Abstract
In vertebrates the development and function of the nervous system is regulated by neurotrophic factors (NTFs). Despite extensive searches no neurotrophic factors have been found in invertebrates. However, cell ablation studies in Drosophila suggest trophic interaction between neurons and glia. Here we report the invertebrate neurotrophic factor in Drosophila, DmMANF, homologous to mammalian MANF and CDNF. DmMANF is expressed in glia and essential for maintenance of dopamine positive neurites and dopamine levels. The abolishment of both maternal and zygotic DmMANF leads to the degeneration of axonal bundles in the embryonic central nervous system and subsequent nonapoptotic cell death. The rescue experiments confirm DmMANF as a functional ortholog of the human MANF gene thus opening the window for comparative studies of this protein family with potential for the treatment of Parkinson's disease.
Keywords: development, dopamine, Drosophila, glia, neurite
Neurotrophic factors (NTFs) are small secretory proteins that by binding to their cognate receptors regulate the development, maturation, and survival of neurons (1). In particular, NTFs control the number of neurons, neurite branching, synaptogenesis, adult synaptic plasticity, and maturation of neuronal phenotype. NTFs also have prominent functions outside the nervous system and they have significant therapeutic potential for the treatment of various chronic neurological disorders (2).
So far no NTFs have been found in nematodes and arthropods and the absence has been explained by fundamental differences in development and maintenance of vertebrate and insect nervous systems. Nevertheless, an increasing amount of sequencing data from different genomes has revealed that at least at sequence level neurotrophins and their receptor homologs exist in primitive deuterosomes, like sea urchin and sea squirts (3). In protostomes neurotrophin receptor homologs also exist such as LTrk in molluscans (4). Interestingly enough, Drosophila has an orphan receptor homologous to the mammalian NTF glial cell-derived neurotrophic factor (GDNF) receptor Ret (5), but no homologs to GDNF family ligands (6). Recently, NTF family homologous to neurotrophin's cystein-knot motif, were found in Drosophila. However, their receptor(s) remains unidentified (7).
During development neurons and glia are overproduced in both vertebrates and invertebrates. In vertebrates the excess of neurons is removed during target innervation by programmed cell death (PCD) controlled by NTFs. In Drosophila there are 2 ways to regulate the cell numbers in early neural development. One is the cell autonomous control during neurogenesis just after the cell fate determination (8–11). The other, the cell nonautonomous control is less documented. In Drosophila the embryonic glial survival is determined by their interactions with neurons and epidermal growth factor receptor ligands secreted by neurons (12, 13). Importantly, there is also evidence for neurotrophic support from glia (14, 15). However, the proteins underlying the neurotrophic support in Drosophila have remained elusive.
The mammalian mesencephalic astrocyte-derived neurotrophic factor (MANF, also known as Armet) selectively promotes the survival of DA neurons in vitro (16). We have recently discovered a paralogous gene for MANF in vertebrate genomes, conserved dopamine neurotrophic factor (CDNF). In the rat 6-OHDA lesion model of Parkinson's disease, CDNF protects and repairs the nigrostriatal DA system (17). In vitro cell culture studies indicate MANF being upregulated in unfolded protein response (UPR) and inhibiting endoplasmic reticulum (ER) stress-induced cell death (18). MANF and CDNF form the first family of NTFs with well-conserved protein sequences among multicellular organisms from Caenorhabditis elegans to human. Here we report the homologous gene in Drosophila—DmMANF.
We demonstrate that DmMANF is required at the end of Drosophila embryogenesis for the maturation of the nervous system. Analysis of DmMANF maternal and zygotic null mutants revealed a total loss of dopaminergic neurites and drastic reduction in dopamine levels followed by degeneration of axonal bundles and subsequent nonapoptotic cell death. In larval zygotic mutants before their death specific and significant reduction of dopaminergic neurites occurs. These results suggest an evolutionarily conserved role for NTFs. Finally, we prove that human MANF is the ortholog of the Drosophila DmMANF gene.
Results
DmMANF Is an Evolutionarily Conserved Secreted Protein.
The amino acid sequence identity between Drosophila and the vertebrate MANF family is on average 50% (Fig. 1A and supporting information (SI) Fig. S1, Table S1). Corresponding amino acid sequences of several insect species are more related to mammalian MANFs (over 50% identities) than to the CDNF sequence (47% identity on average) (Table S1). In all proteins of the MANF family the spacing of all of the 8 cysteines is strictly conserved (Fig. 1 A and B), indicating a significant structural similarity. According to Drosophila genome annotation, the fly homolog to human MANF and CDNF is CG7013 (Fig. 1C), also known as arginine-rich protein-like (ARP-like). We suggest here the name DmMANF for CG7013 because it lacks the amino-terminal arginine tract originally described for human Armet. DmMANF is a secreted protein, as it was found in the medium of Schneider-2 cells transfected with cDNA construct of DmMANF (Fig. 1D). To study whether it also holds true in vivo, we took advantage of somatic mutant mosaic null clones in large ovarian follicular cells expressing DmMANF. Indeed, DmMANF is secreted by large ovarian follicle cells to neighboring null clones lacking all endogenous DmMANF production (Fig. 1E).
Fig. 1.
