Abstract
We used a systems biology-based approach to investigate the basis of cell-specific expression of the water channel aquaporin-2 (AQP2) in the renal collecting duct. Computational analysis of the 5′-flanking region of the AQP2 gene (Genomatix) revealed 2 conserved clusters of putative transcriptional regulator (TR) binding elements (BEs) centered at −513 bp (corresponding to the SF1, NFAT, and FKHD TR families) and −224 bp (corresponding to the AP2, SRF, CREB, GATA, and HOX TR families). Three other conserved motifs corresponded to the ETS, EBOX, and RXR TR families. To identify TRs that potentially bind to these BEs, we carried out mRNA profiling (Affymetrix) in mouse mpkCCDc14 collecting duct cells, revealing expression of 25 TRs that are also expressed in native inner medullary collecting duct. One showed a significant positive correlation with AQP2 mRNA abundance among mpkCCD subclones (Ets1), and 2 showed a significant negative correlation (Elf1 and an orphan nuclear receptor Nr1h2). Transcriptomic profiling in native proximal tubules (PT), medullary thick ascending limbs (MTAL), and IMCDs from kidney identified 14 TRs (including Ets1 and HoxD3) expressed in the IMCD but not PT or MTAL (candidate AQP2 enhancer roles), and 5 TRs (including HoxA5, HoxA9 and HoxA10) expressed in PT and MTAL but not in IMCD (candidate AQP2 repressor roles). In luciferase reporter assays, overexpression of 3 ETS family TRs transactivated the mouse proximal AQP2 promoter. The results implicate ETS family TRs in cell-specific expression of AQP2 and point to HOX, RXR, CREB and GATA family TRs as playing likely additional roles.
Keywords: aquaporin 2, kidney, microarrays, transcription, vasopressin
Renal water excretion is tightly regulated chiefly through effects of vasopressin on the molecular water channel, aquaporin-2 (AQP2) (1). AQP2 gene expression in the kidney is restricted to collecting duct principal cells and connecting tubule cells (2, 3). Aside from control of trafficking of AQP2-containing vesicles (1), AQP2 is regulated through changes in the total abundance of the AQP2 protein in collecting duct cells. Vasopressin increases the renal abundance of the AQP2 protein (4) via changes in AQP2 mRNA levels (5), in part by transcriptional regulation. Studies in transgenic mice in which 14–15 kb of the 5′-flanking region of the AQP2 gene was coupled to reporters established that cell-specific expression of the AQP2 gene in the collecting duct is dependent on cis-elements in this region (6, 7). Altered AQP2 protein abundance in the renal collecting duct is largely responsible for water balance abnormalities associated with diverse clinical states including lithium-induced diabetes insipidus, congestive heart failure, and the syndrome of inappropriate antidiuresis (1). Understanding the roles of AQP2 in these clinical states hinges largely on understanding the mechanism of cell-specific expression of the AQP2 gene.
Sequencing of the 5′-flanking region of the AQP2 gene revealed several putative cis-binding element (BE) motifs including a cAMP-response element (CRE) and an SP-1 site (8, 9). Subsequent studies of the CRE confirmed the importance of this cis-element in vasopressin-stimulated AQP2 transcription (10–12). A GATA site has also been reported (9, 13). Hozawa et al. (10) provided evidence for an AP2 site and Yasui et al. (12) for an AP1 site in the 5′-flanking region of the AQP2 gene. Finally, in a mouse collecting duct cell line, mpkCCDc14, that expresses AQP2 mRNA and protein (14), the nuclear factor of activated T cells (NFAT) family of transcriptional regulators (TRs) was found to be critical for tonicity-regulated AQP2 expression (15, 16).
