stn1 makes cells highly sensitive to the replication inhibitors hydroxyurea (HU) and methyl-methane sulfonate (MMS). Unexpectedly, this sensitivity corresponds with Stn1 OP blocking most, if not all, aspects of the S phase checkpoint. The checkpoint kinase Rad53 is phosphorylated with normal timing in Stn1 OP cells, indicating Stn1 does not interfere with signaling steps involved in activating the checkpoint. Part of the role of Stn1 in telomere integrity is mediated through the Pol12 subunit of DNA polymerase α (Polα). We show that overproduced Stn1 generally associates with chromosomes in HU treated and untreated cells, and, remarkably, Stn1 chromosome binding and OP checkpoint defects are rescued in pol12 mutants. We propose Stn1 normally promotes Polα activity at telomeres but can be recruited through Pol12 to nontelomeric sites when overproduced. During replication stress, the mislocalized Stn1 may inappropriately promote Polα in a manner that interferes with Rad53 effector mechanisms controlling replication fork integrity.
Keywords: polymerase α, POL12, RAD53
Telomere-binding proteins have a critical role facilitating linear chromosome maintenance, forming complexes that not only protect the chromosome ends, but also regulate extension of the G-rich telomere repeats by telomerase (1). Telomere-binding proteins may also impact terminal replication forks or postreplicative synthesis of the telomere C-strand (2, 3). Although less well understood, roles for telomere proteins in telomere replication are likely to be crucial, because failure to fully duplicate the chromosome termini can compromise genome stability. Telomeres that lose the function of their protective protein complexes are unmasked and activate DNA damage checkpoint signaling pathways. Failure to block inappropriate nuclease action or to duplicate fully the chromosome termini can compromise genome stability.
In the budding yeast Saccharomyces cerevisiae, Cdc13 binds to the single-strand G-rich telomere repeats and collaborates with 2 interacting proteins, Stn1 and Ten1, to protect chromosome ends (4), preventing generation of extensive telomere-proximal single-stranded (ss) DNA during S and G2/M phases of the cell cycle (5, 6). A consensus model is that Cdc13, Stn1, and Ten1 function together to bind telomeric DNA, forming a physical cap that blocks nuclease activity. The situation, however, may be more complex. In particular, simultaneously increasing the level of the N-terminal putative OB-fold domain of Stn1 together with Ten1 allows telomere capping even in the complete absence of Cdc13, indicating redundant or alternate Cdc13-independent means for achieving a protected state (7). Moreover, in Schizosaccharomyces pombe, the Stn1 and Ten1 homologs apparently function independent from Pot1, which binds the ss telomere G-rich DNA (8).
In addition to telomere capping, Cdc13, Stn1 and Ten1 are involved in telomere replication. Cdc13 has been shown to be important in allowing telomerase to access chromosome ends to synthesize the G-rich telomere strand (4). Furthermore, after telomerase extension, completion of telomere replication requires that Polα synthesize the telomere C-rich strand, using the G-rich ss region as template. Based in part on interactions of Cdc13 and Stn1 with Polα, Cdc13, Stn1, and Ten1 are suggested to promote Polα activity at telomeres (9, 10). In some way that is not yet understood, Polα is critical for Stn1 to promote Cdc13-independent capping, suggesting a relationship between the capping and replicative aspects of Stn1 function (7).
Here, we demonstrate that increased Stn1 levels, and in particular, the Stn1 C terminus, strongly interfere with the S phase checkpoint response to DNA replication stress. This interference occurs at a step downstream of activation of Rad53, the central kinase responsible for transducing the S phase checkpoint signal, and affects multiple checkpoint controls, including regulation of late origin firing and replication fork progression. Overproduced Stn1 is broadly distributed on chromosomal DNA, and mutations in the Pol12 regulatory subunit of Polα block Stn1 chromosome association. Critically, these same pol12 alleles also restore the S phase checkpoint in Stn1 OP cells. Based on these findings, we discuss a model in which Stn1 and Pol12 normally interact to promote Polα replicative activity at telomeres. When Stn1 levels are increased, however, Pol12 redirects Stn1 to other chromosomal sites, potentially misregulating Polα in a way that overrides S phase checkpoint-mediated replication fork control.