DmMANF is an evolutionarily conserved secreted protein. (A) The alignment of amino acid sequences of MANF homologs among 6 species: D.m., fruit fly; D.p., North American fruit fly; A.m., honeybee; H.s., human; M.m., mouse; D.r., zebrafish. Signal peptides in rectangular boxes, 8 conserved cysteines in yellow (arrowheads). Identical amino acids are colored. (B) Scheme of MANF and CDNF proteins. Bars indicate conserved cysteines. Numbers of amino acids are shown. Pre indicates signal sequence cleaved off before secretion. (C) Genomic organization of DmMANF gene and P-element insertions (triangles). Arrows show direction of transcription. After excision of the P-element KG03250 deletions DmMANFΔ96 and DmMANFΔ112 were obtained. Exons in black, untranslated regions in white. (D) Western analysis of the medium and cell lysates of Schneider-2 cells transfected with DmManf cDNA construct. (E) DmMANF is secreted from DmMANF positive ovarian follicle cells to neighboring mosaic somatic DmMANFΔ96 null clones. Green, anti-DmMANF; dashed line marks the border of an egg chamber; arrows indicate DmMANF positive dots located on the predicted DmMANFΔ96 cell borders. Two-dimensional confocal image of 0.972-μm thick area.
Analysis of DmMANF Expression.
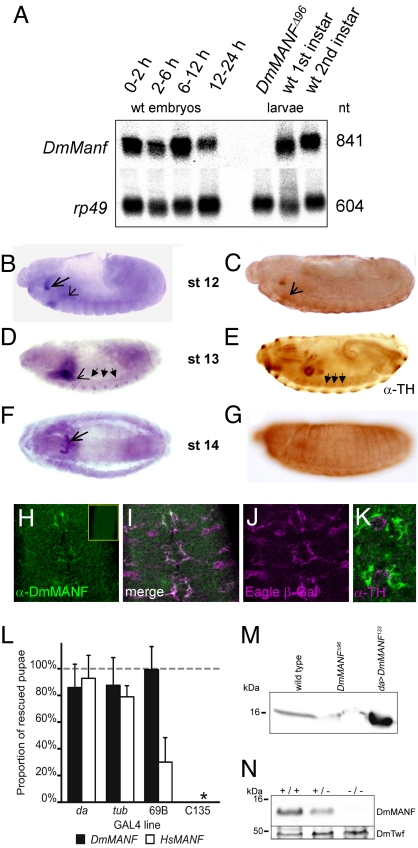
During Drosophila development DmManf mRNA was detectable at all developmental stages (Fig. 2A and Fig. S2). A high level of DmManf mRNA was already present in embryos less than 2 hours old, indicating a strong maternal contribution. DmManf transcription was detected at stage 12 in the stomodeum and the salivary gland primordia and weakly in the mesoderm (Fig. 2B). From stage 13 onward, DmManf mRNA localized in garland cells, the outer wall of the proventriculus, and the salivary glands (Fig. 2 D and F). Simultaneously, the punctuate expression in the ventral nerve cord (VNC) appeared (Fig. 2D). In accordance with mRNA localization, DmMANF antibody (Fig. 2N) recognized the same tissues (Fig. 2C) and in addition at stage 15 the epidermis (Fig. 2G). Interestingly, the expression patterns of DmManf mRNA and tyrosine hydroxylase (TH, the rate limiting enzyme for dopamine synthesis) protein resembled each other at stage 13 embryos: TH is expressed at the inner wall of the proventriculus, in VNC, and in the epidermis as well (Fig. 2 D and E), as insects use dopamine-derived quinones to build their cuticle (19). We took a closer look at DmMANF VNC expression and found DmMANF in eagle-positive cell body glia (20) (Fig. 2 H–J), of which the medialmost cell body glia also surround TH-positive midline neurons (Fig. 2K). Weaker expression of DmMANF was detected in longitudinal glia positive for transcription factor Prospero and in Engrailed-positive channel glia (Fig. S3 D–F). During embryogenesis DmMANF displayed no colocalization with any general or specific neuronal markers (Elav, TH) (Fig. S3 and Fig. 2K). Taken together, these data suggest that DmMANF is expressed in glial cells comparable to mammalian astrocytes (21) nearby dopaminergic neurons i.e., in the predicted localization for a putative NTF in Drosophila.
Fig. 2.
During development DmMANF is expressed in nonneuronal tissues and embryonic VNC cell body glia around DA neurons. (A) Northern analysis of staged embryos and larvae. mRNA from wild-type staged embryos (Left, various hours after egg laying) and from larvae (Right). DmMANFΔ96 larvae were late first instars. rp49 probe is loading control. (B–G) Expression analysis of wild-type embryos: in situ mRNA hybridization (B, D, and F), immunohistochemistry with anti-DmMANF (C and G), and anti-TH (E). The short arrows show salivary gland primordia and salivary glands, open arrow points to garland cell expression, and full arrows to VNC expression. (H–J) Colocalization of anti-DmMANF (green) with Eagle (magenta) at stage 16 embryonic VNC of eagle-LacZ/+. Three thoracic and the first abdominal segments are shown. Anti-DmMANF (H), anti-β-Gal merged with anti-DmMANF (I), and anti-β-Gal (J) staining. In DmMANFΔ96mz VNC no DmMANF is detected (H, box). (Scale bar, 15 μm.) (K) Ventral view of VNC at embryonic stage 16. Anti-TH (magenta) in the ventral midline DA neurons; DmMANF-positive glia (green) surround the DA neurons. (L) DmMANFΔ96 larval lethality is rescued by ectopic DmMANF and HsMANF expression. GAL4 driver lines used: ubiquitously expressed daughterless (da) and tubulin (tub), 69B with epidermal and CNS expression, and C135 in proventriculus. The proportion of rescued pupae relative to all pupae is presented; dotted line indicates the maximum expected value of complete rescue estimated by Mendelian inheritance. *, not determined. Mean ± SD. (M) Western analysis of larval extracts show DmMANF expression in 3 genotypes. In first instar DmMANFΔ96 larvae the remnants of maternally contributed DmMANF persist. da>DmMANF133-ubiquitous DmMANF overexpression under daughterless promoter. (N) Western analysis of embryonic extracts shows DmMANF expression confirming the lack of DmMANF protein in DmMANFΔ96mz (−/−) and the specificity of the antibody. Twinfilin (DmTwf) serves as loading control.
