Abstract
Genetic data has become an essential part of ecological studies, because the analyses of diversity within and among natural populations may grant access to previously overlooked ecological and evolutionary causalities, especially among cryptic species. Here, we present an example of how phylogenetic analysis of molecular data obtained within a DNA barcoding study, in combination with morphological and ecological data from the field and laboratory experiments, unraveled a striking predator-prey interaction between aquatic organisms. The “crown of thorns,” a conspicuous morphological feature among water fleas of the Daphnia atkinsoni species complex (Crustacea: Cladocera), is considered to represent a species-specific trait. However, our study, initiated by the analysis of sequence variation in 2 mitochondrial genes, shows that this feature is phenotypically plastic and is induced by chemical cues released by Triops cancriformis, the tadpole shrimp (Notostraca). The trait acts as an effective antipredator defense, and is found in several Daphnia lineages coexisting with notostracans. These facts suggest that the “crown of thorns” evolved in coexistence with this ancient predator group.
Keywords: DNA barcoding, phenotypic plasticity, predator-prey interactions, Cladocera, Notostraca
Phenotypic plasticity in defensive traits has evolved as an adaptation to heterogeneity in predation risk. Inducible defenses affect predator–prey relationships, competitive interactions, and potentially also ecological processes and ecosystem functions. Therefore, this phenomenon is interesting from both ecological and evolutionary points of view. Inducibility of traits is favored under the following conditions: spatial or temporal variation in predation risks, the availability of reliable cues indicating a threat, the ability to form effective defenses within a relatively short time span, and an association of costs that can be saved when defenses are not required (1). Inducible defenses are found in a variety of taxa spanning from bacteria and algae to ciliates, rotifers, crustaceans, insect larvae, and even vertebrates (1). In freshwater ecosystems, prey organisms often sense predator-released chemical cues, the so-called kairomones. These signals, which provide reliable information on the actual level of predation, have been shown to induce behavioral, life history, or morphological changes in the prey (1).
An intensively studied group of model organisms in ecology, evolutionary biology, and environmental sciences are water fleas of the genus Daphnia (Crustacea: Cladocera), among which various remarkable morphological defenses can be found. However, until now, only helmet-like (e.g., helmets, crests) or spike-like (e.g., tail-spines, neck-teeth) structures have been attributed to defense (2). The functions of various other conspicuous morphological features exhibited by daphnids remain unknown, although understanding them may provide further important insights into ecological and evolutionary processes. Here, we demonstrate how a study integrating genetic, ecological, and morphological data revealed the function of an inducible morphological structure in a cryptic Daphnia species complex.
Members of the Daphnia atkinsoni complex are typical for the Mediterranean region, although the range of the species complex extends to Central Asia, North-West Europe, and even Iceland and Greenland. A characteristic morphological feature shared by these Daphnia is a dorsal extension of the carapace into the head shield that forms a heart-shaped lobe (Fig. 1). In some populations, these lobes cover a large portion of the head and are armed by long spines on the edge, forming a veritable “crown of thorns”. Most forms of the D. atkinsoni complex described in the past have been synonymized with the nominate species. Nevertheless, 2 distinct morphs occurring in the Mediterranean, D. atkinsoni (Baird, 1859) and D. bolivari (Richard, 1888), are regarded as distinct species (3, 4). Their separation has been based on the armament and size of the carapace extension, growth allometry, and ecological requirements (3, 5). D. bolivari, the form with a wide and spined lobe, is reported to occur in the typical community of temporary waters, coexisting with large phyllopod crustaceans, such as fairy shrimps (Anostraca) or tadpole shrimps (Notostraca) (3). Notostracans are particularly interesting, because these omnivores, extant with unchanged morphology for 220 million years (6), may feed on Daphnia (7) and can play a key role in structuring the temporary water macroinvertebrate communities (8).
