Abstract
We have shown that smoking impacts bronchial airway gene expression and that heterogeneity in this response associates with smoking-related disease risk. In this study, we sought to determine whether microRNAs (miRNAs) play a role in regulating the airway gene expression response to smoking. We examined whole-genome miRNA and mRNA expression in bronchial airway epithelium from current and never smokers (n = 20) and found 28 miRNAs to be differentially expressed (P < 0.05) with the majority being down-regulated in smokers. We further identified a number of mRNAs whose expression level is highly inversely correlated with miRNA expression in vivo. Many of these mRNAs contain potential binding sites for the differentially expressed miRNAs in their 3′-untranslated region (UTR) and are themselves affected by smoking. We found that either increasing or decreasing the levels of mir-218 (a miRNA that is strongly affected by smoking) in both primary bronchial epithelial cells and H1299 cells was sufficient to cause a corresponding decrease or increase in the expression of predicted mir-218 mRNA targets, respectively. Further, mir-218 expression is reduced in primary bronchial epithelium exposed to cigarette smoke condensate (CSC), and alteration of mir-218 levels in these cells diminishes the induction of the predicted mir-218 target MAFG in response to CSC. These data indicate that mir-218 levels modulate the airway epithelial gene expression response to cigarette smoke and support a role for miRNAs in regulating host response to environmental toxins.
Keywords: cigarette smoke, mir-218, bronchial airway epithelium
Approximately 1.3 billion people smoke cigarettes worldwide, which contributes to 5 million preventable deaths per year (1). Smoking is a significant risk factor for lung cancer (2), the leading cause of cancer-related death in the United States, and chronic obstructive pulmonary disease (COPD), the fourth leading cause of death worldwide. Cigarette smoking has been found to induce a number of genetic and molecular changes in the respiratory tract, including cellular atypia (3), loss of heterozygosity (4, 5), and promoter hypermethylation (6), which can occur in cytologically normal airway epithelium (5, 7). We have characterized the effects of smoking on the human airway epithelial transcriptome and found that smoking induces expression of airway genes involved in regulation of oxidant stress, xenobiotic metabolism, and oncogenesis while suppressing those involved in regulation of inflammation and tumor suppression (8). In addition, we have described a pattern of gene expression in cytologically normal large airway epithelial cells that can serve as a biomarker for lung cancer elsewhere in the lung (9). These studies suggest that gene expression changes in airway epithelium reflect host response to and damage from cigarette smoke; however, the regulatory mechanisms mediating these gene expression changes are unclear.
MicroRNAs (miRNA) are small noncoding RNAs that serve to down-regulate gene expression by suppression of translation or messenger RNA (mRNA) degradation. Thirty percent of protein-coding genes are predicted to be regulated by miRNAs (10). Hundreds of human miRNAs have been cloned (11), with each of them predicted to regulate hundreds of genes by complementary binding to the 3′-untranslated region (UTR) of target mRNAs (12). A number of miRNAs have been shown to play important roles in developmental timing, cell death, carcinogenesis, and stress responses in vitro (13–16), such as hypoxia (17). Whereas miRNA expression has been examined in lung tumors (18–24), neither the expression of miRNA in bronchial epithelium nor the effect of smoking on bronchial airway miRNA expression has been characterized, and it is unclear what role miRNAs play in regulating the physiological response to environmental exposures such as tobacco smoke.
In this study, we have profiled the miRNAs expressed in a relatively pure population of bronchial airway epithelial cells and identified those that are differentially expressed with smoking. Additionally, by whole-genome profiling of both miRNA and mRNA from the same in vivo samples, we have identified mRNAs that are potentially regulated by smoking-induced changes in miRNA expression. Through in vitro transfection experiments, we have shown that modulating the expression of one such miRNA (mir-218) is sufficient to alter the expression of a subset of the mRNAs that are both predicted targets of this miRNA and altered by smoking in vivo. Further, we have shown that cigarette smoke condensate (CSC) suppresses the expression of mir-218 in primary bronchial epithelial cells and that altering the levels of mir-218 in these cells attenuates the CSC-dependent induction of mir-218 target mRNAs. These studies suggest that smoking-dependent changes in miRNA expression levels mediate some of the smoking-induced gene expression changes in airway epithelium and that miRNAs might therefore play a role in the host response to environmental exposures and the pathogenesis of smoking-related lung disease.
Results
Smoking Affects Expression of miRNAs in Bronchial Epithelium.