Regulation of gene expression often occurs in a combinatorial fashion involving multiple TRs that bind to multiple closely spaced BEs organized into so-called cis-regulatory modules (CRMs) (17). The TRs can be placed in at least 2 classes: (i) signal-specific TRs whose abundance and activity in the nucleus is regulated chiefly by posttranslational modification, regulated degradation, or ligand binding; and (ii) tissue- and cell-specific TRs that are generally regulated at a transcriptional level. Because of the combinatorial nature of gene regulatory networks, it may be necessary to use a “systems” approach to understand transcriptional regulation of AQP2, looking at all possible transcriptional regulators in parallel. Here, we use such an approach employing bioinformatic analysis of the 5′-flanking region of the AQP2 gene and transcriptomic profiling with Affymetrix microarrays to identify putative cis-regulatory elements and TRs involved in cell-specific expression and transcriptional regulation of the AQP2 gene.
Results
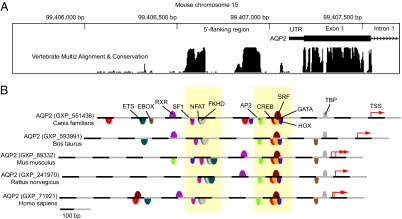
Fig. 1A maps sequence conservation among 5 mammalian species (dog, cow, mouse, rat, and human) for 1,000 bp of the 5′-flanking region of the AQP2 gene (referenced to mouse chromosome 15; specific sequences in Fig. S1). Two highly conserved regions centered at −224 bp and −513 bp upstream from the transcription start site are evident. We analyzed 1,000 bp of the 5′-flanking region using the Frameworker program in the Genomatix software suite to locate possible conserved TRBEs in these conserved regions (Fig. 1B), revealing a conserved cluster of 3 BE motifs in the upstream conserved region and a cluster of 5 BE motifs in the downstream conserved region. These 8 BE motifs include 4 that were identified and studied (see Introduction) corresponding to distinct TR families: NFAT, activator protein-2 (AP2), CREB, and GATA. In addition, putative BEs corresponding to steroidogenic factors (SF1), forkhead domain factors (FKHD), serum response element-binding factors (SRF), and homeobox-binding factors (HOX) were present in the conserved regions. Four other conserved TRBEs outside of these clusters were identified in all 5 species (Fig. 1B), including the TATA box (TBP), Ets (ETS), retinoid X receptor (RXR), and E-box (EBOX). Sequences corresponding to these TRBEs are listed in Fig. S2.
Fig. 1.
Bioinformatic analysis of 5′-flanking region of AQP2 gene. (A) Sequence conservation analysis for 1,000 bp of 5′-flanking region of AQP2 gene (http://genome.ucsc.edu). Conserved regions are centered 513 and 224 bp upstream from transcription start site. (B) Identification of conserved TR-binding element motifs in 1,000 bp of 5′-flanking region of AQP2 gene based on conserved sequence among 5 species (Genomatix).
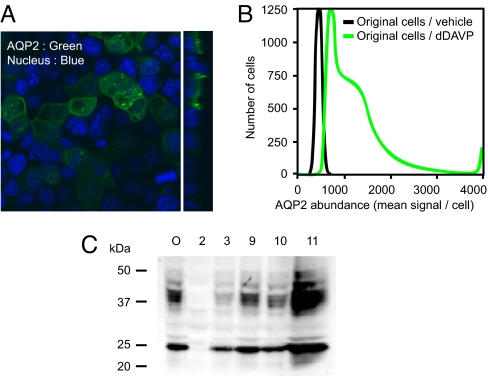
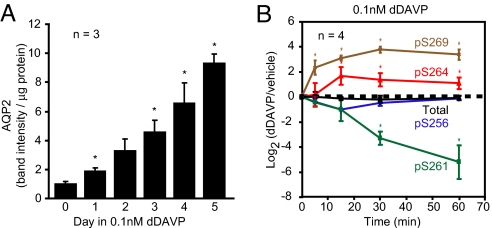
To address what TRs corresponding to the conserved TRBEs are expressed in collecting duct cells, we used transcriptomic profiling of mpkCCDc14 mouse collecting duct cells (Affymetrix). Initial experiments using confocal immunofluorescence labeling of AQP2 revealed heterogeneity of AQP2 protein abundance among mpkCCDc14 cells in confluent, polarized monolayers (Fig. 2A). Laser-scanning cytometry revealed a biphasic distribution of AQP2 immunofluorescence (Fig. 2B). To obtain homogeneous cells for study, we cloned several cell lines from the original mpkCCDc14 cells, each expressing different levels of AQP2 protein when grown in the presence of the vasopressin analog dDAVP (0.1 nM) (Fig. 2C). The lowest abundance of AQP2 was found in clone 2 and the highest in clone 11. Clone 11 expressed AQP2 protein at a level approximately equal to that seen in native inner medulla (Fig. S3). Clone 11 was characterized further, showing a significant increase in AQP2 protein abundance in response to dDAVP (0.1 nM for 1, 3, 4, or 5 days, Fig. 3A) and vasopressin-dependent changes in AQP2 phosphorylation similar to those seen in native collecting ducts (Fig. 3B).