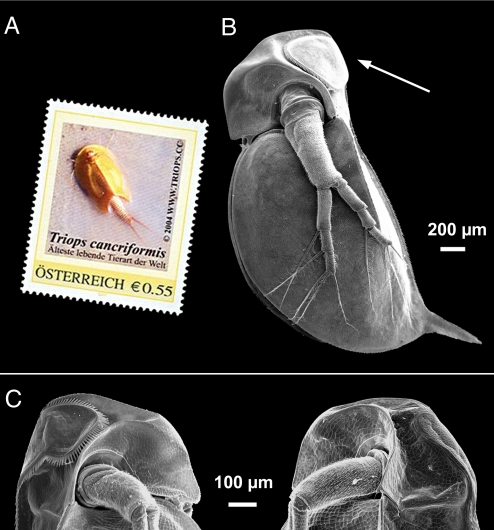
Fig. 1.
Induction of the “crown of thorns” in the D. atkinsoni species complex exposed to chemical cues released by Triops cancriformis. This notostracan is portrayed on an Austrian stamp as “the most ancient extant animal species”. (A) Induced Daphnia show a distinctly enlarged carapace extension into the head shield, forming heart-shaped lobes lined with strong spines (B: lineage 1, whole body SEM image; C Left: lineage 3, head). Noninduced individuals exhibit inconspicuous lobes without thorns (C Right: identical clone of lineage 3).
In this study, we benefited from the use of molecular data assembled for the purpose of species identification, the so-called molecular barcodes (9), which we used to assess the diversity of the daphniid subgenus Ctenodaphnia in the Western Palaearctic region. Despite a number of limitations in such a single-locus approach (10, 11), barcoding has proven to unravel a number of basic questions related to biodiversity, systematics, and taxonomy (12), and may serve as a convenient tool for ecological research.
We characterized, by molecular barcodes, individuals from several populations of the D. atkinsoni complex, and subsequently subjected them to a phylogenetic analysis. We found that the “crown of thorns,” allegedly a species-specific trait, was not linked to DNA-based grouping but rather showed correlation with habitat conditions. This discrepancy raised the question as to what extent the spined lobe is phenotypically plastic and whether it can be induced by chemical cues released by the key predators of the habitat—tadpole shrimps. In a second step, we verified the adaptive value of this trait by using predation experiments. The combination of a phylogenetic analysis and ecological experiments allowed us to unravel an ecological interaction between Daphnia and tadpole shrimps, and to explain the function of a conspicuous inducible morphological trait, the “crown of thorns.”
Results
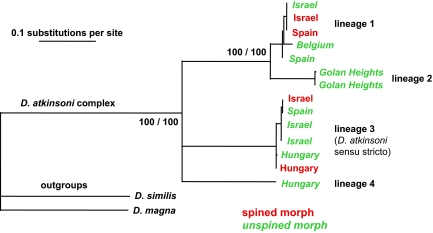
We observed substantial diversity of mtDNA lineages in the subgenus Ctenodaphnia across the Western Palaearctic region, in particular in the temporary waters of the Mediterranean. Four distinct lineages, putatively distinct species (13) because of their substantial divergence (minimum between-group sequence divergence in the range of 11.0–22.5% at COI, and 3.0–10.3% at the gene for 12S rRNA), were observed within the D. atkinsoni species complex [this grouping into a species complex, apart from morphological similarity, agrees with the concept suggesting a divergence cutoff of 14% at 12S] (13, 14). From these, 2 lineages (1 and 3, the latter corresponding to D. atkinsoni in a strict sense) were widespread: Both cooccur in the Eastern and Western Mediterranean, and their distribution area also extends into Central or Western Europe (lineage 1 was found in Belgium and lineage 3 in Hungary). The remaining 2 lineages of the complex are apparently rarer: One was restricted to localities in the Golan Heights (lineage 2), and the other was found at a single site in Hungary (lineage 4).