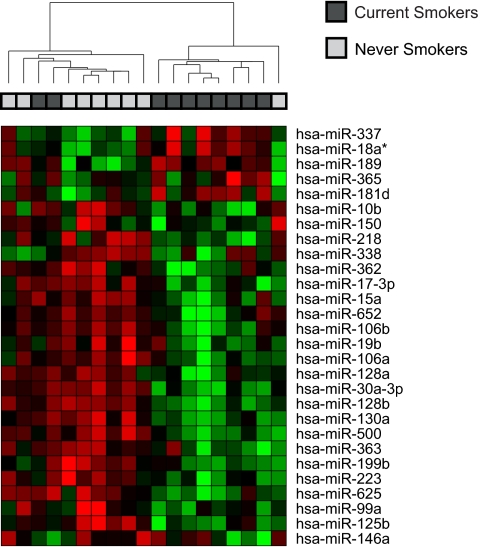
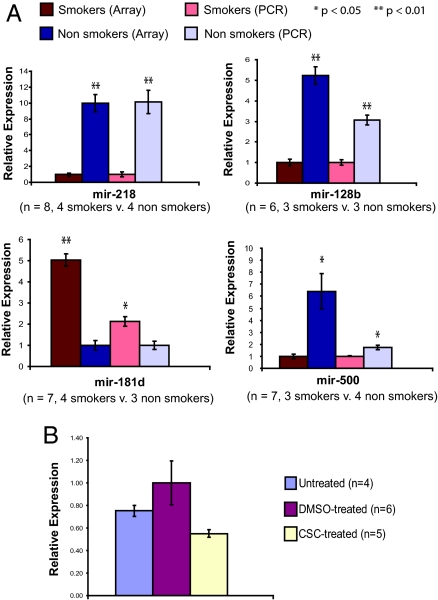
MicroRNA and mRNA expression levels in bronchial epithelial RNA collected from 20 volunteers (10 current smokers, 10 never smokers) recruited for this study were determined by using microarrays [supporting information (SI) Fig. S1]. Demographic data for these subjects are presented in Table 1. After miRNA microarray data preprocessing (Fig. S2A), 232 miRNAs were detected above background and the expression of these miRNAs was analyzed further. Temporal (3 months) and technical replicates run on miRNA arrays were highly reproducible across these 232 detected miRNAs, with the temporal replicates having an R2 = 0.91 and technical replicates having an R2 = 0.94. Principal component analysis of all 232 detected miRNAs demonstrates modest separation of subjects by smoking status (Fig. S3), suggesting that smoking impacts miRNA expression in bronchial epithelium. Using a Welch's t test, 28 miRNAs were found to be differentially expressed between current and never smokers (P < 0.05), which is ≈3 times more than is expected by chance at that threshold (Fig. 1, Table S1). Of these 28 miRNAs, 23 were down-regulated in smokers. The most significantly altered miRNA was mir-218 (P = 5 × 10−4), which was down-regulated 4-fold in smokers. Four of the top differentially expressed miRNAs (mir-218, mir-128b, mir-500, and mir-181d) were validated in a subset of bronchial samples by using quantitative (q)RT-PCR (Fig. 2A), with fold changes comparable to those found on the miRNA arrays.
Table 1.
Patient demographics
| Current smokers | Never smokers | P value | |
|---|---|---|---|
| n | 10 | 10 | |
| Age | 37.2 ± 9.9 | 31.2 ± 10 | 0.20 |
| Race | 5 AFA, 4 HIS, 1 CAU | 3 AFA, 2 HIS, 5 CAU | 0.15 |
| Sex | 7 M, 3 F | 3 M, 7 F | 0.07 |
| Pack years | 18.8 ± 16 | 0 | 0.0002 |
Demographics for individuals recruited for bronchoscopy are listed above. P values for sex (M, male; F, female) and race (AFA, African-American; HIS, Hispanic; CAU, Caucasian) were calculated by using Fisher's exact test. Standard deviations for age and pack years are listed. P values for age and pack years were calculated by using Student's t test.
Fig. 1.
Unsupervised hierarchical clustering of 28 differentially expressed microRNAs across bronchial epithelium of smokers and nonsmokers. Red indicates higher expression, green indicates lower expression. In general, most miRNAs that are altered by smoking show decreased expression in the airway epithelium of smokers. A small number of current smokers group with never smoker samples on the basis of expression of these miRNAs, suggesting heterogeneity in the response to smoking.
Fig. 2.
(A) qRT-PCR validation of 4 differentially expressed miRNAs, mir-218, mir-128b, mir-181d, and mir-500 in bronchial epithelial samples. Expression of these miRNAs changes in the same direction between smokers and nonsmokers both in microarray and in PCR analysis. (B) Expression of mir-218 in primary human bronchial epithelial cells that were untreated (n = 4), DMSO treated (n = 6), and cigarette smoke condensate treated (CSC, n = 5). Mir-218 expression was reduced in CSC-treated cells with respect to untreated (P = 0.008) cells. In both A and B, error bars indicate standard error, and P values were determined using Student's t test.
mRNA and miRNA Expression Are Correlated in Vivo.
We have shown that smoking alters mRNA expression in airway epithelial cells (8, 25). To determine whether miRNAs might be modulators of mRNAs that are differentially expressed in smokers (25), we first examined whether the mRNAs that we previously identified as being affected by smoking are enriched for predicted targets of each of the human miRNAs in MSigDb (26) (see SI Methods). Interestingly, 26% of the mRNAs that are differentially expressed in smokers are predicted to be targets of just 20 miRNAs/miRNA families (Table S2). This suggests that alterations in the levels of a relatively small number of miRNAs in response to smoking could potentially contribute to a number of the smoking-associated changes in mRNA expression and that modulation of specific miRNAs might therefore represent a mechanism that contributes to the overall host response to tobacco smoke exposure.
