Abstract
Heparanase is an endoglycosidase that degrades heparan sulfate (HS) at the cell surface and in the extracellular matrix. Heparanase is expressed mainly by cancer cells, and its expression is correlated with increased tumor aggressiveness, metastasis, and angiogenesis. Here, we report the cloning of a unique splice variant (splice 36) of heparanase from the subterranean blind mole rat (Spalax). This splice variant results from skipping part of exon 3, exons 4 and 5, and part of exon 6 and functions as a dominant negative to the wild-type enzyme. It inhibits HS degradation, suppresses glioma tumor growth, and decreases experimental B16–BL6 lung colonization in a mouse model. Intriguingly, Spalax splice variant 7 of heparanase (which results from skipping of exon 7) is devoid of enzymatic activity, but unlike splice 36 it enhances tumor growth. Our results demonstrate that alternative splicing of heparanase regulates its enzymatic activity and might adapt the heparanase function to the fluctuating normoxic–hypoxic subterranean environment that Spalax experiences. Development of anticancer drugs designed to suppress tumor growth, angiogenesis, and metastasis is a major challenge, of which heparanase inhibition is a promising approach. We anticipate that the heparanase splicing model, evolved during 40 million years of Spalacid adaptation to underground life, would pave the way for the development of heparanase-based therapeutic modalities directed against angiogenesis, tumor growth, and metastasis.
Keywords: alternative splicing, angiogenesis, blind mole rat, cancer, heparan sulfate
Heparanase is a mammalian endoglycosidase that degrades heparan sulfate (HS) at the cell surface and in the extracellular matrix (ECM) (1–7). Consequently, it facilitates migration of inflammatory and tumor cells, releases growth factors bound to HS in the ECM, and induces new blood vessel formation (angiogenesis) (3–8). Heparanase expression in tumor cells correlates with disease severity, and its overexpression in experimental tumor models results in accelerated tumor growth and metastases formation (3–10). Heparanase up-regulation was noted in an increasing number of primary human cancers, correlating with reduced postoperative survival of cancer patients (5, 9). Elevated levels of heparanase were detected in the urine and plasma of patients with aggressive metastatic disease (11). Recently, we cloned a splice variant of human heparanase lacking exon 5 whose function has yet to be unraveled (12). Moreover, we cloned a splice variant from Spalax that lacks exon 7 (splice 7) (13).
Spalax is a mammal that lives its whole life, averaging 3 years, in sealed underground tunnels (14, 15). Life in darkness resulted in atrophic eyes, and continuous digging led to short extremities and strong neck muscles (15). Spalax can survive extremely low oxygen levels, which may exist in its underground burrows mainly during rainy weather and heavy flooding (16). Under laboratory conditions, terminal pO2 of Spalax ranges from 18 to 28 Torr (17). Blood vessel density in Spalax tissues is higher than that found in rats (18, 19). VEGF levels in Spalax muscles are constitutively high regardless of oxygen level, similar to its expression in highly tumorigenic cells (19). The p53 gene in healthy Spalax individuals possesses 2 amino acid substitutions in its DNA binding domain, which are identical to mutations found in human tumors (20, 21), resulting in up-regulation of p53-targeted genes involved in DNA repair and inhibition of p53-targeted genes associated with apoptosis. Special characteristics of Spalax hemoglobin, myoglobin, haptoglobin, and neuroglobin have been reviewed elsewhere (14, 15). Here, we describe a unique splice variant of heparanase (splice 36) cloned from Spalax, which lacks enzymatic activity, and functions as a dominant negative protein to the wild-type heparanase enzyme.
Results
Cloning Splice Variant 36 of Spalax Heparanase.
We have reported the cloning of wild-type Spalax heparanase and a splice variant of it lacking exon 7 (splice 7), which has led us to search for other splice variants of heparanase. Using PCR and a set of primer pairs, we screened different Spalax tissues' cDNA for the presence of additional bands suggestive of splice variants. PCR on cDNA of kidney from Spalax judaei (15) exposed to a pulse of hypoxia (6% O2, 3 h) led to the cloning of splice variant 36. Sequence analysis revealed that splice 36 of Spalax heparanase results from skipping part of exon 3, exons 4 and 5, and part of exon 6 (Fig. 1A). Compared with the wild-type cDNA, this splice variant exhibits a deletion of 372 bp, without any shift in the translation frame. The donor site in exon 3 and the acceptor site in exon 6 share the nucleotide sequence AAGAAGG, suggesting it as a possible signal to the splicing machinery (Fig. 1B). This nucleotide sequence is conserved in exons 3 and 6 of human heparanase.
