Abstract
The MYC family of cellular oncogenes includes c-Myc, N-myc, and L-myc, which encode transcriptional regulators involved in the control of cell proliferation and death. Accordingly, these genes become aberrantly activated and expressed in specific types of cancers. For example, c-Myc translocations occur frequently in human B lymphoid tumors, while N-myc gene amplification is frequent in human neuroblastomas. The observed association between aberrations in particular MYC family genes and specific subsets of malignancies might reflect, at least in part, tissue-specific differences in expression or function of a given MYC gene. Since c-Myc and N-myc share substantial functional redundancy, another factor that could influence tumor-specific gene activation would be mechanisms that target aberrations (e.g., translocations) in a given MYC gene in a particular tumor progenitor cell type. We have previously shown that mice deficient for the DNA Ligase4 (Lig4) nonhomologous DNA end-joining factor and the p53 tumor suppressor routinely develop progenitor (pro)-B cell lymphomas that harbor translocations leading to c-Myc amplification. Here, we report that a modified allele in which the c-Myc coding sequence is replaced by N-myc coding sequence (NCR allele) competes well with the wild-type c-Myc allele as a target for oncogenic translocations and amplifications in the Lig4/p53-deficient pro-B cell lymphoma model. Tumor onset, type, and cytological aberrations are similar in tumors harboring either the wild-type c-Myc gene or the NCR allele. Our results support the notion that particular features of the c-Myc locus select it as a preferential translocation/amplification target, compared to the endogenous N-myc locus, in Lig4/p53-deficient pro-B cell lymphomas.
Keywords: nonhomologous end joining, pro-B cell lymphoma, genomic instability, immunoglobulin genes
The MYC gene family encodes highly related basic helix–loop–helix/leucine zipper transcription factors that function in the control of cell proliferation, apoptosis, and transformation (1, 2). MYC family members include the c-Myc, N-myc, and L-myc proto-oncogenes, all of which have been implicated as oncogenes in particular types of human malignancies (3–5). MYC transcription factors bind to the same DNA target sequences and show a substantial functional redundancy (1, 2, 6), although specific functions are suggested by their different expression patterns (4, 7, 8). As all 3 MYC family members are abundantly expressed in developing embryos, the late embryonic lethality of knockout mice for individual MYC genes suggests that different Myc proteins might compensate for each other in earlier developmental stages (2). In adults, c-Myc is expressed in most proliferating cells, while N-myc expression becomes restricted to specific differentiated cell types, including developing B and T lineage cells and also cells of the brain, testis, and heart (9–11).
Some studies have indicated that c-Myc, N-myc, and L-myc proteins have differences in their ability to induce apoptosis and to modulate specific target genes. On the other hand, cell culture studies suggested that c-Myc and N-myc possess similar ability to induce cell proliferation and transformation; although c-Myc may be more effective in some contexts (12–14). In mice, homozygous replacement of the c-Myc coding exons with the corresponding region of the N-myc gene, N-myc/c-Myc replacement allele (NCR allele), allows normal development, indicating that N-myc can compensate for c-Myc loss when appropriately expressed (15). Mouse embryonic fibroblasts (MEFs) homozygous for the NCR allele show growth characteristics that are not substantially impaired compared to those of wild-type (WT) MEFs. Similarly, lymphocyte development in NCR mice is normal and purified B and T cells in culture respond to different mitogens only slightly less robustly than WT B and T cells (15).