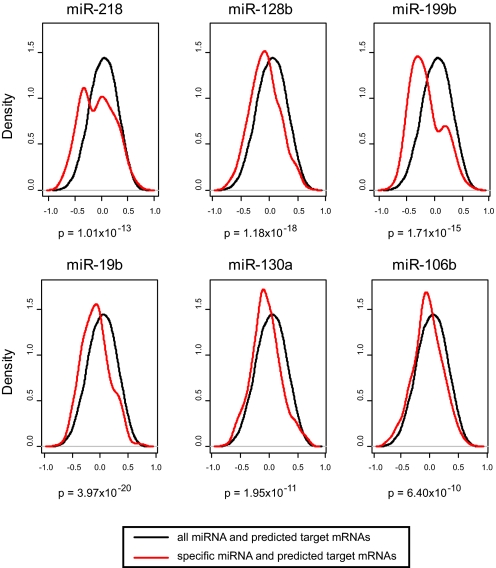
To explore this possibility, we compared the Pearson correlation coefficients for each of the differentially expressed miRNAs with their putative mRNA targets (as defined by publicly available databases of predicted miRNA targets) to the background distribution of the Pearson correlation coefficients of all miRNAs and their predicted mRNA targets (see Fig. S2B). We found that many of the 28 differentially expressed miRNAs were highly inversely correlated with their predicted targets compared with all miRNAs and their predicted mRNA targets (P < 1 × 10−15). Density plots of 6 miRNAs with the highest number of negatively correlated targets (at least 20 mRNAs with Pearson correlation, P < 0.05) are shown in Fig. 3. Density plots for the remaining differentially expressed miRNAs that had predicted targets in either the PicTar or the Targetscan databases are shown in Fig. S4. Gene ontology analysis of the anticorrelated targets of these miRNAs reveals these genes are involved in cell–cell adhesion, cellular signaling, and cytoskeletal structure (Table S3).
Fig. 3.
Relationship between the expression of miRNAs and their predicted mRNA targets in airway epithelium of current and never smokers. The distribution of Pearson correlation coefficients calculated between the miRNA and predicted target mRNA expression for 6 of the 28 differentially expressed miRNAs is shown in red. All 232 miRNAs and their predicted targets are in black. There is a significant increase in the degree of negative correlation between differentially expressed miRNAs and their targets relative to all miRNAs and their targets, which reflects the inhibitory effects of miRNAs on target gene expression. These 6 miRNAs have the highest number of predicted mRNA targets with significantly anticorrelated expression in vivo.
Regulation of mir-218 in Response to Acute in Vitro Cigarette Smoke Exposure.
To further investigate whether cigarette smoke is responsible for the miRNA expression differences seen in airway epithelial cells from smokers and nonsmokers, we performed 2 experiments. First, a publicly available mRNA microarray expression dataset (GSE10700) of normal human bronchial epithelial (NHBE) cells exposed to cigarette smoke was analyzed using Gene Set Enrichment Analysis (GSEA) (26). We found that there was significant overrepresentation of mir-218 targets among the genes most up-regulated by cigarette smoke exposure (SI Methods, Table S4). This indicates that the targets of mir-218 are induced upon acute exposure to cigarette smoke and suggests that mir-218 expression likely decreases upon such exposure. To confirm this, we next exposed NHBE cells to 20 μg/ml CSC. The concentration of CSC used was similar to that in other studies that exposed NHBE cells to CSC (27, 28). We found an almost 2-fold reduction in mir-218 levels in cells exposed to CSC at 24 h relative to DMSO-treated controls (Fig. 2B). Thymidine incorporation assays confirmed that the 20 μg/ml CSC dose used does not inhibit cell proliferation or DNA synthesis in NHBE cells (data not shown), suggesting that the mir-218 changes observed are unlikely due to growth inhibition or toxicity.
Overexpressing mir-218 in Vitro Preferentially Suppresses Its Predicted Targets.
To explore the functional significance of the inverse correlation between the expression of miRNAs and their predicted targets, we performed a series of in vitro transfection experiments (see Fig. S2C). First, we increased mir-218 levels in H1299 cells by transient transfection with a synthetic RNA oligonucleotide mimic of mir-218 precursor (premir-218) or a scrambled sequence that served as a negative control. After confirming that this resulted in increased levels of mature mir-218 by real-time PCR (data not shown), Affymetrix all-exon microarrays were run to examine mRNA expression in these transfectants relative to the negative control. GSEA showed a significant enrichment of predicted mir-218 targets among the genes most down-regulated in H1299 cells transfected with mir-218 (P < 0.01), confirming that overexpression of mir-218 results in the down-regulation of many of its predicted targets. GSEA was then performed to determine whether any literature-defined pathways are altered in response to increased mir-218 levels in vitro. Several groups of genes involved in various cell cycle pathways and the p53, apoptosis, and Rho/Rac/Ras pathways (false discovery rate, FDR < 0.17) were enriched. Targets of mir-218 involved in apoptosis pathways were also anticorrelated to its expression in vivo (Table S3), suggesting that mir-218 might either directly or indirectly affect genes involved in these processes.