Assignment of taxon names based on morphological characters did not agree with the phylogenetic reconstruction based on molecular data (Fig. 2). First, a substantial mtDNA variation was discovered among individuals carrying an unspined lobe (referred to as “unspined morphs” in the text). Second, monophyly of the alleged species Daphnia bolivari, i.e., the morphotype carrying the “crown of thorns,” was rejected, and no lineage could be reliably synonymized with this name. Spined morphs (i.e., those with spined lobes), recorded in Spain, Israel, and Hungary, were grouped with unspined populations within both common clades of the complex (lineages 1 and 3; Fig. 2). Because the putative D. bolivari-specific characters—the size of the heart-shaped lobe and the presence of spines forming the “crown”—were variable within 2 divergent clades, they cannot be regarded as taxonomically relevant. However, these characters were strongly linked to habitat conditions, in particular, to predator presence. Spined morphs were found in temporary pools, in a number of which notostracans of the genus Triops were observed during the sampling. We did not observe notostracans in coexistence with unspined morphs of the D. atkinsoni complex.
Fig. 2.
Consensus tree showing the phylogenetic relations among analyzed individuals of the D. atkinsoni complex, based on the maximum likelihood analysis of mitochondrial genes for 12S ribosomal DNA and cytochrome c oxidase subunit I. Each branch is indicated by the region of origin; labels in red denote individuals possessing the “crown of thorns” collected in the wild; unspined Daphnia are marked by a green italic font. Numbers at selected branches indicate bootstrap values for maximum likelihood analysis/posterior probability values from the Bayesian inference of phylogeny. The exact relationship of lineage 4 to others could not be determined based on the sequence data available.
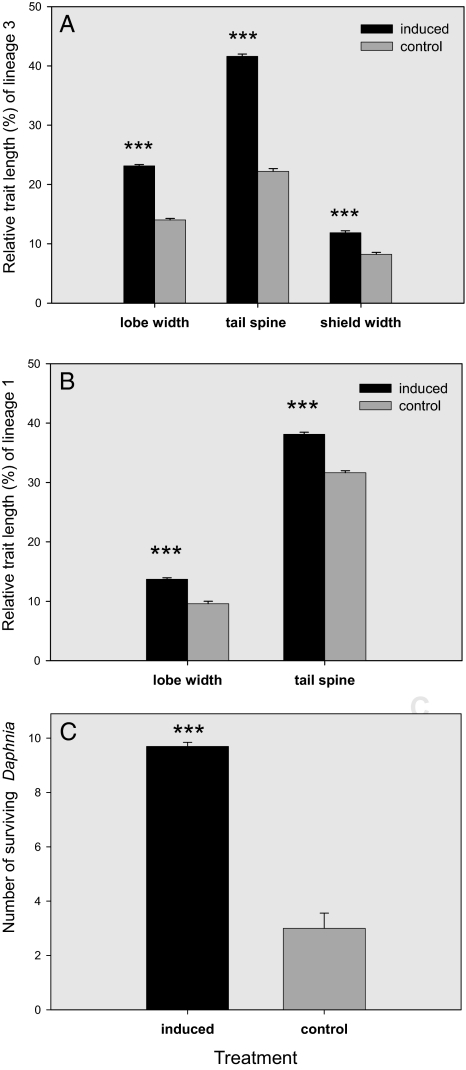
Inducibility of the “crown of thorns” by chemical cues released by predatory tadpole shrimps was tested by incubating unspined females of both lineages in net cages placed in vessels containing Triops cancriformis. Daphnia exposed to predator cues indeed developed the typical wide, heavily spined lobe (Fig. 3 A and B; nested ANOVA: lineage 3: F1,8.7 = 105.59, P < 0.001; lineage 1: F1,15 = 24.95, P < 0.001) and a longer tail spine (Fig. 3 A and B; nested ANOVA: lineage 3: F1,8.6 = 205.66, P < 0.001; lineage 1: F1,24 = 14.38, P = 0.001). Moreover, the shoulder-shield, which protects the base of the swimming antennae, was significantly enlarged in the induced morph of lineage 3, i.e., D. atkinsoni in the sense of stricto (s.str.) (Fig. 3A; nested ANOVA: F1,8.7 = 1120.52, P < 0.001). Predation trials revealed a significantly higher survival (97%) of individuals of the induced morph of this lineage in the presence of T. cancriformis compared with the noninduced morph (30%) of the same clone (Fig. 3C; paired Wilcoxon test for related samples, n = 10, Z = −3.8, P < 0.001).