Results
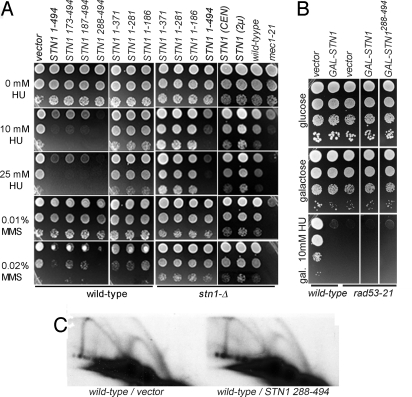
We previously showed that the Stn1 C terminus provides only modest dosage suppression of cdc13-1 temperature sensitivity and cannot bypass CDC13 essential function in a manner similar to the Stn1 N terminus (7). In investigating these observations, we found that Stn1 OP in wild-type cells leads to extreme sensitivity to DNA replication stress induced by either HU or MMS (Fig. 1A). This toxicity requires Stn1 OP, because strains expressing STN1 from its native promoter on either high-copy or low-copy plasmids show similar HU- and MMS-resistance as controls (Fig. 1A). The severity of this phenotype was remarkable. Full-length Stn1 (Stn11-494) and the nonessential Stn1 C terminus (Stn1173-494, Stn1187-494, Stn1288-494) confer sensitivity to 10 mM HU, although Stn1288-494 sensitizes cells to concentrations of HU as low as 5 mM [supporting information (SI) Fig. S1]. On the other hand, the Stn1 N terminus has a significantly reduced impact (Fig. 1A) (7). Thus, OP of a C-terminal fragment of Stn1 is sufficient to induce extreme sensitivity to DNA replication stress. This region contains no obvious protein motifs, but interacts with Pol12 and Cdc13 (7, 10).
Fig. 1.
Stn1-overproducing cells are HU and MMS sensitive. (A) Twentyfold serial dilutions of the indicated strains were stamped on HU- or MMS-containing media. Wild-type cells bear empty vector (pLX416) or high-copy plasmids expressing full-length or truncated STN1 from the ADH promoter (OP plasmids: pCN418-pCN424). stn1-Δ cells are kept viable by OP plasmids or plasmids expressing STN1 from its native promoter (pCN1, pVL1066). Wild-type and mec1–21 strains are shown for comparison. (B) Stn1 OP is not toxic in rad53–21. Tenfold serial dilutions of wild-type and rad53–21 cultures with vector, pGAL-STN1 (pVL1051) or pGAL-STN1288–494 (pPC33) were stamped on the indicated media. (C) Subtelomere Y′ replication intermediates in pADH-STN1288–494 (pCN186)-expressing cells. EcoR1-digested DNA was fractionated on 2D gels and probed for an ARS within the a telomere Y′ element (32).
There are at least 2 general interpretations for the sensitivity of Stn1-overproducing cells to replication stress. First, Stn1 OP might cause DNA damage independently of exogenous stress or otherwise act to increase the potency of HU or MMS. However, Rad53 is not required to maintain viability in cells overexpressing STN1 from a galactose-inducible promoter in the absence of HU (Fig. 1B). Thus, Stn1 OP alone does not impose a requirement for DNA damage checkpoint-surveillance mechanisms. In addition, neither aberrant ss DNA nor an altered replication fork pattern is observed in cells overexpressing Stn1288-494 (Fig. 1C, data not shown), suggesting that Stn1 OP did not obviously cause DNA replication stress on its own.
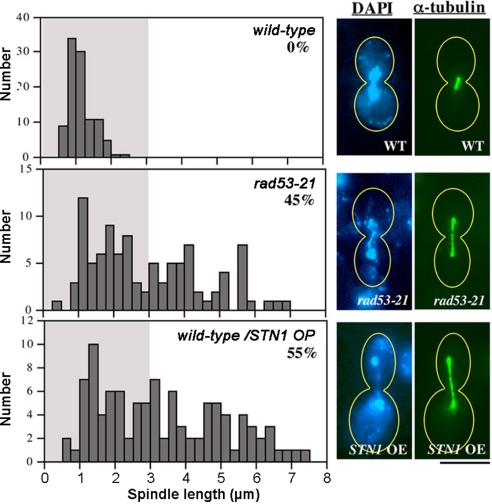
A second interpretation of the HU sensitivity of Stn1 OP strains is that the S phase checkpoint is defective. In budding yeast, a defining phenotype of S phase checkpoint mutants is that they exhibit premature extension of the mitotic spindle after HU treatment, leading to abortive segregation of partially replicated chromosomes (11–13). S phase checkpoint proficient cells, on the other hand, arrest with short spindles. We observed that the Stn1 overproducing cells show a severe uncoupling of DNA replication and spindle extension in the presence of HU, exhibiting a percentage of cells with abnormally extended spindles comparable to HU-treated rad53-21 mutants (Fig. 2).