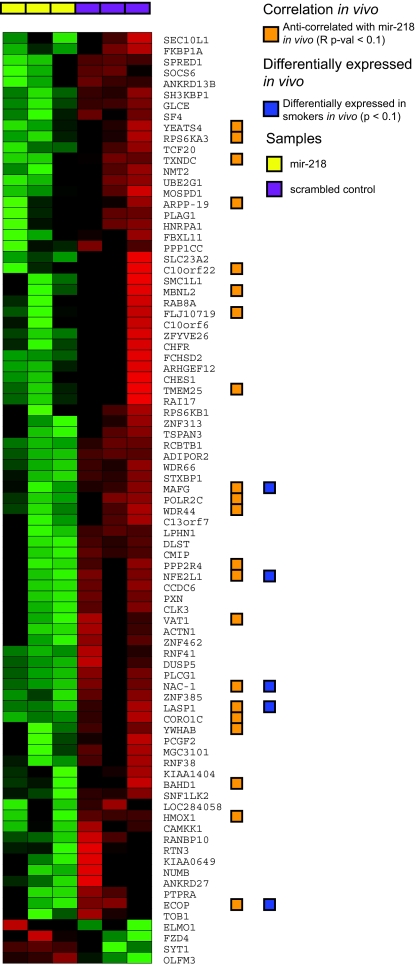
A Welch's t test yielded 1235 genes that changed with mir-218 overexpression (P < 0.05) relative to the negative control transfectants, with the large majority (≈83%) being down-regulated by elevated mir-218 levels. Of these 1235 genes, 85 were database-predicted targets of mir-218. A χ2 test confirmed (P = 0.0001) that predicted targets of mir-218 were more likely to be differentially expressed with its overexpression compared with nontargets. Almost all (≈95%) of these 85 predicted target genes were down-regulated with overexpression of mir-218 (Fig. 4), indicating that among the genes that are altered by increasing mir-218 levels, its predicted targets are more likely to have lower expression relative to controls than nontargets (Fisher's exact test, P = 0.002).
Fig. 4.
Expression of genes differentially expressed by increasing mir-218 levels in vitro (P < 0.05) that are predicted to be targets of mir-218. The orange blocks designate genes that are also anticorrelated with mir-218 expression in vivo (Pearson correlation, P < 0.1). Blue blocks indicate genes that are also differentially expressed in smokers (P < 0.1). Red indicates higher expression, green indicates lower expression.
We next sought to determine the relationship between genes that have altered expression in response to elevated mir-218 in vitro and the genes that are associated either with mir-218 levels or with smoking in vivo. For this analysis, we examined the in vivo expression patterns of the 85 putative mir-218 targets that are altered in response to elevated mir-218 levels in vitro. The expression of 21 of these 85 genes is highly inversely correlated (Pearson correlation, P < 0.1) with mir-218 levels in vivo (see Fig. 4). This is significant (chi-square P = 0.01) as expression of only 16% of the predicted mir-218 targets examined in vivo is significantly inversely correlated with mir-218 expression. Five of these 21 inversely correlated genes are differentially expressed in smokers (P < 0.1) and each of the 5 shows significantly lower expression (P < 0.05) in the presence of elevated mir-218 in vitro and higher expression in smokers (where mir-218 expression is repressed). These 5 genes (MAFG, ECOP, LASP1, NAC-1, and NFE2L1) thus are candidates for being regulated by changing mir-218 levels in response to cigarette smoke. Using qRT-PCR, we validated the effects of overexpressing mir-218 on the levels of these 5 genes in both H1299 and primary NHBE cells. We found that increasing mir-218 levels in H1299 cells caused significant decreases in all 5 genes, but in NHBE cells only 3 (ECOP, NAC-1, and LASP1) of the 5 genes were significantly repressed (Fig. S5A and Fig. S6A). The remaining two targets (MAFG, NFE2L1) are expressed at very low levels at baseline in NHBE cells compared with H1299 cells.
Knockdown of mir-218 Increases Expression of mir-218 Target Genes.