Fig. 3.
Predator cue induction experiments. (A) Mean relative lobe width, tail spine length and shield width (%), and SE of a clone of D. atkinsoni s.str. (lineage 3) raised with (induction) and without (control) chemical cues released from Triops cancriformis. (B) Mean relative lobe width and tail spine length of a clone of lineage 1 under the same experimental conditions. Asterisks indicate highly significant differences (P < 0.001). Survivorship. (C) The number of surviving animals of each of the morphs (induced = spined; control = unspined) of D. atkinsoni s.str. (lineage 3) after a 30-min predation experiment with Triops cancriformis. Ten animals of each morph were placed together with 1 predator at the beginning of the experiment. Means and standard errors from 10 replicates are shown. Asterisks indicate a highly significant difference (P < 0.001).
Discussion
The discovery of the inducibility of the “crown of thorns,” and experimental proof of its protective mechanism, extends well beyond DNA taxonomy and species delimitation. The collected sequence data per se are relevant for Daphnia taxonomy: (i) the discovery of yet-unknown cryptic lineages warrants a taxonomic revision of European Ctenodaphnia, in particular the D. atkinsoni complex; (ii) the mismatch between morphological and molecular data questions the validity of the species D. bolivari within the D. atkinsoni species complex and stresses the need for a reassessment of identification characters. The added value of our dataset, however, is in elucidating previously unknown ecological interactions between Daphnia and tadpole shrimps.
Our results show that the “crown of thorns,” formerly regarded as a species-specific character, represents a phenotypically plastic trait and is therefore not suitable for taxonomical purposes. The identification of phenotypic plasticity in apparently ecologically relevant traits allows us to challenge current hypotheses on the morphological and ecological differentiation and biogeographical patterns among members of the D. atkinsoni complex. Morphotypes exhibiting the ”crown of thorns” have a disjunctive distribution: They were recorded in temperate areas spanning from the Mediterranean to Central Europe and Central Asia; however, they also have been shown to occur under entirely different climatic conditions in Greenland (15). This unusual presence of a spined morph of the complex in the Arctic environment may be explained by the coexistence with another notostracan species, Lepidurus arcticus, common in shallow arctic ponds and known to feed on Daphnia (7). The most geographically isolated morph of Daphnia exhibiting the “crown of thorns,” named after this feature, is Daphnia coronata Sars found in South Africa (16). The relationship of these geographically isolated populations to other phenotypically similar Daphnia is a matter of current taxonomic studies. Nevertheless, D. coronata occurs in temporary waters and coexists with notostracans. It may therefore be anticipated that its “crown“ serves the same purpose as in the D. atkinsoni complex, and may represent a case of parallel character evolution under strong selection pressure from these invertebrate predators.
Predation is a crucial selective agent structuring planktonic communities. In many freshwater habitats, predation risk may substantially vary temporally because of predator ontogenetic development. Biota in temporary ponds are constrained by the length of the hydroperiod, and only predators showing either high dispersal abilities or exhibiting the potential to overcome dry periods via diapause are able to cope with such fluctuating environments (17). Hence, the impact of predation in temporary ponds was considered to be low (18). Although notostracans have long been considered as potential biological control agents of mosquitoes (19, 20), their role as effective predators has still been widely ignored. Only recently has it been shown that tadpole shrimps have a significant impact on community structure in temporary ponds (21). Because they occasionally reach high densities (22), prey species are forced to either alter their diapausing strategies or to develop behavioral or morphological defense mechanisms. If these reduce the fitness of organisms in the absence of predators, selection may favor lineages which alter their behavior or phenotype according to current environmental conditions (phenotypic plasticity). However, this strategy will only become established if the prey organisms detect the presence of predators in time. Infochemicals released by predators, the so-called kairomones, have been found to provide such reliable signals. They are exclusively advantageous to the receiver in an interspecific information-transfer context and enable prey organisms to exhibit predator-specific defenses (1).