Overexpression of specific MYC family genes is frequently associated with particular types of human tumors. N-myc deregulation is almost exclusively associated with solid tumors and only rarely observed in lymphomas. N-myc is amplified in ≈30% of cases of neuroblastoma and at lower frequencies in other tumors of neuroectodermal origin (glioma, medulloblastoma, retinoblastoma, small cell lung carcinoma) (3, 4, 16). Amplification of the L-myc gene is frequently observed in small cell lung carcinomas (3, 5). The c-Myc gene is commonly translocated in human B cell malignancies and amplified and/or overexpressed in a variety of solid tumors, including breast, ovarian, and gastrointestinal cancers (3, 5). Translocations of c-Myc in B cell lymphomas typically fuse c-Myc to the Ig heavy chain (IgH) or, less frequently, to the Ig light chain (IgL) loci and are believed to mediate c-Myc overexpression by bringing it into close proximity with strong Ig transcriptional enhancers (17–19). In developing B and T lineage cells, the recombination activating gene (RAG)-1 and -2 proteins initiate V(D)J recombination by introducing DNA double-strand breaks (DSBs) at target V, D, and J segments in the Ig and Tcell receptor loci, which are then fused by the nonhomologous DNA end-joining pathway (NHEJ) to make complete V(D)J exons (20). In the IgH locus of mature B cells, the activation-induced cytidine deaminase (AID) initiates formation of DSBs in large repetitive S region sequences that flank different sets of IgH constant region exons, with the breaks subsequently being joined by NHEJ or alternative end joining to fuse 2 different S regions to effect class switch recombination (CSR) (21). Oncogenic c-Myc/Ig translocations in B cell lymphomas can be associated with errors in either V(D)J recombination or CSR (17, 18, 22, 23).
Mice that are deficient for the tumor suppressor p53 and for certain NHEJ factors, such as DNA Ligase 4 (Lig4), invariably develop pro-B cell lymphomas harboring 2 characteristic translocations between chromosome 12 (which contains the IgH locus) and chromosome 15 (which contains the c-Myc locus) (24–28). One complex 12;15 translocation (c12;15) contains coamplified c-Myc and IgH sequences. Such complex translocations with associated gene amplifications have been referred to as “complicons.” A second 12;15 translocation (t12;15) in these NHEJ/p53 double-deficient pro-B lymphomas appears to be a by-product of the c12;15 and contains IgH sequences but lacks c-Myc sequences. On the basis of the structure of the translocations and other studies, IgH/Myc complicon formation in Lig4/p53-deficient (LP) pro-B cell lymphomas was found to be initiated by aberrant joining of RAG-generated DSBs at the J region of the IgH locus (JH) to sequences far downstream (50–700 kb) of c-Myc, generating a dicentric 12;15 chromosome. In the p53-deficient background, this dicentric 12;15 inevitably leads to c-Myc amplification via a breakage–fusion–bridge (BFB) mechanism (26, 28).
Even though N-myc amplification is rarely observed in LP pro-B lymphomas, both N-myc and c-Myc are expressed in pro-B cells (9). In addition, N-myc can support normal B cell development in the absence of c-Myc (15), and overexpression of either c-Myc or N-myc under the control of the B cell-specific Eμ enhancer results in development of pro-B cell lymphomas (29–32). Finally, complex N-myc/IgH translocations frequently arise in mice deficient for p53 and the Artemis NHEJ factor, showing that, in this genetic background, the endogenous N-myc gene can compete with c-Myc as a pro-B cell oncogenic translocation/amplification target (33). Together, this set of findings is consistent with the notion that mechanistic aspects of translocation targeting, rather than cellular selection per se, may influence the choice of c-Myc vs. N-myc activation in particular tumor types and/or genetic backgrounds. To further test this possibility, we bred the NCR allele into the LP background and asked whether N-myc, in the place of c-Myc, serves as an effective translocation target in pro-B cell lymphomas.
Results
LP Mice Heterozygous or Homozygous for the NCR Allele Develop Pro-B Cell Lymphoma.
We previously generated the NCR allele by employing gene-targeted mutation to replace the c-Myc coding region (exons 2 and 3) with the corresponding coding region of N-myc (15). NCR homozygous (N/N) mice are born at sub-Mendelian rate but show relatively normal development and lymphocyte functions (15). The NCR allele was bred into the LP background, to generate LPMycN/+ and LPMycN/N mice. As previously observed for LP mice (25), LPMycN/+ and LPMycN/N mice were born at reduced frequency and were smaller than WT and heterozygous littermates. LPMycN/+ and LPMycN/N cohorts were monitored for tumor development. Both LPMycN/+ and LPMycN/N animals became moribund by 8–12 weeks of age (Fig. 1A), similar to what we previously reported for LP mice (25, 28). As also found for LP mice, most of the LPMycN/+ (20/21) and LPMycN/N (8/9) mice succumbed to B220+/IgM− pro-B cell lymphoma (Table 1 and Fig. 1B) that presented mostly in the peripheral lymph nodes with infiltration of the spleen and thymus, but rarely with involvement of mesenteric lymph nodes. These results demonstrate that replacing c-Myc with N-myc does not alter the kinetics and the spectrum of tumor development in the LP model.