Fig. 1.
Structure of heparanase splice variant 36. (A) Schematic structure of exons composing the wild-type heparanase gene (Top), splice 7 (lacking exon 7) (Middle), and splice 36 (resulting from skipping part of exon 3, exons 4 and 5, and part of exon 6) (Bottom). (B) Exons 3–6 of Spalax heparanase. Exons 3 and 5 are in bold. The nucleotides missing in splice 36 are boxed; note that the donor site in exon 3 and the acceptor site in exon 6 share the nucleotide sequence AAGAAGG (shaded), suggesting this sequence as a possible signal to the splicing machinery.
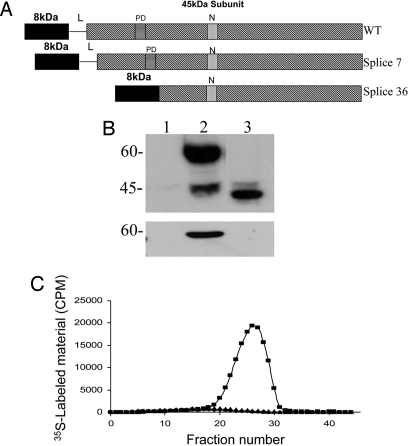
The ORF of splice 36 consists of 1,230 bp that encode for a polypeptide of 410 aa, compared with 1,602 bp and 534 aa in the wild-type Spalax heparanase. The protein structure of splice 36 combines the 8-kDa subunit to the N terminus of the truncated 45-kDa subunit (excluding the linker and part of the 45-kDa subunit, compared with the wild-type heparanase) (Fig. 2A) and lacks 2 of the 3 putative N-glycosylation sites found in the wild-type enzyme (13).
Fig. 2.
Expression of Spalax heparanase splice 36. (A) Schematic structure of the heparanase protein. (Top) Wild-type Spalax heparanase. (Middle) Splice 7 lacking 16 aa between the proton donor (PD) and the nucleophile (N). (Bottom) Splice 36 combining the 8-kDa subunit to the truncated 45-kDa subunit. (B) Western blot of lysates (Upper) and conditioned media (Lower) of 293HEK cells transfected with mock (lane 1), Spalax heparanase wild type (lane 2), or splice 36 (lane 3). Cell lysates and conditioned media were subjected to Western blot analysis using anti-heparanase antibodies 1453. (C) Heparanase enzymatic activity. Lysates of 293HEK cells stably transfected with pcDNA3 vectors containing Spalax wild type (■), splice 36 (▴), or insert free plasmid alone (◆) were incubated (4 h, 37 °C, pH 6.0) with 35S-labeled ECM. Labeled degradation fragments released into the incubation media were analyzed by gel filtration on Sepharose 6B. A peak representing HS degradation products was obtained only with cells transfected with the wild-type Spalax heparanase (fractions 20–30).
Functional Expression of Splice 36 in Mammalian Cells.
The full-length Spalax heparanase cDNA and splice 36 cDNAs were subcloned into the expression vector pcDNA3 and transfected into HEK293 cells. Western blot analysis of splice 36 partially purified from cell lysates revealed a protein band of ≈40 kDa (Fig. 2B, lane 3). Wild-type Spalax heparanase appeared as 60- and 45-kDa protein bands, corresponding to the latent and active forms of the enzyme (Fig. 2B, lane 2). Splice 36 [similar to splice 7 (13)] was not detected in the medium of cultured cells, regardless of the presence of heparin (Fig. 2B Lower). In contrast, wild-type latent heparanase is secreted into the culture medium and accumulates upon the addition of heparin (22). The lack of heparanase splice variants 7 and 36 in the incubation media compared with the wild-type enzyme is probably caused by structural changes in these variants that result in deficient secretion of the corresponding proteins.
Heparanase Enzymatic Activity.