Our results show that chemical cues released by Triops cancriformis induce the “crown of thorns,” a unique but up to now unrecognized morphological defense shared among several lineages of the D. atkinsoni species complex. This “armor” consists of rigid cuticular shields, armed with long spikes, to protect the especially vulnerable body parts, the head and the base of the swimming antennae. Similarly, it has been shown that a stronger cuticle protects Daphnia against invertebrate predation (23, 24). The “crown of thorns” likely acts synergistically with the elongation of the tail spine, which is known to be an efficient defense mechanism in Daphnia against some predatory invertebrates (e.g., 25). Our predation trials showed a significant protective effect from induced morphological alterations, suggesting that the “crown of thorns” is a specific adaptation in the D. atkinsoni species complex. This trait represents one of the most specific antipredator adaptations in aquatic habitats.
Materials and Methods
Molecular Analyses
Marker Selection.
Two mitochondrial genes, one encoding the cytochrome c oxidase subunit I [COI; standard locus for DNA barcoding] (9) and the second for the small ribosomal subunit (12S rRNA), frequently used in Daphnia (26–28), were sequenced to identify species and to reconstruct phylogenetic relationships. The within-species variation in these 2 genes is substantially lower than divergence among species in most Daphnia species complexes, therefore allowing unambiguous species identification.
Sample Collection.
Among other Western Palaearctic Ctenodaphnia, samples from 14 populations of the D. atkinsoni complex were obtained at various localities in Europe (Belgium, Hungary, and Spain) and the Eastern Mediterranean (Israel and the Golan Heights) (see supporting information (SI) Table S1). Animals were collected using plankton nets and preserved in 96% ethanol; alternatively, living adult Daphnia females were used to establish laboratory clonal cultures. All analyzed individuals were characterized by their 12S rDNA sequences, and 1 or 2 representatives of each divergent mtDNA lineages (putative species) by COI. Both markers were also sequenced for 2 other Ctenodaphnia species, D. similis and D. magna, which were used as outgroups in the phylogenetic analysis.
DNA Extraction and Amplification.
DNA was extracted from single Daphnia individuals preserved in ethanol or originating from laboratory cultures by proteinase K digestion (29). Fragments of 12S rDNA and COI genes were amplified by using standard protocols (26), the PCR product was purified by column chromatography and sequenced on ABI automatic capillary sequencers (series 377 and 3700). Sequences were aligned using ClustalW (30) and the alignment subsequently checked manually in MEGA version 4 (31).
Sequence Analyses.
We identified divergent lineages potentially representing cryptic species by grouping individuals according to sequence similarity of the analyzed genes. The phylogenetic relationships within the D. atkinsoni complex were subsequently analyzed by using the total evidence approach based on both COI and 12S rDNA regions [combinability of both genes being confirmed by the test for homogeneity] (32). We used Modeltest 3.7 (33) to select the best model of nucleotide substitution by using the Akaike Information Criterion, and subsequently assessed the phylogeny using the Maximum Likelihood analysis (ML) in PAUP* 4.0b10 (34) and Bayesian inference (BI) in MrBayes version 3.1.2 (35). In ML, heuristic searches were conducted with tree bisection-reconnection branch swapping and 10 random sequence taxon additions; branch support was evaluated by nonparametric bootstrapping with 100 pseudoreplicates. BI used 2 parallel runs of 4 Monte Carlo Markov chains run for 3 million generations, trees sampled every 100 generations but the first 20% of trees containing the burn-in phase were discarded; parameters for both genes were estimated separately.
Induction and Predation Experiments.
We used laboratory-cultured clonal lines of lineage 1 and 3 of the D. atkinsoni complex for our experiments. Lineage 1 was isolated from a flooded field south of Tel Ashdod, Israel (31°45′06.0′′N, 34°39′06.6′′E), inhabited by a population of a “spined” morphotype, coexisting with Triops sp.; lineage 3 (D. atkinsoni s.str.) was hatched from resting eggs from a locality with unspined individuals—temporary puddles on a meadow in the Hungarian plains, northwest of Hajdúböszörmény (47°43′06.9′′N, 21°23′17.6′′E). No notostracans were observed at this latter locality. A culture of T. cancriformis was provided by Dr. E. Eder (Zoological Institute, University of Vienna) and kept in a temperature-controlled room at 20 ± 1 °C.