Fig. 1.
LPMycN/+ and LPMycN/N mice develop pro-B cell lymphomas. (A) Kaplan–Meier curves of the LPMycN/+ (n = 21) and LPMycN/N (n = 9) cohorts are shown. The curves represent tumor-free survival. (B) Surface marker expression in LPMycN/+ and LPMycN/N tumors was analyzed by cytofluorimetry with B220/IgM (Top) or CD4/CD8 (Bottom) antibodies. C, control; spl, spleen; pLN, peripheral lymph nodes; thy, thymus. Representative cases are shown.
Table 1.
Summary of LPMycN/+ and LPMycN/N tumors
| Mouse | Tumor | Affected organs | Myc locus alteration | Weeks | FISH | |||
|---|---|---|---|---|---|---|---|---|
| pLN | Spl | Thy | mLN | |||||
| LPMycN/+ | ||||||||
| 633 | Pro-B | Yes | NA | NA | c-Myc amplified | 7 | ||
| 399 | Pro-B | NA | Part. | Yes | c-Myc amplified | 7 | t12;15, c12;15 | |
| 455 | Pro-B | Yes | NA | Yes | c-Myc amplified | 7 | t12;15, c12;15 | |
| 407 | Pro-B | Yes | Part. | Yes | c-Myc amplified | 11 | t12;15, c12;15 | |
| 515 | Pro-B | Yes | Part. | p + th | c-Myc amplified | 10 | ||
| 530 | Pro-B | Yes | Yes | Yes | c-Myc amplified | 9 | ||
| 531 | Pro-B | Yes | Part. | Yes | c-Myc amplified | 7.5 | t12;15, c12;15 | |
| 563 | Pro-B | Yes | Part. | p + th | c-Myc amplified | 10 | ||
| 572 | Pro-B | Part. | Part. | Part. | c-Myc amplified | 12 | ||
| 573 | Pro-B | Yes | Part. | Yes | c-Myc amplified | 7.5 | ||
| 618 | Pro-B | Yes | No | Part. | yes | c-Myc amplified | 11 | |
| 620 | Pro-B | Yes | Part. | Yes | c-Myc amplified | 12.5 | ||
| 654 | Pro-B | Yes | Part. | Yes | yes | c-Myc amplified | 7 | t12;15, c12;15 |
| 501 | Pro-B | Yes | Part. | Yes | NCR amplified | 8 | t12;15, c12;15 | |
| 580 | Pro-B | Part. | Part. | Yes | NCR amplified | 8 | t12;15, c12;15 | |
| 591 | Pro-B | Yes | Part. | Yes | NCR amplified | 6 | t12;15, c12;15 | |
| 635 | Pro-B | Yes | Yes | p + th | NCR amplified | 14 | t12;15, c12;15 | |
| 788 | Pro-B | Yes | Part. | NCR amplified | 8.5 | t12;15, c12;15 | ||
| 617 | Pro-B | Yes | Part. | p + th | No detectable amplification | 15 | t12;15, c12;15 | |
| 619 | Pro-B | Yes | Part. | Yes | No detectable amplification | 11.5 | t16;4 | |
| 404b | Thymic | None | 14 | |||||
| LPMycN/N | ||||||||
| 215 | Pro-B | Yes | Part. | Yes | No alteration | 9 | t 17;12 | |
| 404 | Pro-B | Yes | NA | NA | NCR amplified | 11 | ||
| 871 | Pro-B | Yes | p + th | NCR rearranged | 10 | |||
| 15 | Pro-B | Yes | Yes | Yes | NCR amplified | 9 | ||
| 233 | Pro-B | Yes | Yes | Yes | NCR amplified | 8 | ||
| 234 | Pro-B | NA | Part. | NA | NCR amplified | 12 | t12;15, c12;15 | |
| 426 | Pro-B | Yes | NA | NA | NCR rearranged | 8 | ||
| 504 | Pro-B | Yes | Yes | Part. | NCR amplified | 11 | t12;15, c12;15 | |
| 797 | Thymic | None | 16 | |||||
Part, partial; p + th, partial pro-B and thymic tumors; NA, not analyzed.