Fig. 2.
Spindle elongation in HU-treated Stn1 OP cells. G1-arrested cells were released into 200 mM HU, fixed after 3 h, and stained with α-tubulin. Histograms show the distribution of spindle lengths in wild-type, rad53–21, or pADH-STN1 cells (100 each). The percentage of cells with spindles >3 μm is displayed, along with images of DNA and spindle staining.
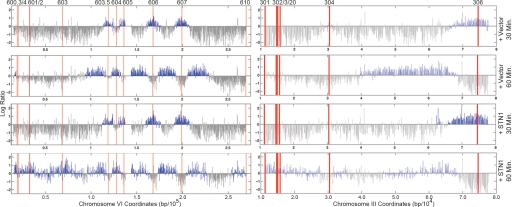
In addition to blocking spindle extension, the S phase checkpoint also acts to delay late replication origin firing and stabilize active replication forks from potentially lethal collapse (14). To determine whether these aspects of the S phase checkpoint were also deregulated by Stn1, we analyzed origin firing and fork progression in Stn1 OP cells in the presence of MMS, which activates the S phase checkpoint but permits limited replication. To do this, BrdU was allowed to incorporate into DNA for defined periods of time. The labeled DNA was recovered by immunoprecipitation (IP) with anti-BrdU antibodies and hybridized to a microarray to reveal regions of active DNA synthesis (15). As evaluated by this method, late replication origins such as ARS601/2 and ARS603 can be seen to fire inappropriately in Stn1 OP cells during the 30- to 60-min BrdU pulse after release from α-factor (Fig. 3A). Increased DNA synthesis is also observed within this time frame at the telomere proximal ARS610. No such firing of these origins was observed in vector controls. This analysis also showed that replication forks appear to progress less efficiently in the MMS-treated Stn1-overproducing cells. For example, in comparing replication fork progression from ARS306 or ARS607 during the 30- to 60-min interval, the DNA synthesis associated with the progressing fork is less extensive in Stn1 OP than the vector samples (Fig. 3). Consistent with this data, and similar to cells with checkpoint defects (15), FACS analysis shows that Stn1 OP cells traverse S phase more quickly than control cells in the presence of MMS (e.g., 75 min or 90 min) and show normal cell cycle timing in the absence of exogenous damage (Fig. S2). In the checkpoint-deficient strains, the additional forks that emanate from the fired late replication origins are thought to compensate for the slower progression of individual forks.
Fig. 3.
Replication origin firing and fork progression in MMS by BrdU-IP-chip. G1 synchronized cells expressing vector or pADH-STN1 (pLX421) were released into media containing 0.033% MMS. For each strain, 1 aliquot was harvested after incubation with BrdU from 0 to 30 min after release, and a second aliquot was harvested after BrdU exposure from 30 to 60 min. Replicated DNA was isolated by α-BrdU IP, labeled, and hybridized to a tiling array covering sequences on chromosome VI and the left arm of chromosome III. ARS608, ARS609, and ARS305 are deleted in this strain. Total DNA from G1-arrested cells was used as a reference control. Blue lines represent enriched regions. The scale of the X axis differs for chromosomes VI and III.