Predator-cue induction experiments were carried out in 12-liter glass aquaria. The bottom of each aquarium was covered with sand that had been sterilized before the experiments. The aquaria were filled with 10 L of an artificial medium (36). One third of the medium was exchanged weekly. Both induction experiments were conducted at 20 ± 1 °C in a temperature-controlled room under fluorescent light with a constant day-night rhythm (16h:8h). We cultured age-synchronized cohorts of both Daphnia lineages in 30-liter plastic buckets. The predator-cue induction experiments were started by randomly placing 20 ovigerous daphnids of a single clone originating from the third clutch of the age-synchronized cohorts into each aquarium. Three juvenile T. cancriformis (4000–5000 μm) with body sizes too small to feed on even neonate daphnids were introduced into each aquarium, serving as the induction treatment. After reaching a size of ≈8000 μm, the T. cancriformis were replaced by smaller animals to prevent strong predation effects on the daphnids, but to still guarantee a sufficient amount of predator-released chemicals. Fish food (1 g/d), which was tested to be ineffective in inducing morphological changes in Daphnia in preliminary experiments, was used as food source for the omnivorous T. cancriformis. The same amount of fish food was also placed into the control-treatment aquaria. Each aquarium was cleaned of exuviae, feces, and remaining fish food every day. The daphnids were fed daily by adding Scenedesmus obliquus at a concentration of 1.5 mg of C/L into each aquarium. Each experiment was replicated 6 times in the experiment with lineage 1 and 5 times in the experiment with lineage 3. Mothers of both the F1 and the F2 generation were removed after releasing their clutch. Mature Daphnia (1200–2000 μm) of the F3 generation were then used for analysis to include possible transgenerational effects (37). The following morphological parameters were recorded for both Daphnia species by using a digital image-analysis system (Soft Imaging System, Analysis Pro): body length, tail spine length, and lobe width. Additionally, the “shoulder”-shield width (distance between secondary fornices, i.e., lateral extensions of the carapace) was recorded in lineage 3 (D. atkinsoni s.str.). Statistics were calculated by using the software package SPSS V12.0 (SPSS Inc.). To compensate for size-dependent changes, a relative value was calculated for each trait. Arcsin-square-root-transformed data (38) were then tested for normal distribution and a nested ANOVA, with the replicates as the random factor, was performed for both experiments to analyze for treatment effects between control Daphnia and daphnids exposed to chemical cues released by T. cancriformis.
Predation trials with T. cancriformis hunting on lineage 3 (D. atkinsoni s.str.) were conducted in a temperature-controlled room at 20 ± 1 °C in 500-ml glass beakers under daylight conditions. The body length of T. cancriformis used for predation trials, measured from the top of the carapace to the caudal part of the body, was 27–30 mm. Ten mature Daphnia individuals (1200–2000 μm) of both morphs, Triops-induced (spined) and noninduced (unspined), were introduced into each beaker. The experiment started at the time when a single predator was placed into the beaker and launched its first attack on the daphnids. After 30 min the number of killed and surviving animals was recorded, and the surviving Daphnia were classified as predator-induced or noninduced using a stereo-microscope. The predation trial was replicated 10 times, and a paired Wilcoxon test for related samples was used to analyze this dataset.
Supplementary Material
Acknowledgments.