The NCR Allele Is Often Amplified in LP Pro-B Cell Tumors.
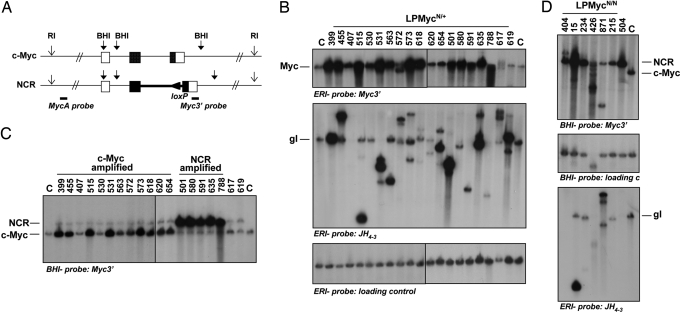
Pro-B cell tumors that arise in LP mice routinely amplify c-Myc and IgH as a result of 12;15 translocations and subsequent BFB cycles (28). To determine whether a similar mechanism takes place in LPMycN/+ lymphomas, we performed Southern blot analysis on DNA extracted from the various tumor samples, using the EcoRI enzyme, which generates a 20-kb fragment encompassing the c-Myc or NCR (referred to as “Myc”) locus (Fig. 2A). Almost all LPMycN/+ tumors analyzed showed significant amplification of the Myc locus (Fig. 2B Top), as judged by comparing with normal spleen control DNA. Loading of a similar amount of DNA in each lane was confirmed by reprobing the same membrane with a control probe (Fig. 2B Bottom). Most of the amplified Myc fragments did not show rearrangements, indicating that potential translocation breakpoints giving rise to the amplification were located outside of the EcoRI fragment, either upstream or downstream of the c-Myc locus, as previously found in LP tumors (28).
Fig. 2.
The NCR locus is amplified in LPMycN/+ and LPMycN/N tumors. (A) Schematic representation of the endogenous c-Myc and targeted NCR loci. Positions of relevant restriction sites and probes used are shown. (B) Southern blot analysis of LPMycN/+ tumor DNA digested by EcoRI (ERI) restriction enzyme. Probes used are indicated at the bottom of each panel. Positions of the WT c-Myc (Top) and germ-line IgH (Middle) bands are indicated. Loading of an equal amount of DNA in each lane was confirmed by using a control probe (Bottom) hybridizing to the MDC1 gene. (C) Southern blot analysis of LPMycN/+ tumor DNA digested by BamHI (BHI) restriction enzyme and analyzed with Myc 3′ probe. Positions of the WT c-Myc and NCR bands are indicated. (D) Southern blot analysis of LPMycN/N tumor DNA. Restriction enzymes and probes used are indicated at the bottom of each panel. Positions of the WT c-Myc and NCR bands (Top) and germ-line IgH band (Bottom) are indicated.
We analyzed the same panel of lymphomas with a probe hybridizing downstream of the JH region (JH4–3) and found that most samples contained distinct JH rearranged bands, demonstrating clonality of the tumor cell population (Fig. 2B Middle). The low-level germ-line band detected in some samples likely derives from non-B lineage cells within the tumor. As previously shown for LP tumors (24, 25), amplification of the JH region is frequent (e.g., tumors 399, 455, 531, and 635) but not invariably detected by this probe. In the latter context, lack of functional NHEJ during V(D)J recombination has been shown to result in extensive deletions in the region of the D-J join extending into the region represented by the JH probe (34), likely explaining lack of rearranged JH bands in some tumor samples (e.g., tumors 407 and 530).
We next asked whether the NCR allele contributed to the translocations and amplifications in some LPMycN/+ tumors. The targeting strategy used for the N-myc/c-Myc replacement allowed us to distinguish the NCR allele from the WT c-Myc allele by Southern blotting (see Fig. 2A). We analyzed DNA from LPMycN/+ tumors by digestion with BamHI enzyme and hybridization with a probe from the 3′ end of the c-Myc locus, which can recognize both the WT c-Myc and the NCR allele. This analysis allowed us to verify that amplification took place on the WT c-Myc allele in 13/20 LPMycN/+ tumors and on the NCR allele in 5/20 LPMycN/+ tumors (Fig. 2C and data not shown). Two LPMycN/+ tumors (617 and 619) did not show detectable amplification of either Myc allele (see below). These results clearly demonstrate that the NCR allele can substitute for and compete with the WT c-Myc allele as a target of oncogenic translocations and amplifications in pro-B cell lymphomas, suggesting that the preference for c-Myc vs. N-myc amplification in the LP background may involve differential translocation targeting to the specific chromosomal locations. This conclusion was further reinforced by the observation that lymphomas arising in LPMycN/N mice also show high frequency of amplification in the Myc and IgH loci (Fig. 2D and Table 1).