Generation and Verification of DmMANF Mutant Alleles.
A P-element insertion P{SUPor-P}KG03250 (22), hereafter referred to as KG03250, is located 157 bp upstream from DmMANF (CG7013) 5′UTR (Fig. 1C). DmMANF expression remains unaltered in this homozygous viable line (Fig. S2). Expression of the upstream neighboring gene CG14879 was partially abolished in KG03250 (Fig. S2). However, the line KG03250 is fully homozygous viable. We used KG03250 in a P-element mobilization screen to create deletions removing regions of the DmMANF gene. Two of the obtained lines were verified by sequencing—Δ96 was deficient in 278 bp of the DmMANF ORF lacking the first 2 exons and part of the third exon, and Δ112 missed 15 bp of 5′UTR of DmMANF (Fig. 1C). Both mutant lines were larval lethal with similar observations—homozygotes died as late first instar larvae or immediately after the delayed first molt with defects in the larval nervous system. First these larvae atypically wander away from the food, then move more slowly, and finally freeze, immobilized but still responding to touch. The embryonic neural development of DmMANFΔ96 mutants proceeded normally as verified by neuronal markers Fasciclin II (Fas II), Futsch, and BP102 (Fig. 4B and data not shown) probably because DmMANF gene products were maternally contributed (Fig. 2 A and M). In first instar DmMANFΔ96 mutant larvae, traces of maternal DmMANF protein are still present (Fig. 2M). No DmManf mRNA was detected in DmMANFΔ96 larvae before their death as late first instars (Fig. 2A), neither was any DmMANF protein present in embryos lacking both maternal and zygotic product—DmMANFΔ96mz (Fig. 2N). We verified that DmMANFΔ96 mutant lethality was solely the result of the DmMANF deletion because ubiquitous and ectopic expression of DmMANF in the mutant DmMANFΔ96 background resulted in complete larval rescue into pupal stage (Fig. 2L). Furthermore, by using 69B-GAL4 driving the expression in epidermis and CNS (23, and our observations) we were able to maintain the DmMANFΔ96 mutant flies by DmMANF ectopic expression only, further verifying the lethality was caused by deletion of DmMANF and proving the importance of DmMANF expression in both the epidermis and CNS.
Fig. 4.
Mutants lacking maternal and zygotic DmMANF show severely aberrant neuronal phenotype, decomposition of neuropile, and cell death. A–D, mAb BP102 recognizing CNS and VNC neurite bundles. (A) Wild type. (B) DmMANFΔ96. (C–D) DmMANFΔ96mz at stage 16 (C) and stage 17 (D). (E–L) Ultrastructural analysis of DmMANFΔ96mz. Transverse images taken from the VNC thorax and abdominal border area. (E) Localization of TEM images (F–J) shown in boxes on low magnification of transverse section of VNC. Np, neuropile; ns, neuronal somae; dotted line marks the borders of neuropile and VNC. (F–H) Same area of VNC. (F and I) Wild type, stage 17. (H) DmMANFΔ96mz, late stage 16. (G and J) Late stage 17 DmMANFΔ96mz shows cell death and decomposition of neuropile (open arrows). Closed arrows point to glia, open arrows to nonapoptotic cell death. [Scale bar, 1 μm (F–I); 2 μm (J).] (K and L) Ultrastructure of the cuticle of stage 17 embryos. (K) Wild type. (L) DmMANFΔ96mz, all cuticular layers are disorganized. (Scale bars, 200 nm.)
DmMANF Mutants Display Diminished Volumes of Dopamine Positive Neurites and Lowered Dopamine Levels.
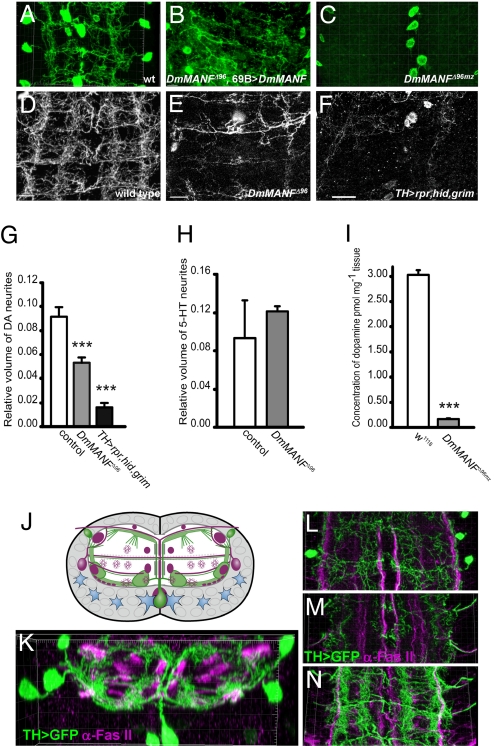
We generated embryos lacking both maternal and zygotic DmMANF (24). In wild-type embryos, the expression of TH in neuronal somae is first detectable at late stage 16 and before hatching neurites also become TH-positive (Fig. 3A). We visualized the neurites of DA neurons by membrane-targeted GFP construct expressed under TH promoter (25) (TH>mCD8-GFP) (Fig. 3 A, C–F, and K–N). In DmMANFΔ96mz mutants the neuronal somae expressed TH as well but almost no TH-positive neurites were detected (Fig. 3C). The absence of TH-positive neurites and embryonic lethality was completely rescued by ectopic overexpression of DmMANF under 69B-GAL4 driver in DmMANFΔ96 background (Fig. 3B), supporting the notion that the phenotype is caused by the lack of DmMANF only. The number of glial cells in DmMANFΔ96mz VNC remained unaltered (Fig. S4).