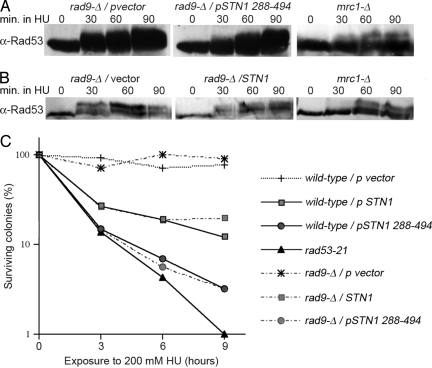
Together, these data indicate that Stn1 OP induces a virtually complete override of multiple aspects of the S phase checkpoint that are important in maintaining genome stability. Therefore, we next evaluated hypotheses for how this checkpoint interference might occur. The simplest hypothesis is that Stn1 short-circuits an upstream step in the checkpoint signaling pathway. Because the loss of STN1 function can lead to excessive ss DNA at telomeres (16, 17), one hypothesis to explain these observations is that Stn1 OP acts in an opposite way, reducing ss DNA at stalled forks. In theory, this could lead to an attenuation of signaling structures responsible for activating the checkpoint. However, we found that in response to HU or MMS treatment, the extent of the Rad53 phosphorylation shift, which provides a reliable indicator of checkpoint activation (18–20), does not appear to be affected by OP of Stn1 or Stn1288-494 (Fig. 4 A and B and Fig. S3).
Fig. 4.
Activation of Rad53. (A and B) Time course of Rad53 activation by HU treatment. Cultures of rad9-Δ cells bearing vector (pLX416), pADH-STN1 (pLX421), or pADH-STN1288–494 (pLX423) were synchronized in G1 and released into 200 mM HU. At the indicated times, protein extracts were prepared and analyzed by α-Rad53 Western blot. Rad53 phosphorylation in a mrc1-Δ strain is shown for comparison. (C) Cell survival after HU exposure. rad53–21or wild-type and rad9-Δ strains with pLX416, pLX421, or pLX423 were exposed to 200 mM HU. At the indicated times, the percentage of cells able to form colonies on plates lacking HU was determined. The averages from 3 experiments are plotted.
It has been shown that Rad53 can be activated through 2 different mediators of checkpoint signaling, Mrc1 and Rad9. In response to activation of the S phase checkpoint by HU treatment, Rad53 is normally activated by Mrc1 (21). When Mrc1 is defective, however, Rad53 can still be activated, albeit with delayed kinetics, through the Rad9-dependent DNA damage pathway (21). From these observations, it remained possible that Stn1 OP might interfere with the S phase checkpoint by blocking Rad53 activation through Mrc1, in essence acting like a mrc1 mutant. In this case, Stn1 OP in a rad9 strain should lead to a failure to activate Rad53, similar to mrc1 rad9 mutants (21). Therefore, we examined Rad53 phosphorylation in rad9-Δ cells (Fig. 4 A and B and Fig. S3). Although we reproduced the delay and partial reduction in Rad53 activation in HU-treated mrc1-Δ mutants, the extent and kinetics of Rad53 phosphorylation appear unaltered in rad9-Δ cells overproducing Stn1. Furthermore, Stn1 OP is distinct from what has been previously demonstrated for mrc1 loss because inactivation of RAD9 does not influence the viability or HU sensitivity of Stn1 OP cells (Fig. 4C and data not shown), and Stn1 OP cells are much more sensitive to HU than mrc1 (Figs. 1A and 4C) (21). Thus, Stn1 OP is not equivalent to inactivating Mrc1.
After Rad53 activation, the levels of ribonucleotide reductase (RNR) increase, counteracting the impact of HU on dNTP pools and allowing rad53-Δ strains to be viable (22). Thus, one way that Stn1 OP might antagonize S phase checkpoint regulation is by preventing dNTP accumulation. However, disrupting sml1, encoding a RNR inhibitor, and overexpressing RNR1 do not alter the HU sensitivity of Stn1 OP strains (Fig. S4). This indicates that Stn1 OP is not perturbing S phase checkpoint control by interfering with RNR.
As Stn1 OP does not obviously interfere with Rad53 activation, another possibility is that Stn1 generally antagonizes the ability of activated forms of Rad53 to execute checkpoint functions. As one test of this, we asked whether the DNA damage checkpoint remains intact in Stn1 OP cells in which a conditional DSB is generated by inducing expression of the HO endonuclease (23) (Table S1). After induction of the DSB, the Stn1 OP cells delayed cell division in a manner similar to vector controls, indicating that Stn1 overproducing cells remain proficient for the G2/M DNA damage checkpoint. Thus, if Stn1 interferes with Rad53, it must do so in a way that specifically deregulates targeting of checkpoint substrates involved in the response to HU or MMS treatment. On the whole, our results are most consistent with the view that Stn1 OP overrides the S phase checkpoint downstream of Rad53, interfering with checkpoint effector mechanisms.