LP/NCR Pro-B Cell Lymphomas Harbor Characteristic Chromosomal Aberrations.
To better characterize the lymphomas arising in LPMycN/+ and LPMycN/N mice, we obtained metaphase spreads from short-term tumor cell cultures and analyzed them by chromosomal paints and fluorescence in situ hybridization (FISH). As previously shown for pro-B cell lymphomas from NHEJ/p53-deficient mice (28), all analyzed LPMycN/+ samples contained the characteristic complicon with coamplified signals for a 3′IgH locus BAC probe and a c-Myc locus BAC probe (Fig. 3A Left and B). Sequential reprobing of the same slides with 5′IgH and 3′IgH BACs (Fig. 3A Middle) and chromosome 12 (ch12) and 15 (ch15) paints (Fig. 3 A Right and B) clearly allowed us to identify the different chromosomal species involved in translocations. LPMycN/+ and LPMycN/N tumors invariably harbored the classical Myc/IgH complicon (c12;15) and frequently a 12;15 translocation, which also contains sequences represented in the 3′IgH BAC but does not contain Myc locus sequences (t12;15). Also, a normal ch12 (positive for both 5′IgH and 3′IgH BACs) and 1 or 2 normal ch15's (positive to Myc BAC) were present (Fig. 3A).
Fig. 3.
LPMycN/+ and LPMycN/N tumors harbor characteristic c12;15 and t12;15 translocations. (A) FISH and chromosome paint analysis of metaphases from selected LPMycN/+ and LPMycN/N tumors. The same metaphase was sequentially analyzed with a different set of probes as indicated at the top of each panel. Note that the red signal from the second hybridization was not efficiently stripped and still shows in the third set of images. Results from chromosome paint hybridization were confirmed on an independent set of metaphases for all tumors analyzed (not shown). A schematic of the different chromosomal species detected is shown at the Top. (B) Summary of FISH and chromosome paint analyses on additional LPMycN/+ tumors. Only chromosomes involved in translocations are shown. Sequential hybridization with the set of probes indicated at the top was performed. A graphic representation of the translocations observed in each case is shown.
Notably, no significant differences in translocation patterns were evident between tumors that amplified the WT c-Myc allele (407, 455, and 399, see Fig. 3; and 531 and 654, data not shown) vs. the NCR allele (504, 635, 580, and 591, see Fig. 3; and 788, data not shown). Tumor 399 (c-Myc amplified) and 635 (NCR amplified) lacked the t12;15, but still contained the 12;15 complicon. Tumor 635 also harbored another complex 12;15 translocation, not associated with amplification in ≈50% of the metaphases (Fig. 3B). These results indicate that the NCR allele can support the same type of translocations as the unmodified, endogenous c-Myc allele.
Characterization of LPMyc Tumors Lacking Myc Amplification.
Some LPMycN/+ and LPMycN/N tumors lacked amplification of either Myc allele (Fig. 2). Tumor 871 (LPMycN/N) had a rearrangement of the NCR allele not associated with amplification of downstream sequences (Fig. 2D, hybridization with Myc 3′ probe). Such translocations have been shown in a minor subset of LP and XRCC4/p53 double-deficient pro-B cell tumors (24, 25). In tumor 871, amplification of a 5′ Myc probe (MycA) was detected (Fig. 4A), suggesting that the translocation breakpoint is upstream of the BamHI site downstream of exon 3 (see Fig. 2A). Tumor 617 (LPMycN/+) showed no significantly amplified bands with 3′ Myc (Fig. 2B) and 5′ Myc (Fig. 4A) probes. Spectral karyotyping (SKY) and FISH showed this tumor to harbor 12;15 translocations and complex translocations involving ch12 and ch16 (Fig. 4B), but additional analyses suggested ongoing instability/rearrangements, with only a fraction of the metaphases showing c-Myc amplification (not shown), explaining lack of Myc amplification as detected by Southern blot. Tumors 619 and 215 also lacked amplification or rearrangements involving the c-Myc locus (Fig. 2 B and D), but in these cases SKY analyses also showed the tumors to lack 12;15 translocations. Tumor 619 harbors a clonal 16;4 nonreciprocal translocation (Fig. 4C); while tumor 215 has a clonal 17;12 translocation (data not shown) and WT N-myc locus amplification as detected by Southern blotting (Fig. 4A).