Fig. 3.
DmMANF mutants show diminished volume of DA neurites and reduced dopamine level. (A–C) Longitudinal views of DA neurites of late stage 17 VNC, maximal projections of 2 thoracic and abdominal segments visualized by TH>mCD8-GFP (A and C), and with anti-TH (B). (A) Wild type. (B) Rescue of DmMANFΔ96mz by 69B>DmMANF. (C) DmMANFΔ96mz. (D–F) Longitudinal views of DA neurites of late first instar larval VNC, maximal projections of 2 thoracic and 2 abdominal segments. (D) Wild-type TH>mCD8-GFP with 4 longitudinal ventral bundles of DA neurites. (E) DmMANFΔ96 mutant larvae before death: TH>mCD8-GFP positive neurites show clear degeneration, especially in abdominal region. (F) Larvae with proapoptotic proteins Rpr, Hid, and Grim targeted to DA neurons show severe loss of TH>mCD8-GFP neurites with several DA neuronal somae still visible. [Scale bar, 10 μm (D and E); 20 μm (F).] (G) Volume quantification of DA neurites. In y axis relative volume of neurites to total volume of the neuropile. Analyzed genotypes: wild type (n = 17), DmMANFΔ96, TH>mCD8-GFP (n = 18), and TH>mCD8-GFP>rpr;hid;grim (n = 5). Mean ± SEM. ***, P < 0.001 versus wild type, Student's t-test. (H) Quantification of serotonergic (5-HT) neurite volumes shows no statistical difference between DmMANFΔ96 (n = 3) and wild type (n = 5). (I) DmMANFΔ96mz mutants show significantly lowered levels of dopamine compared to wild type of the late stage 17. Interaction bar plot for dopamine pmol mg−1; ***, P < 0.001 versus wild type, Student's t-test. (J–N) VNC of TH>mCD8-GFP late first instar larvae, anti-Fas II (magenta). (J) Scheme of transversal view represents neuropile (white), cell body glia (blue), Fas II landmarks (magenta), and DA neurites and bundles (green). (K) Transversal view of 3D image, 2 abdominal segments. Dorsal (L), mid- (M), and ventral projections (N) of Fas II and DA neurites.
Next we studied the fate of DA neurons in DmMANFΔ96 mutant larvae with maternally contributed DmMANF. The localization of the DA neuronal somae has been characterized in third instar larvae (26). We observed all predicted DA neuronal somae present in DmMANFΔ96 late first instar larvae (data not shown). In wild-type larvae DA neurites assemble a complex massive network all over the tightly packed neurites termed the neuropile (Fig. 3 D, J, and K). To further map the location of DA axons we used Fas II landmarks (27) (Fig. 3 J–N). The ventral unpaired midline DA interneurons send 1 axon toward the neuropile, which bifurcates and turns in a longitudinal direction (28). There are 2 main longitudinal DA bundles alongside the Fas II-positive ventral fascicles (Fig. 3 J, K, and N). Dorsolaterally located TH-positive neurons send axons to lateral parts of the neuropile, which branch extensively (Fig. 3 J–N). In cross section, the DA neurites form a lemniscate shape around the neuropile (Fig. 3 J and K).
We observed that in DmMANFΔ96 mutants the volume of DA neurites was significantly diminished (Fig. 3 E and G) whereas the neuronal somae persisted. The neuritic degeneration appears in the abdominal area of VNC mostly. We noticed similar degeneration of DA neurites when apoptosis was induced in the DA neurons by activation of proapoptotic genes reaper, hid, and grim (SMAC/Diablo homologs in Drosophila) (Fig. 3F). Surprisingly, despite the massive loss of GFP-positive neurites, several DA neuronal somae were still visible in these larvae (Fig. 3 F and G) dying as late first instars at the same time window as DmMANFΔ96. In DmMANFΔ96 VNC there were no significant changes in neurite volume of serotonergic neurons (Fig. 3H) or in the volume of motoneuron subpopulation (Fig. S5), verifying that neurite loss occurred specifically in the DA neurons.
Because we observed massive degeneration of DA axons resembling the pathology in Parkinson's disease we asked whether the dopamine levels are also altered in the mutant flies. We measured DA content of DmMANFΔ96mz and wild-type embryos of exactly the same stage 17. Indeed, the dopamine levels of DmMANFΔ96mz embryos were dramatically reduced (Fig. 3I).