In the second experiment to determine whether mir-218 might regulate the expression of smoking-related genes, we sought to determine whether the endogenous levels of mir-218 serve to repress the expression of mir-218-targeted mRNAs. To test this hypothesis, we decreased the level of mir-218 in both H1299 and NHBE cells by transfection of an antisense oligonucleotide to mir-218 (anti-mir-218), using a scrambled-sequence oligo as a control. After finding that transfection with anti-mir-218 resulted in a 4-fold reduction in mir-218 levels in H1299 cells and a 10-fold reduction in NHBE cells, we subsequently measured expression of MAFG, ECOP, LASP1, NAC-1, and NFE2L1 by qRT-PCR, 48-h posttransfection. Decreasing mir-218 expression resulted in moderate increases in expression of each gene in H1299 cells, although not all at statistically significant levels (Fig. S5B). Similar trends were observed in NHBE cells where MAFG and ECOP expression increased in response to decreased mir-218 levels (Fig. S6B). These results suggest that mir-218 may serve to repress expression of these genes in vitro, lending further support to the model that mir-218 levels modulate the expression of these genes.
MAFG, a transcription factor, was one of our most significantly differentially expressed genes both in vivo and in vitro. By using the ROVER algorithm (29), we found transcription factor binding sites for MAFG were significantly overrepresented (P < 0.003) among all genes that were altered in smokers. This suggests that mir-218 may play a role in altering nontarget genes in response to tobacco exposure through regulation of the transcription factor MAFG.
Modulating mir-218 Levels Attenuates the Impact of Cigarette Smoke on MAFG Expression.
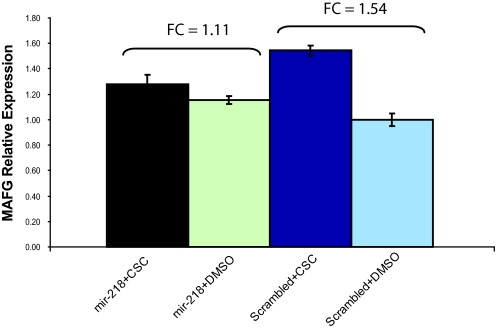
To determine whether the repression of mir-218 by smoking is required for the induction of mir-218 targets in response to cigarette smoke, we determined the effects of CSC exposure on the expression of MAFG in NHBE cells when the CSC-dependent repression of mir-218 was attenuated either by overexpressing or knocking down mir-218. Exposure to cigarette smoke condensate alone caused MAFG to increase by 1.54-fold (Fig. 5). When the levels of mir-218 are increased by transfection with the premir-218 oligo, the levels of mature mir-218 are increased ≈1000-fold (data not shown) and the modulation of mir-218 levels by CSC is completely inhibited. Under these conditions CSC induces MAFG by only 1.11-fold (Fig. 5), which is significantly less than the CSC-dependent modulation of MAFG in the presence of endogenous mir-218 levels (2-way ANOVA interaction effect, P = 0.0001). This result demonstrates that attenuating the modulation of mir-218 by CSC attenuates the CSC-dependent increase in the mir-218 target MAFG. When the levels of mir-218 are decreased by transfection with anti-mir-218 oligo, MAFG levels are induced by ≈20%, and the CSC-dependent increase in MAFG levels is, similar to the overexpression experiments, attenuated (Fig. S7). These data suggest that the modulation of mir-218 levels by smoke exposure contributes to the smoking-dependent regulation of MAFG expression levels.
Fig. 5.
Overexpressing mir-218 attenuates the induction of MAFG expression by cigarette smoke condensate in NHBE. CSC induces MAFG expression 1.54-fold (Scrambled + DMSO vs. Scrambled + CSC). However, when mir-218 is overexpressed, CSC induces MAFG expression only 1.11-fold (mir-218 + CSC vs. mir-218 + DMSO). The significantly reduced induction of MAFG by cigarette smoke condensate in the presence of high levels of mir-218 (2-way ANOVA interaction effect, P = 0.000003) suggests that modulation of mir-218 contributes to the CSC-dependent regulation of MAFG expression.
Discussion
In vitro studies have shown that miRNAs play a role in cellular response to stress as well as cell growth and death (17, 30, 31). The purpose of this study was to determine whether airway miRNA expression is altered in response to smoking and whether these changes contribute to smoking-induced mRNA expression changes in airway epithelial cells (8, 25). We found 28 miRNAs that are differentially expressed, with most (82%) down-regulated in smokers. These findings are similar to prior studies of miRNAs in cancer. One study, which profiled 217 miRNAs across multiple different types of cancer tissues, found that most miRNAs were down-regulated in tumors compared with normal tissues, regardless of cell type (32). This may be because miRNA levels are tightly linked to degree of cellular differentiation and that reduced expression of miRNAs in cancer is associated with more dedifferentiated tumors. Consistent with this model, the down-regulation of miRNA expression in airway epithelial cells with smoking may be a consequence of repetitive airway injury that results in accumulation of proliferating and dedifferentiated cells.