Fig. 4.
Characterization of LPMycN/+ and LPMycN/N tumors lacking Myc amplification. (A) Southern blot analysis of DNA from tumors 617, 619 (LPMycN/+) and 871, 215 (LPMycN/N) with probes specific for the 5′ region of the c-Myc locus (MycA, Left panel) and for the N-myc locus (Right panel). Positions of the WT c-Myc, WT N-myc, and NCR bands are indicated. Loading of an equal amount of DNA in each lane was confirmed by using a control probe (Right Bottom panel) hybridizing to the MDC1 gene. (B) SKY analysis on tumor 617. One representative metaphase is shown. The arrows indicate chromosomes involved in clonal translocations. A detailed view of these chromosomes is presented in the panels on the Right, showing DAPI, spectral, and computer-classified staining for each chromosome. (C) SKY analysis on tumor 619. One representative metaphase is shown. The arrow indicates the chromosome involved in clonal translocation. The complete karyotype (computer-classified staining) and details of the 16;4 translocations are shown.
Discussion
Cellular selection for oncogenic activity plays a major role in the appearance of recurrent translocations that activate dominant cellular oncogenes in particular tumors. In this context, overexpression of c-Myc or N-myc in normal cells can generate a p53-dependent oncogenic stress response that promotes cellular apoptosis (2, 35). Therefore, p53 deficiency confers a strong survival advantage to various cell types exposed to activated c-Myc or N-myc expression, providing a rationale for why translocations/amplifications of c-Myc or N-myc are so frequently observed in tumors that arise in the context of dual deficiency for NHEJ factors and p53 (24, 28, 33, 36). Thus, NHEJ deficiency predisposes to increased levels of translocations and p53 deficiency allows tumor progenitor cells to tolerate translocations that activate Myc gene expression. This scenario alone, however, does not explain why certain NHEJ/p53-deficient tumors almost exclusively activate c-Myc and others commonly activate N-myc. In particular, most LP pro-B tumors, and pro-B tumors in mice deficient for p53 plus Xrcc4, Ku70, Ku80, or DNAPKcs (24, 26, 28, 37), routinely show c-Myc activation via complicons that involve IgH. Yet, selection for c-Myc activation is not specific to pro-B cell tumors, because N-myc is frequently amplified in Artemis/p53-deficient pro-B cell lymphomas (33). Our current findings establish that the N-myc coding region, when inserted in place of that of c-Myc, indeed substitutes for c-Myc as a target of oncogenic translocations in LP pro-B cell lymphomas. Therefore, we conclude that features of the c-Myc locus may mechanistically contribute to preferential appearance of this locus in complicons of LP mice. Our findings also raise the possibility that mechanistic aspects of the translocation process (e.g., breaks or proximity) might influence tissue and stage specificity of other oncogene translocations in other cell types, developmental stages, or genetic backgrounds.