We assessed the ability of splice 36 of Spalax heparanase to degrade HS in intact ECM. For this purpose, lysates of HEK293 cells stably transfected with splice 36 were incubated (4 h, 37 °C, pH 6.0) with intact, naturally-produced, sulfate-labeled ECM. Cells transfected with full-length Spalax heparanase, splice 7, or mock (empty) vector were used as controls. Labeled degradation fragments released into the incubation medium were then analyzed by gel filtration on Sepharose 6B. Lysates of splice 36 transfected cells failed to release degradation products of HS (Fig. 2C). Similar results were obtained with splice 7 (13) and mock-transfected cells. In contrast, incubation of the ECM with lysates of cells transfected with the wild-type Spalax heparanase resulted in release of low molecular mass-labeled degradation fragments eluted toward the Vt of the column (fractions 20–30; 0.5 < Kav < 0.8) (Fig. 2C). These fragments were shown to be degradation products of HS as they were (i) 5- to 6-fold smaller than intact HS side chains, (ii) resistant to further digestion with papain and chondroitinase ABC, and (iii) susceptible to deamination by nitrous acid (23).
Dominant Negative Effect of Splice 36 on Wild-Type Heparanase.
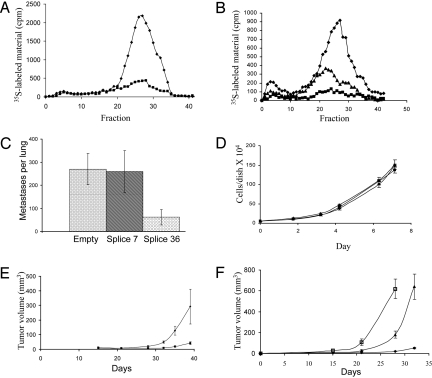
B16 melanoma cells express high endogenous levels of heparanase and readily degrade HS chains in intact ECM (24). B16 melanoma cells were transfected with plasmids containing splice 36 or splice 7 or with insert free plasmid. Cell lysates were incubated with sulfate-labeled ECM, and the incubation media were analyzed by gel filtration as described above. Cells transfected with insert free plasmid degraded the ECM HS, as reflected by a prominent peak of HS degradation fragments released into the incubation medium (Fig. 3A). In contrast, cells transfected with splice 36 yielded a nearly 80% lower HS degradation peak (Fig. 3A). Splice 7 minimally affected the ability of B16 melanoma cells to degrade HS, and the elution profile of HS degradation fragments was similar or mildly lower than that of control mock-transfected cells (data not shown). To test the ability of intact cells to degrade HS in the ECM, B16 melanoma cells were grown on sulfate-labeled ECM, and HS degradation fragments released into the incubation medium were analyzed after 48 h of incubation. As demonstrated in Fig. 3B, splice 36 transfected cells yielded a much lower level of HS degradation fragments as compared with control cells transfected with empty plasmid. Splice 7 decreased the degradation of HS by B16 melanoma cells albeit to a lower extent than that of splice 36 (Fig. 3B).
Fig. 3.
Dominant negative effect of splice 36 on wild-type heparanase. (A) Cell lysates of B16–BL6 murine melanoma cells transfected with pcDNA3 plasmids containing splice 36 (■) or an empty control (◆) were assessed for their ability to degrade HS in intact ECM. Note that splice 36 inhibited the ability of B16–BL6 cells to degrade HS. (B) B16–BL6 cells transfected with pcDNA3 plasmids containing splice 36 (■), splice 7 (▴), or an empty control (◆) were grown on sulfate-labeled ECM, and HS degradation fragments released into the incubation media were analyzed after 48 h of incubation. (C) B16–BL6 melanoma cells electroporated with splice 7, splice 36, or an empty pcDNA3 vector were injected into the tail vein of C57BL/6 mice (0.4 × 106 cell per mouse). After 15 days, mice were killed, and their lungs were fixed and examined for the number of melanoma colonies on the lung surface; note that lung colonization was significantly lower in the splice 36 group. (D) Growth curve (in vitro) of U87 cells transfected with Spalax heparanase splice 36 (■), splice 7 (▴), or an empty vector control (◆). (E) Tumorigenicity. U87 cells (5 × 106/0.2 mL) transfected with Spalax heparanase splice 36 (■) or mock-transfected (◆) were injected s.c. to nude mice. Tumor size is presented as a function of time. (F) Wild-type Spalax heparanase and its splice variant 7 enhances tumor growth. U87 cells (5 × 106/0.2 mL) transfected with wild-type Spalax heparanase (□), heparanase splice variant 7 (▴), or a mock (empty) plasmid (◆) were injected s.c. to nude mice. Tumor size is presented as a function of time. Error bars represent SDs.