These results together with the DmMANF localization in the cell body glia surrounding the embryonic DA neurons (Fig. 2K) raise the possibility that glial-derived DmMANF in addition to neuritogenic effect have also survival-promoting trophic function for these neurons.
Embryos Lacking Both Maternal and Zygotic DmMANF Result in Cuticular Defects, Axonal Degradation, and Cell Death.
DmMANFΔ96mz embryos developed normally until late stage 16 (Fig. 4C) but thereafter obtained severe abnormalities (Fig. 4D). From stage 16 onward the VNC overcondensed (Fig. 4D) compared to the wild type (Fig. 4A). To determine the impact of apoptosis, we quantified cells positive for cleaved Caspase-3 in DmMANFΔ96mz VNC at late stage 16 before the appearance of abnormalities. In comparison to the wild type there was no significant increase in the number of apoptotic cells (Fig. S6).
We further analyzed the ultrastructure of DmMANFΔ96mz VNC. At early stage 16 the mutant neuropile still resembled that of the wild type (Fig. 4 F and H). However, by late stage 17 we detected signs of axonal degeneration as evidenced by the degradation of axonal membranes starting at the border of glia and neuropile (Fig. 4G). In addition, cells defined by their location as cell body glia, looked highly electron dense (Fig. 4 I and J) and contained remnants of cell debris (Fig. 4I). In the mutant VNC in the vicinity of the neuropile we detected dying cells with poor and bleached cytoplasm, swollen nuclei, and with dilated ER and other organelles (Fig. 4 J and G), atypical for the conventional apoptosis. Most probably these cells represent neurons by their location. Thus, the absence of DmMANF caused nonapoptotic cell death of Drosophila neurons.
We also noticed that the cuticle of DmMANFΔ96mz mutants was clearly defective as it remained penetrable for antibodies. After the secretion (stage 16) and assembly (stage 17) of the wild-type cuticle it functions as a nonpermeable barrier for antibodies (29). TEM analysis revealed that indeed all layers of the DmMANFΔ96mz mutant cuticle were disorganized (Fig. 4 K and L). We hypothesize here that this cuticular disorganization could be a consequence of extremely low dopamine levels needed for the synthesis of cuticle crosslinkers—quinones. In conclusion the embryonic lethality of DmMANFΔ96mz mutants is the result of the severe cuticle and CNS defects. DmMANF is required during the maturation of the embryonic nervous system for maintenance of neuronal and cuticular connectivity.
DmMANF Is the Ortholog to Human MANF.
Finally to investigate whether human MANF or CDNF is able to compensate for the function of DmMANF, we carried out rescue experiments with UAS-HsMANF and UAS-HsCDNF transgenic flies. Ubiquitous HsMANF expression was able to significantly rescue larval lethality of DmMANFΔ96 mutants (Fig. 2L) whereas HsCDNF gave no rescue despite the HsCDNF protein production in transgenic flies (Fig. S7). These results demonstrate that human MANF is the fly functional ortholog and that fly and human MANF share the yet unknown cognate receptor.
Discussion
In summary, we characterize the unique evolutionarily conserved NTF in invertebrates, DmMANF. Classically, the NTFs determine the number of neurons by supporting survival and antagonizing death. They also control neurite outgrowth and target innervation. CDNF and MANF support the survival of dopaminergic neurons in the rat models of neurotoxicity, preventing both neurite degeneration and neuronal death (17, M. Voutilainen and M. Saarma, unpublished results). However, whether these factors regulate the number of neurons during PCD is not known. DmMANF is clearly required in Drosophila for the maintenance of the DA neurites but not the neurites of serotonergic or the subpopulation of motoneurons. Surprisingly, despite the axonal degeneration in DmMANFΔ96 mutant larvae the somae of DA neurons persist. Moreover, some somae but not neurites of DA neurons persist even when their death was ectopically triggered by overexpression of the proapoptotic proteins. Thus, programmed death in the Drosophila DA neurons seems to follow a “dying-back” pattern where the neurites degenerate first followed by the death of somae (30). Whether DmMANF is a bona fide NTF promoting the survival of DA neurons remains, however, open as the mutant larvae die before it can be judged. However, in the VNC of DmMANFΔ96mz mutants we observed dying cells with nonapoptotic ultrastructure. The exact identity of those cells remains undetermined but their location close to ventral midline suggests they are midline DA neurons dying after the loss of neurites. By TEM analysis, the elimination of DmMANF causes cell death resembling caspase-independent cell death, characterized by swelling of organelles, and the appearance of “empty” spaces (31). All those characteristics including dilated and rounded ER, are observed in the DmMANFΔ96mz mutant VNC. Dilation of ER indicates ER stress and it has been recently shown that during ER stress MANF is upregulated (18). As DA neurons are highly susceptible to ER stress-induced cell death (32) it could possibly explain why these neurons are specifically altered in DmMANFΔ96mz mutants. Also in mutants deficient of both maternal and zygotic DmMANF glia contain cellular debris indicating activation of glial engulfing activity. During metamorphosis glia accumulate highly electron-dense material (33) when clearing axonal debris associated with neuronal remodeling (34, 35). Taken together, we conclude that DmMANF is the first invertebrate NTF required for maturation and maintenance of DA neurites from embryonic stage 16 onward at least to the second instar larval stage.