Several studies have sought to establish regulatory relationships between miRNA and mRNA expression (33–36). By collecting miRNA and mRNA from the same samples, we were able to discover correlations between miRNA and mRNA expression profiles both across patient groups and within individuals. Whereas this represents the first study characterizing the effects of smoking on miRNA expression, several of the miRNAs from our analyses have been implicated in other diseases. Mir-218, which was down-regulated in smokers and by CSC exposure in vitro, is transcribed within the intronic region of the known tumor suppressor gene SLIT2 (11). SLIT2, which is frequently inactivated in lung and breast tumors (37), was also significantly down-regulated in smokers with expression correlating to that of mir-218 in our in vivo samples. A second identical copy of mir-218 occurs within the SLIT3 locus, and expression of SLIT3 has also been shown to be down-regulated in lung cancer (18). Mir-218 levels have also been found to be decreased in cervical carcinoma cells (38), suggesting a potential general role of mir-218 as a tumor suppressor. Three additional miRNAs (mir-15a, mir-199b, and mir-125b) that are down-regulated in smokers have also been shown to be down-regulated in other cancers including chronic lymphocytic lymphoma (CLL) (39), lung (18), and breast cancer (40). These findings suggest that the down-regulation of miRNAs in smokers could be related to the development of tobacco-related cancers. Given the ability of gene expression differences in the airway to serve as a biomarker for the presence of lung cancer elsewhere in the respiratory tract (9), our finding that miRNAs implicated in carcinogenesis are differentially expressed in the airway epithelium of smokers suggests that airway miRNA expression could potentially serve as an indicator of smoking-induced disease processes.
Functional analysis of the predicted targets of mir-218 that are anticorrelated with its expression in vivo reveals an overrepresentation of genes involved in cell structure, cell–cell adhesion, and cell signaling and ion transport pathways. A subset of these targets is also altered with smoking. These include transcriptional regulators such as MAFG and NAC-1, signaling genes such as ECOP (NFkB pathway), and genes involved in cellular structure (LASP1). A predicted mir-218 target of particular interest is the transcription factor MAFG, which has been shown previously (8), as well as in this study to be up-regulated in the bronchial epithelium of smokers and on exposure of cells to CSC in vitro. Binding sites for MAFG are overrepresented among genes that are differentially expressed in the epithelial cells of smokers, suggesting that alteration of mir-218 levels could indirectly influence the expression of MAFG targets.
The argument for the smoking-dependent regulation of mir-218 contributing to the regulation of smoking-dependent changes in mRNA expression rests on our in vitro perturbation experiments. In H1299 and NHBE cell lines, transient transfections both increasing and decreasing mir-218 levels lead to decreased and increased target expression, respectively, suggesting that altered levels of this miRNA are sufficient to regulate expression of these genes. Furthermore, blocking the CSC-dependent down-regulation of mir-218 by overexpressing mir-218 attenuates the CSC-dependent increase in MAFG expression. A similar, but less dramatic attenuation of the CSC-dependent increase in MAFG expression is observed when mir-218 levels are knocked down. These data suggest that the smoking-dependent modulation of mir-218 contributes to the regulation of mir-218 targets. However, because CSC still has an appreciable effect on MAFG expression even when mir-218 levels are experimentally manipulated, it is likely that other mechanisms contribute to the regulation of smoking-modulated gene expression.
Although we were able to identify miRNAs that are differentially expressed in smokers and correlate with mRNA changes from the same individuals, the small sample size of our study may have limited our power to detect additional smoking-related miRNA and/or mRNA changes. Whereas differentially expressed miRNAs tended to be down-regulated in smokers, we did not explore the mechanisms by which miRNAs are down-regulated. We were able to identify significant levels of anticorrelated expression between miRNAs that are affected by smoking and their target mRNAs, suggesting that miRNAs are modulating mRNA expression in response to smoking. We were further able to establish a role for the levels of mir-218 in regulating the expression of its predicted targets in vitro, using H1299 and NHBE cells. Whereas increasing mir-218 levels in these cells led to significant decreases in expression of many of its targets, down-regulating mir-218 in these cells led to only modest increases in target expression. One possible reason for this disparity is that we were unable to completely abrogate expression of mir-218, and the modest changes could have been a result of residual levels of mir-218 in these cells.
In summary, we have profiled global miRNA expression in the bronchial epithelium of healthy current and never smokers. Our findings suggest that smoking causes many miRNAs to be down-regulated in these epithelial cells. Messenger RNA expression profiles from the same individuals show that expression of putative targets of these miRNAs is generally up-regulated in the airway epithelial cells of smokers. We have further demonstrated that modulating levels of mir-218 in vitro alters the expression of its predicted targets and attenuates the smoking-dependent induction of target mRNA expression. These findings suggest that miRNAs may play a role in regulating the gene expression response to tobacco exposure in airway epithelial cells. MicroRNA profiles obtained from these cells might therefore serve as biomarkers for smoking-related lung diseases and help elucidate the regulatory mechanisms that mediate the host response to tobacco smoke exposure and other environmental agents.
Methods
Study Population.
We recruited never (n = 10) and current (n = 10) smoking volunteers to undergo fiberoptic bronchoscopy at Boston Medical Center. Subjects with significant second-hand environmental cigarette exposure, respiratory symptoms, or regular use of inhaled medications were excluded. For participants, a detailed smoking history was obtained, including cumulative tobacco exposure (measured in pack years), age when they began smoking, and second-hand tobacco exposure. All individuals were screened with routine chest x-ray and spirometry and were excluded if they had evidence of pulmonary pathology. Temporal (3 months) and technical replicates were collected from 2 participants. The study was approved by the Institutional Review Board of Boston Medical Center and all participants provided written informed consent.