Given these considerations, the final hypothesis tested was that Stn1 OP causes telomere-specific functions to be inappropriately redirected to challenged replication forks or the opposite, that proteins critical for challenged forks become misdirected to telomeres. If this were so, one prediction is that critical targets of Stn1 would be components of the replication machinery or telomere binding factors, and blocking the effect of Stn1 OP on its target would restore the checkpoint. We therefore tested whether altering the level or function of proteins that interact with Stn1 could ameliorate Stn1 OP sensitivity to DNA replication stress. Two candidate targets are Cdc13 and Ten1, which are thought to associate with Stn1 to form a heterotrimeric telomere-capping complex. However, our data indicate neither protein is a critical target. First, increasing CDC13 dosage does not suppress Stn1 OP damage sensitivity, although there is a very slight improvement in growth of cells overproducing a Cdc13 protein with a partial deletion of its DNA-binding domain (Fig. S1A). Second, rather than suppressing Stn1 OP, TEN1 overexpression is lethal, even in the absence of exogenous damage (Fig. S1B). This lethality is not necessarily linked to the checkpoint disruption because TEN1 overexpression is toxic only for full-length Stn1 OP, but not for Stn1288-494 OP, which fails to interact with Ten1 (17). Third, neither Cdc13 nor Ten1 OP interferes with S phase checkpoint cell cycle arrest in HU-treated cells (data not shown).
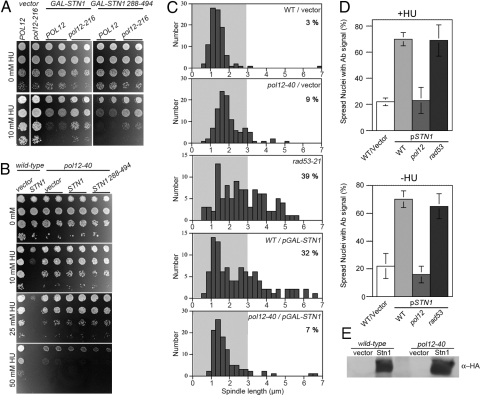
In addition to the telomere capping proteins, Stn1 associates with Pol12, the regulatory subunit of Polα (7, 10). Remarkably, 2 different pol12 loss-of-function mutants, pol12-40 and pol12-216, significantly attenuate both the HU sensitivity and S phase checkpoint spindle extension phenotype associated with increased Stn1. The pol12-40 mutant led to a more dramatic improvement in viability on HU than pol12-216 (Fig. 5 A and B and Fig. S5). However, either allele was able to reverse the spindle elongation defect (Fig. 5C). To test whether any perturbation to Polα function reverses the Stn1 OP phenotypes, we assessed Stn1 OP in cdc17-2 or pol1-236 strains harboring mutations in Pol1, the catalytic subunit of Polα. Unlike the pol12 mutations, neither pol1 mutation suppressed Stn1 OP damage sensitivity (Fig. S6). For cdc17-2, Stn1 OP reduced the maximum permissive temperature of‘ the strain.
Fig. 5.
pol12 mutations reduce Stn1 interference with the S phase checkpoint. (A and B) Tenfold serial dilutions of pol12 cultures overexpressing STN1 or STN1288–494 (pCN177, pPC30) were plated on Ura-galacotose media with varying HU concentrations. (C) Spindle length was measured in G1-arrested cells released into 200 mM HU for 3 h. The percentage of cells with spindles >3 μm is indicated. (D) Chromosome spreads were prepared from the indicated strains transformed with vector (pLX416) or plasmids overproducing HA-tagged Stn1 (pLX421). Cells were harvested 120 min after G1 synchronization/release in the presence or absence of 200 mM HU; 250 nuclei were evaluated for staining above background florescence. (E) HA-Stn1 evaluated in strains used in D by Western blot.