Splice 36 of Spalax Heparanase Inhibits Experimental Metastasis of B16 Melanoma Cells.
B16–BL6 cells electroporated with Spalax heparanase splice 7, splice 36, or empty pcDNA3 vector were injected into the tail vein of C57BL/6 mice (0.4 × 106 cells per mouse). Fifteen days after injection, the mice were killed and their lungs were evaluated for the number of surface metastatic colonies. B16–BL6 melanoma cells transiently electroporated with splice 36 yielded 4- to 5-fold fewer metastatic colonies (mean number of colonies ± SD = 62 ± 34) than B16–BL6 melanoma cells transiently transfected with the empty vector (mean number of colonies ± SD = 270 ± 67; P < 0.001) (Fig. 3C). The number of colonies ± SD produced by B16–BL6 melanoma cells electroporated with splice 7 was 259 ± 92 and was not significantly different from that obtained with the mock-transfected group (P = 0.83) (Fig. 3C).
Cell Proliferation in Vitro.
U87 human glioma cells, which possess endogenous HS-degrading ability, were transfected with plasmids containing splice 36, splice 7, wild-type Spalax heparanase, or empty control. Stable transfected cell populations were seeded (5 × 104 per 35-mm dish) into culture dishes and their number was measured as a function of time. The proliferation rate of splice 36-transfected cells was similar to that of mock-transfected cells (Fig. 3D), cells transfected with splice 7 (Fig. 3D), or cells transfected with wild-type Spalax heparanase (data not shown).
Splice 36 Inhibits Tumor Growth.
U87 cells (5 × 106) stably transfected with either the Spalax heparanase variant 36 or mock insert-free control plasmid were injected s.c. to nude mice. Tumor size was measured twice a week, and its volume was calculated. The tumors, which were similar in size at the beginning of the experiment, developed much more rapidly in the control group, compared with tumors generated by splice 36-transfected cells (Fig. 3E). On day 40, both groups were killed and the tumors were dissected and weighed. The average weight (± SD) of tumors in the control group was 676 ± 184 mg, whereas the splice 36 group developed significantly smaller tumors with an average weight of 120 ± 28 mg (P < 0.0025).
Wild-Type Spalax Heparanase and Its Splice Variant 7 Accelerate Tumor Growth.
Mice harboring U87 glioma cells transfected with either wild-type Spalax heparanase or its splice variant 7 developed tumors at a much faster rate than mock-transfected cells (Fig. 3F). Twenty-eight and 32 days after cell inoculation, tumors produced by the wild-type and splice 7 cells, respectively, were too large for the mice to bear. The mice were killed, and tumors were dissected and weighed. The average weight of the wild-type tumors was 1,193 ± 230 mg, and the average weight of splice 7 tumors was 772 ± 212 mg.
Discussion
Spalax as a Model Organism for Hypoxia Tolerance and Angiogenesis Research.
The subterranean blind mole rat lives in underground environment that is extremely stressful compared with the conditions aboveground (15). Adaptation to this environment resulted in the regression of some tissues and organs and progression of others (14, 15). Among the organs that regressed during Spalax evolution are the eyes that became atrophic and subcutaneous, the tail that is absent in Spalax allowing easier movement in underground tunnels, and short extremities compared with aboveground mammals that allows efficient digging of narrow tunnels (15). Other organs that have developed in Spalax include the heart that has higher myocardial maximal oxygen consumption, the lungs that possesses high diffusion capacity, and the muscles that are more developed and have higher blood vessel density compared with aboveground rats (14, 15). Molecular characteristics of Spalax related to hypoxia tolerance include: constitutively high VEGF expression in Spalax muscles (19), high levels of hypoxia inducible factor 1 α, and elevated levels of erythropoietin compared with rats (25). The high blood vessel density in Spalax tissues led us to focus on the contribution of heparanase to the adaptive evolution of hypoxia tolerance in this mammal.
Spalax Heparanase and Its Splice Variants.