The ability of HsMANF to replace the function of DmMANF suggests that these NTFs should share common signal transduction mechanisms including receptors. This makes the Drosophila model very attractive to study the MANF and CDNF signaling pathways by using the powerful fruit fly genetics. As human MANF and CDNF represent potential drug targets for the treatment of Parkinson's disease, the usage of well-established Drosophila disease models (36, 37) could be extremely important.
Materials and Methods
Fly Strains and Antibodies.
The following fly strains were obtained and maintained at 25 °C: w1118, P{SUPor-P}KG03250 (22), UAS-mCD8-GFP, da-GAL4 (38), tub-GAL4 (39), CQ2>τ-LacZ (40), 69B-GAL4 (23) (from Bloomington); eagle-LacZ289 (21) (from J. Urban), TH-GAL4 (25) (from S. Birman), C135-GAL4 (41) (from L. Hrdlicka), UAS-reaper, hid, and grim (42) (from M. O'Connor). Subsequent antibodies were used: mAb BP102, anti-Repo, mAb 22C10 (anti-Futsch), anti-Elav, anti-Engrailed, mAb 1D4 (anti-Fas II), anti-Wrapper (from Developmental Studies Hybridoma Bank at the University of Iowa), anti-cleaved caspase-3 (Cell Signaling), mouse anti-TH (DiaSorin), rabbit anti-TH (from W. Neckameyer), rabbit anti-CDNF (from P. Lindholm), rabbit anti-β-galactosidase (Cappel), mouse anti-β-galactosidase (Sigma). Images were taken with an Olympus AX70 microscope equipped with an Olympus DP70 camera.
Generation and Purification of the DmMANF Antibody.
The cDNA corresponding to the ORF of DmMANF was cloned into a T7lac vector, expressed in Escherichia coli and purified as previously described (43, 44). Rabbits were immunized and the obtained antiserum was affinity purified (43). Anti-DmMANF antibody was tested in immunohistochemical staining on whole mount embryos as previously described (45); dilutions from 1:1000 to 1:5000 were later used. No DmMANF was detectable in DmMANF-deficient embryos.
Generation of Transgenic Flies.
The DmManf cDNA (LO06293 Berkeley Drosophila Research Center) EcoRI–XhoI, of HsManf XhoI–AsuII, and of HsCdnf XhoI–AsuII fragments (human cDNA subcloned to pMIB, gifts from P. Lindholm) were cloned into pUAST vector. The signal sequences of human MANF and CDNF proteins were replaced with honeybee mellitin signal sequence. Both human MANF and CDNF constructs contained 9 additional amino acids in their C′ terminus and human CDNF construct in addition to 6 amino acids in its N′ terminus. w− embryos were injected. Five genomic insertions for DmManf, 3 insertions for HsCdnf, and 1 for HsManf were recovered.
mRNA Isolation and Characterization by Northern Blot Analyses.
Total RNA was extracted from w1118 and KG03250 strains by RNeasy Mini Kit (Qiagene). mRNA was purified by magnetic particles (Dynabeads Oligo(dT)25, Dynal) according to the instructions of the manufacturer. Total RNA of 30 μg or mRNA of 1.5 μg were separated by electrophoresis on 1.2% formaldehyde agarose gel and capillary blotted to nylon membrane. cDNA used for probes were LO06293 (EcoRI–BglII fragment) and RE20991 (BamHI fragment), rp49 cDNA fragment served as loading control. Hybridizations and washes were carried out at 65 °C as previously described (46).
Immunohistochemistry and RNA in Situ Hybridization Analysis.
RNA in situ and antibody labeling were performed as described (45). Sense probe gave no hybridization (Fig. S8).
Schneider-2 Cell Assay.
cDNA of DmManf (LO06293) was cloned to pMT-V5-His expression vector (Invitrogen). Schneider-2 cells were transiently transfected with DmMANF-pMT according to manufacturer's protocol (Invitrogen). The cells were collected and washed with PBS before lysis. Western blotting was done according to manufacturer's instructions (Amersham Biosciences).
Confocal Microscopy and Image Analysis.
Confocal stacks were acquired in 0.15-μm steps along z axis by confocal laser scanning microscope TCS SP5 AOBS (Leica Microsystems) equipped with 63× HCX PL APO CS glycerol immersion objective (n.a. = 1.3) with 12-bit resolution. Image J (Wright Cell Imaging Facility) for 2D, and Imaris 5.7.1 (Bitplane Inc.) for 3D image analysis were used. For volume measurements by Imaris, isosurfaces were built and for each 3D image relative volume of specific neurites to total volume of the stack was calculated.
Transmission Electron Microscopy Analysis.
Embryos were prepared as previously described (47). After dehydration embryos were mounted in propylene oxide resin. Sections of 70 nm from the thorax and abdominal area of 6 embryos out of 2 separate fixations of each genotype were examined and images taken with Jeol 1200 EX II (Jeol Ltd.) equipped with Gatan Erlangshen ES5000 W, model 782 CCD-camera (Gatan Inc.).
Analysis of Dopamine Concentration.
Approximately 100 embryos were pooled in samples. The samples were homogenized with an ultrasonic processor and analyzed with HPLC using a Spherisorb ODS2 3 μm, 4.6 × 100 mm; column (Waters). Dopamine was quantified using a 12-channel ESA CoulArray Electrode Array Detector system and CoulArray for Windows software (ESA Inc.) (48). In addition to exact retention time, dopamine was identified by its characteristic electrochemical properties.
Rescue Experiments.