Sample Collection.
Bronchial airway epithelial cells were obtained from bronchial brushings of the right mainstem bronchus taken during fiberoptic bronchoscopy with an endoscopic cytobrush (Cellebrity Endoscopic Cytobrush, Boston Scientific). The epithelial cell content of the bronchial brushings performed in our clinic is routinely greater than 90% (8). The brushes were immediately placed in RNAprotect Cell Reagent (QIAGEN) and stored at 4°C until RNA was isolated and fractionated into low molecular weight (LMW) and high molecular weight (HMW) fractions, by using the miRNeasy mini kit (QIAGEN) (see SI Methods).
MicroRNA Microarray Hybridization and Data Analysis.
Three hundred nanograms of LMW RNA were labeled using the FlashTag labeling kit (Genisphere, Inc.), according to manufacturer's protocol (see SI Methods). After labeling, samples were incorporated into a hybridization mix that was then applied to Invitrogen NCode miRNA microarrays containing 1,053 miRNAs from 6 species (467 human miRNAs) printed in triplicate. The arrays were scanned on a GenePix 4000 scanner (Axon/Molecular Devices).
Raw data were extracted by using GenePix Pro 4.0 (Molecular Devices), quantile normalized, and log transformed using the limma package for R (http://www.r-project.org/). On the basis of Principal Component Analysis, 1 nonsmoker sample was removed from further analysis. In addition to our sample filter, only miRNA replicates that were detected at levels 2 standard deviations above the average background in at least 6 samples were considered for further analysis. The median of the remaining replicates for each miRNA was then taken to obtain a single expression value for each miRNA, leaving a total of 232 distinct miRNAs that were analyzed further.
mRNA Microarray Hybridization and Data Analysis.
One microgram of the HMW RNA fraction from each sample was used as starting material for hybridization to Human Exon 1.0 ST GeneChips as described (41). Transcript-level expression estimates for the ≈17,800 empirically supported transcripts were used for our analysis (see SI Methods).
Correlating miRNA and mRNA Expression.
Pearson correlations [and associated P values] were calculated for all miRNA–mRNA pairs across all samples, and the Benjamini-Hochberg false discovery correction (42) was used to adjust for multiple comparisons. Pairs of miRNA–mRNA with statistically significant inverse correlations were then cross-referenced with two target prediction databases [PicTar (43) or TargetScan, version 3.1 (10)] to identify those where the mRNA is a predicted target of the miRNA.
Validating Potential mRNA-miRNA Regulatory Relationships via Transient Transfection of miRNA in H1299 and NHBE Cell Lines.
H1299 cells (an immortalized human lung carcinoma cell line) and primary human bronchial epithelial cells (NHBE, Cell Applications) were cultured in 6-well plates for 24 hours prior to transfection (see SI Methods). MicroRNA-218 was overexpressed or knocked down using synthetic oligonucleotides at a concentration of 30 nM with the appropriate negative controls (n = 5-6 per group). Cells were harvested at 48 hours posttransfection, miRNA/mRNA were isolated and run on Affymetrix Exon arrays (H1299 overexpression group only) and qRT-PCR (all 4 groups).
Cigarette Smoke Exposure in NHBE Cells.
We used a commercially available cigarette smoke condensate (Murty Pharmaceuticals) prepared by smoking University of Kentucky 1R3F research cigarettes on a standard Federal Trade Commission smoking machine. NHBE cells were plated in 6-well plates as described. Cigarette smoke condensate was diluted to 20 μg/ml in growth media and applied to the NHBE cells (n = 6), with DMSO-treated (1 μg/ml, n = 6) and untreated (n = 6) cells serving as controls. The potential toxicity of this concentration of CSC was evaluated using [3H]thymidine incorporation assays (see SI Methods). Cells were harvested 24-h postexposure and RNA was isolated from these cells by using the miRNeasy mini kit according to manufacturer's protocol.
Measuring the Effect of CSC on NHBE Cells with Altered mir-218 Levels.
NHBE cells were plated as described in 6-well plates. Cells were first transfected with either premiR-218 or anti-mir-218 along with the appropriate Cy-3-labeled scrambled oligo (n = 6 in each group), using the PrimeFect reagent. After confirming ≈90% transfection efficiency at 24 h, the transfection media were removed and cigarette smoke condensate was applied to cells at a concentration of 20 μg/ml. The cells were then lysed at 24 h post-CSC exposure and RNA was isolated as described above.
Statistical Analysis of Gene Expression Data.
Differential expression of miRNAs and mRNAs was determined using a Welch's t test. GSEA (26) was performed to determine whether sets of genes comprising either various literature-derived pathways or the sequence-predicted targets of mir-218 targets are perturbed when mir-218 levels are altered in vitro. Hierarchical clustering was performed on z-score-normalized data, using CLUSTER and TREEVIEW software.