Spalax heparanase has 85% homology to the human gene (13). Heparanase is highly expressed in Spalax kidney, liver, heart, brain, and eye, unlike its limited expression in normal human tissues (13). Spalax heparanase has 3 potential N-glycosylation sites, compared with 6 in the human and 4 in the mouse and rat heparanases. Splice 7 of Spalax heparanase possesses all 3 N-glycosylation sites described in the wild-type enzyme (13), whereas splice 36 lacks 2 of them. The 2 alternatively spliced variants of Spalax heparanase (splice 7 and splice 36) lack HS cleavage ability by their own. B16–BL6 murine melanoma cell line possesses high endogenous HS-degrading ability (24). Here, we report that splice 36 functions as dominant negative to the wild-type heparanase enzyme and inhibits ECM degradation by these cells in vitro and metastases formation in vivo. Splice 36 also hinders glioma tumor growth in a mouse model. Intriguingly, Spalax splice variant 7 of heparanase (13) is devoid of enzymatic activity, but unlike splice 36 enhances tumor growth. These results demonstrate that alternative splicing (26, 27) of heparanase regulates its enzymatic activity and might adapt the heparanase function to the fluctuating normoxic–hypoxic subterranean environment that Spalax experiences (16, 18). Development of anticancer drugs directed to suppress angiogenesis is a major challenge (28) of which heparanase inhibition is a promising approach (29, 30).
Alternative Splicing as an Evolutionary Path to Cope with Stress and Increase Genetic Heterogeneity.
The mammalian genome contains ≈25,000 genes. Through alternative splicing each gene can produce several splice variants that results in different proteins (26). Stress is one of the leading catalysts of evolution that foster the processes of adaptation and speciation. Spalax lives under extremely stressful environment underground; one of these stresses is hypoxia. In this study the exposure of Spalax to a pulse of hypoxia resulted in the expression of a variety of splice variants of heparanase in its kidneys. It seems that the stress posed on Spalax elicits the production of these splice variants to cope with the “new” situation. One of the possible roles of these variants could be posttranslation modulation of the effect of the wild-type enzyme on its substrate (HS). These splice variants of heparanase are devoid of HS degradation ability of their own, but seem to possess other nonenzymatic functions (6) that may explain the enhanced tumor growth seen when splice 7 was overexpressed in glioma cells.
Heparanase as a Target for Anticancer Therapy.
Heparanase is a mammalian enzyme that degrades HS in the ECM (3–7). In human, heparanase is expressed mainly in the placenta, platelets, lymphoid organs, and keratinocytes (5–7). Heparanase is overexpressed in nearly every human tumor tested, including breast, pancreas, prostate, colon, and lung cancers, and its overexpression in these malignancies is correlated with aggressive tumor behavior, increased metastasis and angiogenesis, and reduced postoperative patient survival time (5–7). Hence, heparanase is a highly specific target for anticancer therapy; if successfully suppressed, inhibition of tumor growth, metastasis, and angiogenesis is expected to ensue (29, 30). Several inhibitors of heparanase were developed mainly through screening of sulfated compounds, of which PI-88 is being tested in clinical trials (31). A glycol-split N-acetylated heparin (32) has been demonstrated to be highly effective in myeloma tumors (33) and will soon enter clinical trials. Several antiheparanase antibodies have been reported, of which none reached clinical use. Our results show that overexpression of wild-type Spalax heparanase enhances tumor growth, similar to human heparanase. Alternative splicing of heparanase in Spalax results in 2 different splice variants of which splice 36 down-regulates the tumorigenic potential of heparanase and inhibits its ability to degrade HS in the ECM, whereas splice 7 promotes tumor growth. Similarly, the Bcl-x gene has 2 splice variants with opposite effects on apoptosis (27). Recently, we cloned 3 additional splice variants of the Spalax heparanase gene that require additional investigation. Taking into account the significance of heparanase in cancer progression, we anticipate that the ability of heparanase splice variants to inhibit tumor growth will provide important tools for the development of heparanase-inhibiting strategies that will be applied as therapeutic modalities in the treatment of cancer.
Materials and Methods
Animals.