Several features of a given locus could affect the frequency of translocations and associated amplifications. First, DSBs in both participating chromosomal regions are required. In the case of LP pro-B lymphomas complicons, breaks in the IgH locus are introduced by the RAG endonuclease (28). However, the mechanism by which DSBs are generated in c-Myc is unknown. While canonical RAG targets were not obvious in the downstream c-Myc regions where the LP pro-B cell lymphoma translocation breakpoints reside, it has been suggested that RAG may generate nicks, and as a result DSBs, in certain general sequence structures (38). Whatever the origin of the DSBs, sequences and/or transacting factors that could promote the occurrence of and increase the frequency of DSBs around the c-Myc gene relative to N-myc could influence the choice of c-Myc as a preferred translocation partner. Once broken, the 2 translocation target loci must either already lie in close proximity to each other or be brought into proximity to be joined. Therefore, the relative proximity of the broken c-Myc vs. N-myc loci to IgH in LP pro-B cell lymphoma progenitors could be another factor that results in preferential use of c-Myc as a translocation target. Repair pathways active in different cell types, and perhaps the types of ends available to them, might also influence translocation frequency and target sites. In the context of LP pro-B cell lymphomas, a microhomology-based alternative end-joining pathway mediates most translocations (28). In this regard, the finding that Artemis/p53 double-deficient mice frequently develop pro-B cell lymphomas with N-myc/IgH translocations and complicons might somehow reflect the generation of hairpin IgH targets, although it is also possible that the N-myc region is broken more frequently or lies more proximal to IgH in the Artemis-deficient background.
Materials and Methods
Generation of LPMycN/+ and LPMycN/N Mice.
NCR mice were generated previously (15) and crossed into Lig4/p53 germ-line heterozygous mice to obtain triple heterozygous animals. Triple heterozygous and Lig4 heterozygous/p53-null/MycN/+ or MycN/N mice were crossed to obtain the experimental cohorts.
FACS Analysis.
Single-cell suspensions from tumor masses and control organs were stained with CyChrome (CyC)-labeled anti-mouse B220 (eBiosciences), FITC-labeled anti-mouse CD43 (BD Biosciences), and RPE-labeled anti-mouse IgM (Southern Biotech) antibodies or with CyC-labeled anti-mouse CD3e, FITC-labeled anti-mouse CD8, and RPE-labeled anti-mouse CD4 antibodies (all from BD Biosciences).
Data acquisition was performed on a FACScalibur flow cytometer equipped with CellQuest software (Becton Dickinson). Analysis was performed with FlowJo software (Tree Star).
Southern Blotting.
Genomic DNA isolated from tumor masses or normal control tissues was separated on a 0.8% agarose gel and transferred to a Zeta-Probe GT (Bio-Rad) nylon membrane. Hybridization was performed in 50% formamide/SSCPE at 42 °C. The JH4–3 probe was a 1.6-kb HindIII/EcoEI fragment downstream of JH4; the Myc3′ probe was a 1.4-kb genomic fragment (XhoI-KpnI) that contains part of exon 3 and the 3′-untranslated region of c-Myc; as loading control a probe hybridizing to exon 5 of the MDC1 gene was used.
Metaphase Preparation, SKY, and FISH.
Tumor cell suspensions were cultured overnight and colcemid (KaryoMAX Colcemid Solution; GIBCO) was added at the final concentration of 50 ng/ml for 3–5 h. Metaphase spreads were prepared and FISH experiments performed according to standard protocols (39). Sequential hybridization of slides was performed by dehydration through serial ethanols and reprobing. The following BACs were used as probes for FISH: BAC199, covering the 3′ region of the IgH locus encompassing the 3′ IgH enhancer and 100 kb downstream (3′ IgH BAC); BAC207, upstream of the IgH VH region (5′ IgH BAC); and BAC RP23–307D14, containing the c-Myc locus (c-Myc BAC). Whole-chromosome paint specific for mouse chromosomes 12 and 15 was used according to the manufacturer's instructions (Applied Spectral Imaging). Spectral karyotyping was performed with a mouse SKY paint kit (Applied Spectral Imaging), following manufacturer's indications. Images were acquired with a BX61 Microscope (Olympus) equipped with a motorized automatic stage, a cooled-CCD camera, and an interferometer (Applied Spectral Imaging). A 63× objective was used. Analysis was performed with HiSKY and ScanView software (Applied Spectral Imaging). At least 15 metaphases per each sample were analyzed.
Acknowledgments.