Real-Time PCR Validation.
Quantitative real-time PCR was used to confirm the differential expression of a select number of miRNAs in vivo and target mRNAs in vitro (see SI Methods).
Additional Information.
Additional information including raw expression data from microarrays run in vivo and results from correlation analysis can be obtained on the website accompanying this article (http://pulm.bumc.bu.edu/cgi-bin/mirDB/home.cgi). All mRNA and microRNA microarray data have also been deposited in the GEO database under accession number GSE14224.
Supplementary Material
Acknowledgments.
We thank Adam Gower for his assistance in the real-time PCR data analysis. This work was supported by the Doris Duke Charitable Foundation (A.S.), National Institutes of Health/National Cancer Institute Grant R01CA124640 (to A.S., M.E.L., and J.J.C.), National Institutes of Health/National Institute of Environmental Health Sciences Grant U01ES016035 (to A.S. and M.E.L.), and National Institutes of Health/National Heart, Lung, and Blood Institute Grant R01HL095388 (to A.S. and M.E.L.) .
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806383106/DCSupplemental.
References
- 1.WHO. WHO: The Facts About Smoking and Health. Geneva: WHO; 2006. 30 May 2006. [Google Scholar]
- 2.Shields PG. Molecular epidemiology of lung cancer. Ann Oncol. 1999;5(Suppl 10):S7–S11. doi: 10.1093/annonc/10.suppl_5.s7. [DOI] [PubMed] [Google Scholar]
- 3.Franklin WA, et al. Widely dispersed p53 mutation in respiratory epithelium. A novel mechanism for field carcinogenesis. J Clin Invest. 1997;100:2133–2137. doi: 10.1172/JCI119748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell CA, Klares S, O'Connor G, Brody JS. Loss of heterozygosity in epithelial cells obtained by bronchial brushing: Clinical utility in lung cancer. Clin Cancer Res. 1999;5:2025–2034. [PubMed] [Google Scholar]
- 5.Wistuba II, et al. Molecular damage in the bronchial epithelium of current and former smokers. J Natl Cancer Inst. 1997;89:1366–1373. doi: 10.1093/jnci/89.18.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo M, et al. Promoter hypermethylation of resected bronchial margins: A field defect of changes? Clin Cancer Res. 2004;10:5131–5136. doi: 10.1158/1078-0432.CCR-03-0763. [DOI] [PubMed] [Google Scholar]
- 7.Thiberville L, et al. Evidence of cumulative gene losses with progression of premalignant epithelial lesions to carcinoma of the bronchus. Cancer Res. 1995;55:5133–5139. [PubMed] [Google Scholar]
- 8.Spira A, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spira A, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths-Jones S, et al. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar MS, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 15.Leung AK, Sharp PA. microRNAs: A safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Kato M, Slack FJ. microRNAs: Small molecules with big roles - C. elegans to human cancer. Biol Cell. 2008;100:71–81. doi: 10.1042/BC20070078. [DOI] [PubMed] [Google Scholar]
- 17.Kulshreshtha R, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Chiosea S, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 20.Matsubara H, et al. Apoptosis induction by antisense oligonucleotides against miR-17–5p and miR-20a in lung cancers overexpressing miR-17–92. Oncogene. 2007;26:6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 21.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 23.Inamura K, et al. let-7 microRNA expression is reduced in bronchioloalveolar carcinoma, a non-invasive carcinoma, and is not correlated with prognosis. Lung Cancer. 2007;58:392–396. doi: 10.1016/j.lungcan.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Kumar MS, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beane J, et al. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 2007;8:R201. doi: 10.1186/gb-2007-8-9-r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgensen ED, et al. Global gene expression analysis of human bronchial epithelial cells treated with tobacco condensates. Cell Cycle. 2004;3:1154–1168. [PubMed] [Google Scholar]
- 28.Fields WR, et al. Gene expression in normal human bronchial epithelial (NHBE) cells following in vitro exposure to cigarette smoke condensate. Toxicol Sci. 2005;86:84–91. doi: 10.1093/toxsci/kfi179. [DOI] [PubMed] [Google Scholar]
- 29.Haverty PM, Hansen U, Weng Z. Computational inference of transcriptional regulatory networks from expression profiling and transcription factor binding site identification. Nucleic Acids Res. 2004;32:179–188. doi: 10.1093/nar/gkh183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 33.Boren T, et al. MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol Oncol. 2008;110:206–215. doi: 10.1016/j.ygyno.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 34.Marcucci G, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 35.Blower PE, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther. 2007;6:1483–1491. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- 36.Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 2007;8:R166. doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dallol A, et al. SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res. 2002;62:5874–5880. [PubMed] [Google Scholar]
- 38.Martinez I, et al. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott GK, et al. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Liu G, Lenburg ME, Spira A. Comparison of smoking-induced gene expression on affymetrix exon and 3′-based expression arrays. Genome Inform. 2007;18:247–257. [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 43.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.