The animals used for cloning the splice variants of Spalax heparanase belong to the Spalax judaei Anza population (15). Animals were captured in the field and kept in the animal facility at the Institute of Evolution (University of Haifa) for at least 3 months before use. Animals were housed in individual cages. They were kept under controlled conditions at 22–24 °C and fed carrots and apples. Animals used in this study were adults and ranged in weight from 100 to 150 g. Some animals were exposed to a pulse of hypoxia (6% O2, 3 h) and then killed by injection of Ketaset CIII at 5 mg/kg body weight. Whole organs were taken out and immediately frozen in liquid nitrogen. The experiments were approved by the Ethics Committee of the University of Haifa.
RNA and cDNA Preparation.
Total RNA was extracted from tissues by using TRI Reagent (Molecular Research Center) according to the manufacturer's instructions. cDNA was prepared by reverse transcription (M-MLV reverse transcriptase; Promega) of 1 μg of total RNA, by using oligo(dT)15 and random primers (13).
Gene Cloning.
For cloning of Spalax heparanase splice variants, kidney cDNAs were prepared. Spalax-specific primers around different exons were designed (Sigma/Genosys), and PCRs were performed by using TaqDNA polymerase (Qbiogene) and kidney cDNA as a template. Bands corresponding to the splice forms were subcloned into the pGEM-Teasy vector and sequenced by using gene- and vector-specific primers with an automated DNA sequencer (ABI Prism model 310 Genetic Analyzer; PerkinElmer). Full-length Spalax heparanases lacking the spliced-out exons were constructed by means of digestion and ligation by using site-specific restriction enzymes and T4 ligase (Promega), then inserted into the expression vector pcDNA3 (Invitrogen) (13).
Cells and Transfections.
HEK293 and U87 glioma cells were cultured in DMEM (4.5 g glucose per L) containing 10% FCS and antibiotics as described (8). Cells were grown in 60-mm tissue culture dishes and transfected with a total of 1–2 μg of plasmid DNA mixed with 6 μL of FuGene transfection reagent (Roche Applied Science) and 94 μL of DMEM. Transiently-transfected cells were obtained after 24- to 48-h incubation at 37 °C. Stable populations of transfected cells were selected with G418 (Sigma).
B16–BL6 melanoma cells were electroporated with pcDNA3 plasmids containing splice 36, splice 7, or empty vector (4 × 106 cells in 400 μL of medium containing 10 μg of plasmid DNA) by using a single 70-ms pulse at 140 V and an ECM 830 Electro Square porator and disposable cuvettes (model 640, 4-mm gap; BTX) (24). After electroporation, the transfected cells were plated at a density of 0.4 × 106 cells per 100-mm dish and grew for 24–48 h. Efficiency of transfection (≈80%) was evaluated 48 h after electroporation of a vector containing the gene encoding green fluorescent protein by fluorescence microscopy. Stable populations of transfected cells were selected with G418 (Sigma).
Experimental Metastasis.
For the experimental metastasis studies, the lateral tail vein of 6-week-old male C57BL/6 mice was injected with 0.4 mL of a cell suspension containing 0.4 × 106 B16–BL6 melanoma cells transiently electroporated with pcDNA3 plasmids containing splice 36, splice 7, or empty vector. Fifteen days after cell injection, mice were killed and their lungs were removed, fixed in Bouin's solution, and scored under a dissecting microscope for the number of metastatic nodules on the lung surface. Five mice were used per group.
Western Blot Analysis.
Cells (2 × 106) transfected with either splice 36, splice 7, wild-type heparanase, or insert-free pcDNA3 vector alone were lysed in 1 mL of lysis buffer containing 50 mM Tris·HCl (pH 7.4), 150 mM NaCl, 0.5% Triton X-100, and a mixture of protease inhibitors (Roche Applied Science). Heparanase was concentrated by incubating the cell lysate (4 °C, 1 h) with ConA beads (Amersham Biosciences) and washing twice with PBS. The beads were boiled (3 min) in sample buffer and centrifuged and the supernatant was subjected to SDS/PAGE and immunoblot analysis using polyclonal antiheparanase antibodies 1453 (1:2,500), as described (13). Immunoreactive bands were detected by the enhanced chemiluminescence reagent as described (3). For evaluation of secreted heparanase, cells were grown in 100-mm culture dishes to ≈70–80% confluence, and then serum-free medium was applied. Twenty-four hours after incubation the medium was collected and centrifuged, and the supernatant was incubated with ConA beads and subjected to Western blot analysis, performed as described above.
Heparanase Activity.