This work was supported by National Institutes of Health Grant 5P01 CA92625 (to F.W.A.). M.G. is a Leukemia and Lymphoma Society senior fellow. S.R. was supported by a Genentech/IDEC Fellowship from the American Cancer Society and the Dana Farber Cancer Institute Postdoctoral Training Program in Cancer Immunology. F.W.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 2.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 3.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 4.Strieder V, Lutz W. Regulation of N-myc expression in development and disease. Cancer Lett. 2002;180:107–119. doi: 10.1016/s0304-3835(02)00020-4. [DOI] [PubMed] [Google Scholar]
- 5.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Dang CV, et al. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- 7.Zimmerman K, Legouy E, Stewart V, Depinho R, Alt FW. Differential regulation of the N-myc gene in transfected cells and transgenic mice. Mol Cell Biol. 1990;10:2096–2103. doi: 10.1128/mcb.10.5.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downs KM, Martin GR, Bishop JM. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 1989;3:860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- 9.Zimmerman KA, et al. Differential expression of myc family genes during murine development. Nature. 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- 10.Xu L, Morgenbesser SD, DePinho RA. Complex transcriptional regulation of myc family gene expression in the developing mouse brain and liver. Mol Cell Biol. 1991;11:6007–6015. doi: 10.1128/mcb.11.12.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mugrauer G, Alt FW, Ekblom P. N-myc proto-oncogene expression during organogenesis in the developing mouse as revealed by in situ hybridization. J Cell Biol. 1988;107:1325–1335. doi: 10.1083/jcb.107.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nesbit CE, Grove LE, Yin X, Prochownik EV. Differential apoptotic behaviors of c-myc, N-myc, and L-myc oncoproteins. Cell Growth Differ. 1998;9:731–741. [PubMed] [Google Scholar]
- 13.Barrett J, Birrer MJ, Kato GJ, Dosaka-Akita H, Dang CV. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol Cell Biol. 1992;12:3130–3137. doi: 10.1128/mcb.12.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aubry S, Charron J. N-Myc shares cellular functions wiht c-Myc. DNA Cell Biol. 2000;19:353–364. doi: 10.1089/10445490050043326. [DOI] [PubMed] [Google Scholar]
- 15.Malynn BA, et al. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 16.Schwab M. MYCN in neuronal tumours. Cancer Lett. 2004;204:179–187. doi: 10.1016/S0304-3835(03)00454-3. [DOI] [PubMed] [Google Scholar]
- 17.Kuppers R. Mechanism of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 18.Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- 19.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 20.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 22.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 23.Ramiro AR, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Gao Y, et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 25.Frank KM, et al. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 26.Difilippantonio MJ, et al. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J Exp Med. 2002;196:469–480. doi: 10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Difilippantonio MJ, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu C, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 29.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 30.Rosenbaum H, Webb E, Adams JM, Cory S, Harris AW. N-myc transgene promotes B lymphoid proliferation, elicits lymphomas and reveals cross-regulation with c-myc. EMBO J. 1989;8:749–755. doi: 10.1002/j.1460-2075.1989.tb03435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dildrop R, et al. IgH enhancer-mediated deregulation of N-myc gene expression in transgenic mice: generation of lymphoid neoplasias that lack c-myc expression. EMBO J. 1989;8:1121–1128. doi: 10.1002/j.1460-2075.1989.tb03482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheppard RD, Samant SA, Rosenberg M, Silver LM, Cole MD. Transgenic N-myc mouse model for indolent B cell lymphoma: tumor characterization and analysis of genetic alterations in spontaneous and retrovirally accelerated tumors. Oncogene. 1998;17:2073–2085. doi: 10.1038/sj.onc.1202125. [DOI] [PubMed] [Google Scholar]
- 33.Rooney S, et al. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc Natl Acad Sci USA. 2004;101:2410–2415. doi: 10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank KM, et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 35.Luscher B. Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene. 2001;277:1–14. doi: 10.1016/s0378-1119(01)00697-7. [DOI] [PubMed] [Google Scholar]
- 36.Yan CT, et al. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc Natl Acad Sci USA. 2006;103:7378–7383. doi: 10.1073/pnas.0601938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gladdy RA, et al. The RAG-1/2 endonuclease causes genomic instability and controls CNS complications of lymphoblastic leukemia in p53/Prkdc-deficient mice. Cancer Cell. 2003;3:37–50. doi: 10.1016/s1535-6108(02)00236-2. [DOI] [PubMed] [Google Scholar]
- 38.Raghavan SC, Gu J, Swanson PC, Lieber MR. The structure-specific nicking of small heteroduplexes by the RAG complex: implications for lymphoid chromosomal translocations. DNA Repair (Amst) 2007;6:751–759. doi: 10.1016/j.dnarep.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franco S, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]