We thank Tereza Petrusková and Martina Petrů for invaluable assistance during the field sampling, Erich Eder for supplying the Triops culture, László Forró for providing some D. atkinsoni samples from Hungary, N. Jung, M. Kredler, and E. Ossipova for their technical support in this study, and two anonymous referees for their valuable comments on the manuscript. The study was partly supported by the Czech Ministry of Education (project MSM0021620828), the EuroCORES/EuroDIVERSITY project BIOPOOL (through the Czech Science Foundation Grant DIV/06/E007), the German Research Foundation (DFG; SCHW830/7) and the Research Centre Biodiversity and Climate (BiK+F). A.P. acknowledges further support from the German Academic Exchange Service (DAAD). Sampling in Spain was funded by ECODOCA (Access to Research Infrastructure action of the Improving Human Potential Program in Doñana Biological Station).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808075106/DCSupplemental.
References
- 1.Tollrian R, Harvell CD. The Ecology and Evolution of Inducible Defenses. Princeton: Princeton Univ Press; 1999. [Google Scholar]
- 2.Tollrian R, Dodson SI. In: The Ecology and Evolution of Inducible Defenses. Tollrian R, Harvell CD, editors. Princeton: Princeton Univ Press; 1999. pp. 177–202. [Google Scholar]
- 3.Alonso M. Crustacea, Branchiopoda. In: Ramos MA, et al., editors. Fauna Iberica. Vol 7. Madrid: Museo Nacional de Ciencias Naturales – Consejo Superior de Investigaciones Científicas; 1996. p. 486. in Spanish. [Google Scholar]
- 4.Benzie JAH. Cladocera: The genus Daphnia (including Daphniopsis) Ghent, Leiden: Kenobi Productions, Backhuys Publishers; 2005. p. 376. [Google Scholar]
- 5.Alonso M. Nota sobre algunes Ctenodaphnia de la Península Ibèrica [Note on some Ctenodaphnia of the Iberian Peninsula] Butl Inst Catalana Hist Nat. 1980;45:47–53. Sec. Zool., 3. in Catalan. [Google Scholar]
- 6.Kelber K-P. New Triopsids (Crustacea, Notostraca) from the Upper Triassic of Frankonia, Germany. Hallesches Jb Geowiss B, Beih. 1998;5:85–86. [Google Scholar]
- 7.Christoffersen K. Predation on Daphnia pulex by Lepidurus arcticus. Hydrobiologia. 2001;442:223–229. [Google Scholar]
- 8.Yee SH, Willig MR, Moorhead DL. Tadpole shrimp structure macroinvertebrate communities in playa lake microcosms. Hydrobiologia. 2005;541:139–148. [Google Scholar]
- 9.Hebert PDN, Ratnasingham S, deWaard JR. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moritz C, Cicero C. DNA barcoding: Promise and pitfalls. PLoS Biol. 2004;2:1529–1531. doi: 10.1371/journal.pbio.0020354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Will KW, Mishler BD, Wheeler QD. The perils of DNA barcoding and the need for integrative taxonomy. Syst Biol. 2005;54:844–851. doi: 10.1080/10635150500354878. [DOI] [PubMed] [Google Scholar]
- 12.Miller SE. DNA barcoding and the renaissance of taxonomy. Proc Natl Acad Sci USA. 2007;104:4775–4776. doi: 10.1073/pnas.0700466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adamowicz SJ, Colbourne JK, Witt JDS, Hebert PDN. The scale of divergence: A phylogenetic appraisal of intercontinental allopatric speciation in a passively dispersed freshwater zooplankton genus. Mol Phylogenet Evol. 2008 doi: 10.1016/j.ympev.2008.11.026. in press. [DOI] [PubMed] [Google Scholar]
- 14.Colbourne JK, Hebert PDN. The systematics of North American Daphnia (Crustacea: Anomopoda): A molecular phylogenetic approach. Philos Trans R Soc Lond B Biol Sci. 1996;351:349–360. doi: 10.1098/rstb.1996.0028. [DOI] [PubMed] [Google Scholar]
- 15.Wesenberg-Lund C. Gronlads Ferskvandsentomostraca. I. Phyllopoda branchiopoda et cladocera. [Freshwater Entomostraca of Greenland. I. Phyllopoda branchiopoda and cladocera.] Vidensk Medd naturhist Foren Kjobenhavn. 1895;56:82–173. [Google Scholar]