Cell lysates prepared from 1 × 106 cells by 3 cycles of freezing and thawing in heparanase reaction buffer [20 mM phosphate-citrate buffer (pH 6.0), 1 mM DTT, 1 mM CaCl2, and 50 mM NaCl] were incubated (3 h, 37 °C, pH 6.0) with 35S-labeled ECM. The incubation medium containing 35S-labeled HS degradation fragments was analyzed by gel filtration on a Sepharose CL-6B column (3, 8). Fractions (0.2 mL) were eluted with PBS, and their radioactivity was counted in a β-scintillation counter. Degradation fragments of HS side chains were eluted from Sepharose 6B at 0.5 < Kav < 0.8 (fractions 20–30) (3, 8). Each experiment was performed 3 times, and the variation in elution positions (Kav values) did not exceed ± 15% of the mean. For heparanase activity of intact cells, 2 × 106 B16 cells were grown (48 h, 37 °C, pH 6.6) on 35S-labeled ECM, and the incubation medium was analyzed thereafter as described above.
Proliferation Assay.
U87 cells (5 × 104 cells per 35-mm dish) stably transfected with splice 7, splice 36, wild-type or control splice 36, or control vectors were seeded in complete medium into 35-mm culture dishes. The cells were dissociated with trypsin/EDTA and counted in triplicate in a Coulter counter every other day for 7 days.
Tumorigenicity Studies.
U87 cells stably transfected with Spalax heparanase splice 36, splice 7, wild-type, or control insert-free plasmids were injected s.c. to BALB/c athymic nude mice (Harlan). Cell suspension (5 × 106/0.2 mL) was s.c.-inoculated at the right flank. Xenograft sizes were determined weekly by externally measuring tumors in 2 dimensions with a caliper. Tumor volume (V) was determined by the equation: V = L × W2 × 0.5, where L is the length and W is the width of the xenograft. At the end of the experiment, mice were killed by cervical dislocation, and xenografts were dissected and weighed.
Acknowledgments.
We thank Alma Joel for technical assistance. This work was supported by Israel Science Foundation Grant 549/06 (to I.V.), National Cancer Institute Grant RO1-CA106456 (to I.V.), the Israel Cancer Research Fund (I.V.), the Rappaport Family Institute Fund (I.V.), and the Ancell Teicher Research Foundation for Molecular Genetics and Evolution (E.N).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. FM955617).
References
- 1.Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: The sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 2.Fuster MM, Esko JD. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 3.Vlodavsky I, et al. Mammalian heparanase: Gene cloning, expression, and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 4.Hulett MD, et al. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 5.Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341–347. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilan N, Elkin M, Vlodavsky I. Regulation, function, and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Vreys V, David G. Mammalian heparanase: What is the message? J Cell Mol Med. 2007;11:427–452. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldshmidt O, et al. Cell surface expression and secretion of heparanase markedly promote tumor angiogenesis and metastasis. Proc Natl Acad Sci USA. 2002;99:10031–10036. doi: 10.1073/pnas.152070599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parish CR, Freeman C, Hulett MD. Heparanase: A key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–M108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima M, Irimura T, Nicolson GL. Heparanases and tumor metastasis. J Cell Biochem. 1988;36:157–167. doi: 10.1002/jcb.240360207. [DOI] [PubMed] [Google Scholar]
- 11.Shafat I, et al. Heparanase levels are elevated in the plasma of pediatric cancer patients and correlate with response to anticancer treatment. Neoplasia. 2007;9:909–916. doi: 10.1593/neo.07673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasser NJ, Avivi A, Shusy M, Vlodavsky I, Nevo E. Cloning, expression, and characterization of an alternatively spliced variant of human heparanase. Biochem Biophys Res Commun. 2007;354:33–38. doi: 10.1016/j.bbrc.2006.12.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nasser NJ, et al. Adaptive evolution of heparanase in hypoxia-tolerant Spalax: Gene cloning and identification of a unique splice variant. Proc Natl Acad Sci USA. 2005;102:15161–15166. doi: 10.1073/pnas.0507279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nevo E. Mosaic Evolution of Subterranean Mammals: Regression, Progression, and Global Convergence. London: Oxford Univ Press; 1999. [Google Scholar]