One hypothesis to explain why Stn1 override of the S phase checkpoint is sensitive to Pol12 is that the interaction between these proteins causes overproduced Stn1 to become mislocalized to chromosomal regions outside of telomeres. To test this, we examined Stn1 association with chromosomal DNA by spreading analysis. In the presence or absence of HU, overproduced Stn1 localized throughout spread nuclei, showing a tendency to concentrate into punctate foci (Fig. 5D and Fig. S7). As evaluated by Western blot, overproduced Stn1 accumulated to a similar extent in wild-type and pol12-40 cells (Fig. 5E); however, the excess Stn1 largely failed to associate with chromosomes in the pol12-40 strain (Fig. 5D and Fig. S7). Conversely, the Stn1 staining intensity noticeably increased in HU treated rad53 cells (Fig. S7), suggesting that defective S phase checkpoint regulation could stimulate Stn1 chromosome association. These observations show that overproduced Stn1 can associate with chromosomes at nontelomeric sites, and restoring the S checkpoint in pol12 mutants corresponds with reduced Stn1 chromosome binding. Overall, our results suggest interaction between Stn1 and Pol12 on replicating chromosomes is a necessary precondition for Stn1 OP to deregulate the S phase checkpoint.
Discussion
The S phase checkpoint modulates replication fork stability and progression to prevent replication errors and replication fork collapse (14). The mechanisms through which the checkpoint acts on the DNA replication machinery, however, remain poorly defined. Here, we have shown that the telomere maintenance protein Stn1 has an unanticipated ability to interfere with the S phase checkpoint, accompanied by extreme sensitivity to replication inhibitors such as HU and MMS. All aspects of the S phase checkpoint that we examined were deficient in Stn1 OP cells, including the ability of the checkpoint to block late replication origin firing, to maintain progression of stabilized replication forks, and to couple completion of DNA replication to extension of the mitotic spindle. Given that Stn1 OP disrupts multiple checkpoint responses, it is notable that both the timing and extent of Rad53 activation by replication stress appears unaffected by Stn1 OP. In addition, the DNA damage checkpoint, which depends critically on Rad53, remains functional after Stn1 OP. Thus, Stn1 most likely acts downstream of Rad53 to interfere with S phase checkpoint effector mechanisms. The ability of the pol12 mutants to restore HU-resistance and cell cycle arrest supports this interpretation and suggests that Pol12 is an important target of Stn1 checkpoint interference. As discussed below, these findings have implications for how the S phase checkpoint controls replication fork stability and how Stn1 functions at telomeres.
Stn1 and Pol12 in the S Phase Checkpoint.
Polα has previously been implicated in the S phase checkpoint, although a complete understanding of the nature of this involvement has yet to emerge. In fission yeast and Xenopus, primer synthesis by Polα appears to be required to generate a replication stress signal that activates the S phase checkpoint (24, 25). Other studies suggest Polα is a downstream checkpoint target. First, cell cycle-regulated phosphorylation of Pol12 is delayed in a RAD53-dependent manner after DNA damage (20), potentially influencing Pol12 chromatin association or replisome stability (26). Second, analysis of the pri1-M4 allele led to a model where replication of UV- or MMS-damaged DNA templates is controlled by blocking Polα primase activity (27). pri1-M4 mutants were suggested to be immune from this regulation, leading to a dominant G1/S phase checkpoint defect. The pri1-M4 defect is distinct from Stn1 OP, however, because the pri1-M4 strains remain proficient for S phase checkpoint responses induced by HU treatment.
Previous genetic studies have suggested Stn1 and Pol12 collaborate to maintain telomere end protection, with chromosome capping defects synergistically enhanced in double mutants (10). Stn1 and Pol12 may also act together during telomere replication by promoting lagging-strand telomere synthesis. Interestingly, from a genetic standpoint, this interaction parallels our observations after Stn1 OP, with the ability of Stn1 to effect checkpoint override requiring Pol12 function. Although the molecular basis for Stn1 checkpoint abrogation remains to be determined, our observations establish a framework for interpreting this phenomenon. In particular, our data suggest that when Stn1 is present at inappropriate high levels, Pol12 recruits Stn1 to nontelomeric chromosomal sites. The relationship between sites of Stn1 binding, Pol12 localization, and DNA replication are presently unclear. Nonetheless, mislocalized Stn1 might do one of two things. First, it might directly promote priming or another aspect of Polα activity that antagonizes what the checkpoint does to stabilize the replisome. Second, because the Stn1 essential function is thought to be blocking telomere resection (8, 16, 28), it is conceivable that mislocalizing this capping activity to stalled forks has deleterious consequences for checkpoint regulation of fork metabolism. Preventing Stn1 mislocalization by short-circuiting the Stn1–Pol12 interaction would relieve these deleterious effects. Importantly, Stn1 OP does not obviously affect S phase progression or cause DNA damage in the absence of replication stress. Because strains with reduced Polα function are not typically sensitive to HU or MMS, we surmise that Stn1 OP does not normally antagonize Polα, and an impediment to fork progression may be a necessary precondition for Stn1 to interfere with DNA replication.