- 16.Sars GO. The fresh-water Entomostraca of Cape Province. Ann S Afr Mus. 1916;15:303–351. [Google Scholar]
- 17.Williams DD. Environmental constraints in temporary fresh waters and their consequences for the insect fauna. J N Am Benthol Soc. 1996;15:634–650. [Google Scholar]
- 18.Kerfoot WC, Lynch M. In: Predation: Direct and indirect impacts on aquatic communities. Kerfoot WC, Sih A, editors. Hanover: Univ Press of New England; 1987. pp. 367–378. [Google Scholar]
- 19.Maffi M. Triops granarius (Lucas) (Crustacea) as a natural enemy of mosquito larvae. Nature. 1962;195:722. [Google Scholar]
- 20.Kumar R, Hwang JS. Larvicidal efficiency of aquatic predators: A perspective for mosquito biocontrol. Zool Stud. 2006;45:447–466. [Google Scholar]
- 21.Boix D, Sala J, Gascon S, Brucet S. Predation in a temporary pond with special attention to the trophic role of Triops cancriformis (Crustacea: Branchiopoda: Notostraca) Hydrobiologia. 2006;571:341–353. [Google Scholar]
- 22.Boix D, Sala J, Moreno-Amich R. Population dynamics of Triops cancriformis (Crustacea: Branchiopoda: Notostraca) of the Espolla temporary pond in the northeastern Iberian peninsula. Hydrobiologia. 2002;486:175–183. [Google Scholar]
- 23.Laforsch C, Ngwa W, Grill W, Tollrian R. An acoustic microscopy technique reveals hidden morphological defenses in Daphnia. Proc Natl Acad Sci USA. 2004;101:15911–15914. doi: 10.1073/pnas.0404860101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dodson SI. Predation of Heterocope septentrionalis on two species of Daphnia: Morphological defenses and their cost. Ecology. 1984;65:1249–1257. [Google Scholar]
- 25.Caramujo M-J, Boavida M-J. Induction and costs of tail spine elongation in Daphnia hyalina x galeata: Reduction of susceptibility to copepod predation. Freshwater Biol. 2000;45:413–423. [Google Scholar]
- 26.Schwenk K, Posada D, Hebert PDN. Molecular systematics of European Hyalodaphnia: The role of contemporary hybridization in ancient species. Proc R Soc Lond B Biol Sci. 2000;267:1833–1842. doi: 10.1098/rspb.2000.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor DJ, Hebert PDN, Colbourne JK. Phylogenetics and evolution of the Daphnia longispina group (Crustacea) based on 12S rDNA sequence and allozyme variation. Mol Phylogenet Evol. 1996;5:495–510. doi: 10.1006/mpev.1996.0045. [DOI] [PubMed] [Google Scholar]
- 28.Colbourne JK, Wilson CC, Hebert PDN. The systematics of Australian Daphnia and Daphniopsis (Crustacea: Cladocera): A shared phylogenetic history transformed by habitat-specific rates of evolution. Biol J Linn Soc. 2006;89:469–488. [Google Scholar]
- 29.Schwenk K, et al. Genetic markers, genealogies and biogeographic patterns in the cladocera. Aquat Ecol. 1998;32:37–51. [Google Scholar]
- 30.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 32.Farris JS, Kallersjo M, Kluge AG, Bult C. Constructing a significance test for incongruence. Syst Biol. 1995;44:570–572. [Google Scholar]
- 33.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 34.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland: Sinauer Associates; 2002. [Google Scholar]
- 35.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 36.Jeschke JM, Tollrian R. Density-dependent effects of prey defences. Oecologia. 2000;123:391–396. doi: 10.1007/s004420051026. [DOI] [PubMed] [Google Scholar]
- 37.Agrawal AA, Laforsch C, Tollrian R. Transgenerational induction of defences in animals and plants. Nature. 1999;401:60–63. [Google Scholar]
- 38.Sokal RR, Rohlf FJ. Biometry. 3rd Ed. New York: Freeman; 1995. p. 887. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.