Transgenic lines for UAS-DmMANF, UAS-HsMANF and UAS-HsCDNF were generated and recombined together with ubiquitous and specific GAL4 lines into mutant DmMANFΔ96 background. Crosses were carried out at 25 °C. For each experiment 5 independent crosses were made and transferred twice to fresh vials; progeny from 15 vials per each cross was counted.
Supplementary Material
Acknowledgments.
We are indebted to A. Prokop, M. Mende, J. Urban, G. Technau lab, and R. Cantera for advice and technical help. We are grateful for P. Lindholm for the human MANF and CDNF cDNA constructs, Bloomington Stock Center, S. Birman, A. Giangrande, A. Hidalgo, L. Hrdlicka, W. Neckameyer, M. O′Connor, J. Urban and A. Uv for sharing fly lines and reagents. We are grateful to K. Tanhuanpää and M. Molin from LM unit, E. Jokitalo and the EM unit of the Institute of Biotechnology for the help in LM and EM analysis, and T. Päivärinta for the help with the figures. We thank M. Palviainen and M. Vaha for excellent technical assistance. We thank M. S. Airaksinen, J.-O. Andressoo, U. Arumäe, S. Butcher, K. Kaila, P. Lindholm, J. Partanen, C. Samakovlis, and O. Shimmi for critical comments on the manuscript. We acknowledge the support from the Sigrid Jusélius Foundation, the Academy of Finland Neuroscience Program, and the Michael J. Fox Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810996106/DCSupplemental.
References
- 1.Loughlin SE, Fallon JH. Neurotrophic Factors. San Diego: Academic; 1993. p. 599. [Google Scholar]
- 2.Evans JR, Barker RA. Neurotrophic factors as a therapeutic target for Parkinson's disease. Expert Opin Ther Targets. 2008;12:437–447. doi: 10.1517/14728222.12.4.437. [DOI] [PubMed] [Google Scholar]
- 3.Bothwell M. Evolution of the neurotrophin signaling system in invertebrates. Brain Behav Evol. 2006;68:124–132. doi: 10.1159/000094082. [DOI] [PubMed] [Google Scholar]
- 4.Benito-Gutierrez E, Garcia-Fernandez J, Comella JX. Origin and evolution of the Trk family of neurotrophic receptors. Mol Cell Neurosci. 2006;31:179–192. doi: 10.1016/j.mcn.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Sugaya R, Ishimaru S, Hosoya T, Saigo K, Emori Y. A Drosophila homolog of human proto-oncogene ret transiently expressed in embryonic neuronal precursor cells including neuroblasts and CNS cells. Mech Dev. 1994;45:139–145. doi: 10.1016/0925-4773(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 6.Airaksinen MS, Holm L, Hätinen T. Evolution of the GDNF family ligands and receptors. Brain Behav Evol. 2006;68:181–190. doi: 10.1159/000094087. [DOI] [PubMed] [Google Scholar]
- 7.Zhu B, et al. Drosophila neurotrophins reveal a common mechanism for nervous system formation. PLoS Biol. 2008;6:e284. doi: 10.1371/journal.pbio.0060284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prokop A, Bray S, Harrison E, Technau GM. Homeotic regulation of segment-specific differences in neuroblast numbers and proliferation in the Drosophila central nervous system. Mech Dev. 1998;74:99–110. doi: 10.1016/s0925-4773(98)00068-9. [DOI] [PubMed] [Google Scholar]
- 9.Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–219. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- 10.Lundell MJ, Lee HK, Perez E, Chadwell L. The regulation of apoptosis by Numb/Notch signaling in the serotonin lineage of. Drosophila. Development. 2003;130:4109–4121. doi: 10.1242/dev.00593. [DOI] [PubMed] [Google Scholar]
- 11.Miguel-Aliaga I, Thor S. Segment-specific prevention of pioneer neuron apoptosis by cell-autonomous, postmitotic Hox gene activity. Development. 2004;131:6093–6105. doi: 10.1242/dev.01521. [DOI] [PubMed] [Google Scholar]
- 12.Bergmann A, Tugentman M, Shilo BZ, Steller H. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev Cell. 2002;2:159–170. doi: 10.1016/s1534-5807(02)00116-8. [DOI] [PubMed] [Google Scholar]
- 13.Hidalgo A, Kinrade EF, Georgiou M. The Drosophila neuregulin vein maintains glial survival during axon guidance in the CNS. Dev Cell. 2001;1:679–690. doi: 10.1016/s1534-5807(01)00074-0. [DOI] [PubMed] [Google Scholar]
- 14.Sepp KJ, Auld VJ. Reciprocal interactions between neurons and glia are required for Drosophila peripheral nervous system development. J Neurosci. 2003;23:8221–8230. doi: 10.1523/JNEUROSCI.23-23-08221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Booth GE, Kinrade EF, Hidalgo A. Glia maintain follower neuron survival during Drosophila CNS development. Development. 2000;127:237–244. doi: 10.1242/dev.127.2.237. [DOI] [PubMed] [Google Scholar]
- 16.Petrova P, et al. MANF: a new mesencephalic, astrocyte-derived neurotrophic factor with selectivity for dopaminergic neurons. J Mol Neurosci. 2003;20:173–188. doi: 10.1385/jmn:20:2:173. [DOI] [PubMed] [Google Scholar]
- 17.Lindholm P, et al. Novel neurotrophic factor CDNF protects and rescues midbrain dopaminergic neurons. in vivo. Nature. 2007;448:73–77. doi: 10.1038/nature05957. [DOI] [PubMed] [Google Scholar]