- 15.Nevo E, Ivanitskaya I, Beiles A. Adaptive Radiation of Blind Subterranean Mole Rats. Leiden, The Netherlands: Backhuys; 2001. [Google Scholar]
- 16.Shams I, Avivi A, Nevo E. Oxygen and carbon dioxide fluctuations in burrows of subterranean blind mole rats indicate tolerance to hypoxic-hypercapnic stresses. Comp Biochem Physiol A Mol Integr Physiol. 2005;142:376–382. doi: 10.1016/j.cbpa.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Arieli R, Nevo E. Hypoxic survival differs between two mole rat species (Spalax ehrenbergi) of humid and arid habitats. Comp Biochem Physiol A. 1991;100:543–545. doi: 10.1016/0300-9629(91)90367-l. [DOI] [PubMed] [Google Scholar]
- 18.Widmer HR, Hoppeler H, Nevo E, Taylor CR, Weibel ER. Working underground: Respiratory adaptations in the blind mole rat. Proc Natl Acad Sci USA. 1997;94:2062–2067. doi: 10.1073/pnas.94.5.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avivi A, et al. Increased blood vessel density provides the mole rat physiological tolerance to its hypoxic subterranean habitat. FASEB J. 2005;19:1314–1316. doi: 10.1096/fj.04-3414fje. [DOI] [PubMed] [Google Scholar]
- 20.Ashur-Fabian O, et al. Evolution of p53 in hypoxia-stressed Spalax mimics human tumor mutation. Proc Natl Acad Sci USA. 2004;101:12236–12241. doi: 10.1073/pnas.0404998101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avivi A, Ashur-Fabian O, Amariglio N, Nevo E, Rechavi G. p53, a key player in tumoral and evolutionary adaptation: A lesson from the Israeli blind subterranean mole rat. Cell Cycle. 2005;4:368–372. doi: 10.4161/cc.4.3.1534. [DOI] [PubMed] [Google Scholar]
- 22.Levy-Adam F, et al. Identification and characterization of heparin/heparan sulfate binding domains of the endoglycosidase heparanase. J Biol Chem. 2005;280:20457–20466. doi: 10.1074/jbc.M414546200. [DOI] [PubMed] [Google Scholar]
- 23.Vlodavsky I, Fuks Z, Bar-Ner M, Ariav Y, Schirrmacher V. Lymphoma cell-mediated degradation of sulfated proteoglycans in the subendothelial extracellular matrix: Relationship to tumor cell metastasis. Cancer Res. 1983;43:2704–2711. [PubMed] [Google Scholar]
- 24.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- 25.Shams I, Avivi A, Nevo E. Hypoxic stress tolerance of the blind subterranean mole rat: Expression of erythropoietin and hypoxia-inducible factor 1 α. Proc Natl Acad Sci USA. 2004;101:9698–9703. doi: 10.1073/pnas.0403540101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 27.Minn AJ, Boise LH, Thompson CB. Bcl-x(S) anatagonizes the protective effects of Bcl-x(L) J Biol Chem. 1996;271:6306–6312. doi: 10.1074/jbc.271.11.6306. [DOI] [PubMed] [Google Scholar]
- 28.Folkman J. Endogenous angiogenesis inhibitors. Apmis. 2004;112:496–507. doi: 10.1111/j.1600-0463.2004.apm11207-0809.x. [DOI] [PubMed] [Google Scholar]
- 29.Finkel E. Potential target found for antimetastasis drugs. Science. 1999;285:33–34. doi: 10.1126/science.285.5424.33. [DOI] [PubMed] [Google Scholar]
- 30.Fjeldstad K, Kolset SO. Decreasing the metastatic potential in cancers: Targeting the heparan sulfate proteoglycans. Curr Drug Targets. 2005;6:665–682. doi: 10.2174/1389450054863662. [DOI] [PubMed] [Google Scholar]
- 31.Lewis KD, et al. A phase II study of the heparanase inhibitor PI-88 in patients with advanced melanoma. Invest New Drugs. 2008;26:89–94. doi: 10.1007/s10637-007-9080-5. [DOI] [PubMed] [Google Scholar]
- 32.Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: Structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr Pharm Des. 2007;13:2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, et al. The syndecan-1 heparan sulfate proteoglycan is a viable target for myeloma therapy. Blood. 2007;110:2041–2048. doi: 10.1182/blood-2007-04-082495. [DOI] [PMC free article] [PubMed] [Google Scholar]