Fig. 2.
Cloning of mpkCCD-derived cell lines expressing AQP2 at various levels. (A) Confocal immunofluorescence image showing original mpkCCDc14 cells grown in presence of 0.1 nM dDAVP and immunolabeled with AQP2 antibody. (B) Laser-scanning cytometry reveals that the distribution of AQP2 immunofluorescence is biphasic indicating the presence of a heterogeneous cell population. (C) AQP2 immunoblot of homogenates from original mpkCCDc14 cells (O) and 5 clonal lines. Note broad range of AQP2 protein abundance among clonal lines.
Fig. 3.
Characterization of mpkCCD clone 11. (A) AQP2 protein abundance increases after introduction of 0.1 nM dDAVP to basolateral medium. Asterisk indicates a significant difference relative to time 0 (no dDAVP). (B) Time course of phosphorylation changes of AQP2 protein in clone 11 cells based on immunoblotting with phosphospecific antibodies to 4 different phosphorylation sites in the COOH-tail of AQP2.
Comprehensive transcriptomic profiling (Affymetrix Mouse Genome 430 2.0 Arrays) was carried out for the 5 clonal mpkCCD lines shown in Fig. 2C and the original (O) line to assess what TRs are expressed and which of them correlate with AQP2 expression. Table 1 shows a summary of TRs corresponding to the conserved binding site model shown in Fig. 1 that are expressed in both mpkCCD clone 11 cells and in native inner medullary collecting duct (IMCD) cells from rats (IMCD Transcriptome Database, http://dir.nhlbi.nih.gov/papers/lkem/imcdtr) (18). Table S2 summarizes the previously reported TRs (see Introduction) whose putative binding sites are not conserved among all mammalian species examined and thus are not included in Fig. 1. Among the 25 TRs listed in Table 1, only 1 showed a statistically significant positive correlation with AQP2 mRNA abundance, namely E26 avian leukemia oncogene 1 (Ets1). Two others showed a statistically significant negative correlation with AQP2 mRNA, namely E74-like factor 1 (Elf1) and nuclear receptor subfamily 1 group H member 2 (Nr1h2). Among the 12 TRs listed in Table S2, 4 correlated significantly with AQP2 mRNA: Jun-B oncogene (Junb), Kruppel-like factor 9 (Klf9), nuclear factor of kappa light chain gene enhancer in B-cells 1 p105 (Nfkb1), and v-rel reticuloendotheliosis viral oncogene homolog A (Rela). One was negatively correlated, namely fos-like antigen 2 (Fosl2).
Table 1.