Although the relevant substrates are largely unknown, Rad53 is generally thought to control 3 distinct S phase checkpoint effector mechanisms: regulation of origin firing, stabilization of replication forks, and restraint of spindle extension. Alternatively, it has been suggested that the spindle extension defect of HU-treated rad53 mutants is an indirect outcome of defective centromere replication (11), raising the possibility that what appear to be distinct aspects of checkpoint regulation may actually be mechanistically linked. Our observations that perturbations to Pol12, a component of the DNA replication machinery, restore the spindle extension block and restore viability to HU-treated Stn1-overproducing cells is consistent with a coupling of at least the fork stability and spindle extension checkpoint responses; it will be of interest to determine whether the block to late origin firing is similarly restored. Thus, the interaction between Stn1 and Pol12 may have a considerable bearing on how the S phase checkpoint is actually organized.
Stn1 and Pol12 in Telomere Replication.
Despite its interactions with Polα, Stn1 has been considered a telomere-capping protein, protecting the telomere C-rich strand from degradation and having a secondary role in promoting telomere C-rich strand synthesis after telomerase extends the G-rich strand. S. cerevisiae telomeres are ≈350 base pairs long and maintain a short 3′ G-rich terminal overhang, suggesting that the lagging strand can be initiated close to the telomere end. Whether a mechanism exists to facilitate priming close to the terminus during semiconservative replication or to promote complete telomere C-strand synthesis independent of the replication fork is not known. We speculate that when Stn1 levels are increased, the role for Stn1 in promoting telomere C-rich strand synthesis becomes executed in an improper context after DNA replication stress.
In closing, we note that the ability of Stn1 OP to disrupt the S phase checkpoint puts some previous observations in new light. In prior work, we found that OP of Stn11-186 with Ten1 could completely bypass the Cdc13 essential function, but OP of full-length Stn1 was unable to support this Cdc13 bypass (7). Given our current findings, proper S phase checkpoint control may be required to establish or maintain the bypass mechanism, and OP of full-length Stn1 interferes with this process. It is also interesting to consider whether the S phase checkpoint defects associated with increased Stn1 levels contribute to the delayed senescence of strains deficient for both telomerase- and nonsense-mediated mRNA decay (29) or to the reduced telomere length of replication mutants (30). Finally, a putative STN1 homolog, Obfc1, does exist in higher eukaryotes (8). Given that the pathways for maintaining telomeres have shown some flexibility in evolution, it will be of interest to see whether Obfc1 has a critical function in telomere capping, DNA replication, or the response to replication stress.
Materials and Methods
General Genetic Methods.
Serial dilution assays, survival assays after transient exposure to HU, and G1 synchronization/release experiments into media containing 200 mM HU were performed as previously described. To assay cell cycle arrest after an HO-mediated DSB, 49 cells from each strain were micromanipulated to form a grid on glucose or galactose media, incubating at 30 °C. At indicated time points, the number of cell bodies per microcolony was counted.
Protein Methods.
Rad53 phosphorylation was assessed by preparing cell extracts in 20% TCA (18). Fifty microliters of each lysate was loaded and separated on a 10% 30:0.39 acrylamide/bisacrylamide gel. Western Blots were probed with an α-Rad53 antibody (Santa Cruz Biotechnology).
Spindle Measurement.
G1-synchronized/released cells were fixed after a 3-h exposure to 200 mM HU and processed for α-tubulin immunofluorescence as described (11). Metamorph software was used to measure spindle length in at least 100 cells.
Analysis of DNA Replication.