- 18.Apostolou A, Shen Y, Liang Y, Luo J, Fang S. Armet, a UPR-upregulated protein, inhibits cell proliferation and ER stress-induced cell death. Exp Cell Res. 2008;314:2454–2467. doi: 10.1016/j.yexcr.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright TR. The genetics of biogenic amine metabolism, sclerotization, and melanization in. Drosophila melanogaster. Adv Genet. 1987;24:127–222. [PubMed] [Google Scholar]
- 20.Urban J, Technau GM. Cell lineage and cell fate specification in the embryonic CNS of Drosophila. Semin Cell Dev Biol. 1997;8:391–400. doi: 10.1006/scdb.1997.0163. [DOI] [PubMed] [Google Scholar]
- 21.Ito K, Urban J, Technau GM. Distribution, classification and development of Drosophila glial cells during late embryogenesis. Rouxs Arch Dev Biol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- 22.Bellen HJ, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 24.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in. Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friggi-Grelin F, et al. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- 26.Budnik V, Martin-Morris L, White K. Perturbed pattern of catecholamine-containing neurons in mutant Drosophila deficient in the enzyme dopa decarboxylase. J Neurosci. 1986;6:3682–3691. doi: 10.1523/JNEUROSCI.06-12-03682.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landgraf M, Sanchez-Soriano N, Technau GM, Urban J, Prokop A. Charting the Drosophila neuropile: a strategy for the standardised characterisation of genetically amenable neurites. Dev Biol. 2003;260:207–225. doi: 10.1016/s0012-1606(03)00215-x. [DOI] [PubMed] [Google Scholar]
- 28.Budnik V, White K. Catecholamine-containing neurons in Drosophila melanogaster: Distribution and development. J Comp Neurol. 1988;268:400–413. doi: 10.1002/cne.902680309. [DOI] [PubMed] [Google Scholar]
- 29.Payre F. Genetic control of epidermis differentiation in Drosophila. Int J Dev Biol. 2004;48:207–215. doi: 10.1387/ijdb.15272387. [DOI] [PubMed] [Google Scholar]
- 30.Raff M, Whitmore A, Finn J. Axonal self-destruction and neurodegeneration. Science. 2002;296:868–871. doi: 10.1126/science.1068613. [DOI] [PubMed] [Google Scholar]
- 31.Clarke PG. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol. 1990:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- 32.Wang HQ, Takahashi R. Expanding insights on the involvement of endoplasmic reticulum stress in Parkinson's disease. Antioxid Redox Signal. 2007;9:553–561. doi: 10.1089/ars.2006.1524. [DOI] [PubMed] [Google Scholar]
- 33.Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol. 2004;14:678–684. doi: 10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 34.MacDonald JM, et al. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 35.Awasaki T, et al. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 36.Whitworth A, Wes P, Pallanck L. Drosophila models pioneer a new approach to drug discovery for Parkinson's disease. Drug Discov Today. 2006;11:119–126. doi: 10.1016/S1359-6446(05)03693-7. [DOI] [PubMed] [Google Scholar]
- 37.Coulom H, Birman S. Chronic exposure to rotenone models sporadic Parkinson's disease in. Drosophila melanogaster. J Neurosci. 2004;24:10993–10998. doi: 10.1523/JNEUROSCI.2993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of. Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 39.O'Donnell KH, Chen CT, Wensink PC. Insulating DNA directs ubiquitous transcription of the Drosophila melanogaster alpha 1-tubulin gene. Mol Cell Biol. 1994;14:6398–6408. doi: 10.1128/mcb.14.9.6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujioka M, et al. Even-skipped, acting as a repressor, regulates axonal projections in. Drosophila. Development. 2003;130:5385–5400. doi: 10.1242/dev.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hrdlicka L, et al. Analysis of twenty-four Gal4 lines in. Drosophila melanogaster. Genesis. 2002;4:51–57. doi: 10.1002/gene.10125. [DOI] [PubMed] [Google Scholar]
- 42.Zhou L, et al. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc Natl Acad Sci USA. 1997;94:5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peränen J, Furuhjelm J. Expression, purification, and properties of Rab8 function in actin cortical skeleton organization and polarized transport. Methods Enzymol. 2001;329:188–196. doi: 10.1016/s0076-6879(01)29079-x. [DOI] [PubMed] [Google Scholar]
- 44.Peränen J, Rikkonen M, Hyvönen M, Kääriäinen L. T7 vectors with modified T7lac promoter for expression of proteins in. Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- 45.Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- 46.Palgi J, Stumpf E, Otonkoski T. Transcription factor expression and hormone production in pancreatic AR42J cells. Mol Cell Endocrinol. 2000;165:41–49. doi: 10.1016/s0303-7207(00)00265-3. [DOI] [PubMed] [Google Scholar]
- 47.Budnik V, Gorczyca M, Prokop A. Selected methods for the anatomical study of Drosophila embryonic and larval neuromuscular junctions. Int Rev Neurobiol. 2006;75:323–365. doi: 10.1016/S0074-7742(06)75015-2. [DOI] [PubMed] [Google Scholar]
- 48.Airavaara M, et al. In heterozygous GDNF knockout mice the response of striatal dopaminergic system to acute morphine is altered. Synapse. 2006;59:321–329. doi: 10.1002/syn.20245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.