For analysis of telomere proximal replication forks, DNA was purified by cesium chloride density gradient centrifugation as described in ref. 31. The DNA samples were digested with EcoRI before electrophoresis, and the blots were probed as in ref. 32. Two different-sized fragments that contain Y′ ARS were detected in the Southern Blot. The BrdU-IP-chip microarray was conducted and analyzed as described in ref. 15. The procedure is outlined in Fig. S2.
Chromosome Spreads.
Chromosome spreads were prepared and stained as previously described (33) by using DAPI and anti-HA antibody 12CA5 (Roche). From 4 independently prepared slides, 250 nuclei were scored for the presence of staining above background florescence.
Supplementary Material
Acknowledgments.
We thank S. Knott for help with analysis and graphing of the microarray data, and laboratory members for discussion. V. Zakian, D. Gottschling, and S. Elledge generously shared strains. This project was supported by National Institutes of Health Grants R01-GM66190 (to J.B.B.), R01-CA65494 (to O.M.A.), and R01-CA96972 (to C.I.N.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812605106/DCSupplemental.
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Chakhparonian M, Wellinger RJ. Telomere maintenance and DNA replication: How closely are these two connected? Trends Genet. 2003;19:439–446. doi: 10.1016/S0168-9525(03)00135-5. [DOI] [PubMed] [Google Scholar]
- 3.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 4.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 5.Vodenicharov MD, Wellinger RJ. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell. 2006;24:127–137. doi: 10.1016/j.molcel.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 6.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petreaca RC, et al. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol. 2006;8:748–755. doi: 10.1038/ncb1430. [DOI] [PubMed] [Google Scholar]
- 8.Martin V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA. 2007;104:14038–14043. doi: 10.1073/pnas.0705497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 10.Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D. Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 2004;18:992–1006. doi: 10.1101/gad.300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachant J, Jessen SR, Kavanaugh SE, Fielding CS. The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress. J Cell Biol. 2005;168:999–1012. doi: 10.1083/jcb.200412076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 13.Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 14.Branzei D, Foiani M. The DNA damage response during DNA replication. Curr Opin Cell Biol. 2005;17:568–575. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Szyjka SJ, et al. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev. 2008;22:1906–1920. doi: 10.1101/gad.1660408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 17.Petreaca RC, Chiu HC, Nugent CI. The role of Stn1p in Saccharomyces cerevisiae telomere capping can be separated from its interaction with Cdc13p. Genetics. 2007;177:1459–1474. doi: 10.1534/genetics.107.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez Y, et al. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 19.Ma JL, Lee SJ, Duong JK, Stern DF. Activation of the checkpoint kinase Rad53 by the phosphatidyl inositol kinase-like kinase Mec1. J Biol Chem. 2006;281:3954–3963. doi: 10.1074/jbc.M507508200. [DOI] [PubMed] [Google Scholar]
- 20.Pellicioli A, et al. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alcasabas AA, et al. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- 22.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandell LL, Zakian VA. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 24.D'Urso G, Grallert B, Nurse P. DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J Cell Sci. 1995;108(Pt 9):3109–3118. doi: 10.1242/jcs.108.9.3109. [DOI] [PubMed] [Google Scholar]
- 25.Michael WM, Ott R, Fanning E, Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- 26.Desdouets C, et al. Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase alpha. EMBO J. 1998;17:4139–4146. doi: 10.1093/emboj/17.14.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marini F, et al. A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J. 1997;16:639–650. doi: 10.1093/emboj/16.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zubko MK, Lydall D. Linear chromosome maintenance in the absence of essential telomere-capping proteins. Nat Cell Biol. 2006;8:734–740. doi: 10.1038/ncb1428. [DOI] [PubMed] [Google Scholar]
- 29.Enomoto S, Glowczewski L, Lew-Smith J, Berman JG. Telomere cap components influence the rate of senescence in telomerase-deficient yeast cells. Mol Cell Biol. 2004;24:837–845. doi: 10.1128/MCB.24.2.837-845.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brewer BJ, Fangman WL. Mapping replication origins in yeast chromosomes. BioEssays. 1991;13:317–322. doi: 10.1002/bies.950130702. [DOI] [PubMed] [Google Scholar]
- 32.Makovets S, Herskowitz I, Blackburn EH. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein F, et al. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.