Transcriptional regulators with putative binding elements in the AQP2 5′-flanking region that are common to mpkCCD and native IMCD cells
| Gene symbol | Gene name | Ratio (clone 11/clone 2) | Correlation coefficient vs. AQP2 mRNA | P value | mpkCCD signal | IMCD signal |
|---|---|---|---|---|---|---|
| CREB (cAMP-responsive element-binding proteins) | ||||||
| Atf4 | activating transcription factor 4 | 1.11 | 0.48 | 0.33 | 6.90 | 23.41 |
| Atf1 | activating transcription factor 1 | 0.92 | 0.05 | 0.93 | 2.61 | 3.54 |
| Creb3l2 | cAMP responsive element binding protein 3-like 2 | 1.87 | 0.78 | 0.07 | 1.16 | 0.40 |
| Creb3 | cAMP responsive element binding protein 3-like 4 | 1.04 | 0.17 | 0.75 | 0.81 | 1.64 |
| Atf3 | activating transcription factor 3 | 1.13 | 0.19 | 0.72 | 0.46 | 48.17 |
| Crebl1 | cAMP responsive element binding protein-like 1 | 1.20 | 0.58 | 0.22 | 0.45 | 1.43 |
| Atf5 | activating transcription factor 5 | 0.91 | 0.06 | 0.91 | 0.35 | 1.21 |
| EBOX (E-box-binding factors) | ||||||
| Mxi1 | Max interacting protein 1 | 1.05 | −0.18 | 0.73 | 2.16 | 6.37 |
| Myc | myelocytomatosis oncogene | 1.22 | −0.03 | 0.96 | 1.86 | 3.09 |
| Usf2 | upstream transcription factor 2 | 0.86 | −0.57 | 0.23 | 0.54 | 1.45 |
| Mlx | MAX-like protein X | 0.95 | 0.26 | 0.63 | 0.45 | 1.89 |
| ETS (Ets-like factors) | ||||||
| Ets2 | E26 avian leukemia oncogene 2 3' domain | 1.01 | −0.12 | 0.83 | 2.29 | 23.52 |
| Elf3 | E74-like factor 3 | 1.49 | 0.19 | 0.71 | 1.66 | 23.53 |
| Ets1 | E26 avian leukemia oncogene 1 5' domain | 1.57 | 0.99 | 0.00* | 1.62 | 2.42 |
| Elf1 | E74-like factor 1 | 0.72 | −0.90 | 0.01* | 1.41 | 3.27 |
| Elf2 | E74-like factor 2 | 1.08 | −0.18 | 0.73 | 1.30 | 1.31 |
| FKHD (forkhead domain factors) | ||||||
| Foxp1 | forkhead box P1 | 1.06 | −0.46 | 0.36 | 1.03 | 0.62 |
| Foxq1 | forkhead box Q1 | 0.98 | −0.37 | 0.47 | 0.68 | 6.90 |
| GATA (GATA-binding factors) | ||||||
| Gata3 | GATA-binding protein 3 | 1.76 | 0.53 | 0.28 | 2.84 | 13.98 |
| Gata2 | GATA-binding protein 2 | 4.19 | 0.66 | 0.15 | 0.31 | 5.15 |
| HOX (homeobox-binding factors) | ||||||
| Pbx2 | pre B-cell leukemia transcription factor 2 | 1.46 | 0.70 | 0.12 | 0.99 | 0.61 |
| Hoxb8 | homeo box B8 | 2.12 | 0.42 | 0.41 | 0.96 | 0.58 |
| Hoxd3 | homeo box D3 | 3.16 | 0.67 | 0.14 | 0.32 | 2.17 |
| RXR (retinoid X receptor family) | ||||||
| Rxra | retinoid X receptor alpha | 1.73 | 0.66 | 0.15 | 0.73 | 1.62 |
| Nr1 h2 | nuclear receptor subfamily 1 group H member 2 | 0.65 | −0.83 | 0.04* | 0.71 | 2.62 |
Data for mpkCCD transcripts are median-normalized fluorescence readings from 6 Mouse Genome 430 2.0 Arrays (Affymetrix). Full transcriptomic profile for mpkCCD clones is given in Table S1. Data for transcripts in IMCD are taken from IMCD Transcriptome Database (http://dir.nhlbi.nih.gov/papers/lkem/imcdtr).
*Transcript signal significantly correlated with AQP2 mRNA signal (P < 0.05, n = 6).
Full results of the mpkCCD transcript profiling are provided in Table S1 and Fig. S4; 7,983 transcripts were expressed above background in mpkCCD clone 11 cells. This markedly expands the findings of a previous transcript profiling study of mpkCCD cells that used the SAGE technique (19). To provide these data for the general community, we have created a permanent online database on the National Heart, Lung, and Blood Institute (NHLBI) Proteomics and Genomics Database site (http://dir.nhlbi.nih.gov/papers/lkem/mpkccdtr). Raw data can be retrieved from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; GSE13672).
TRs that convey cell-specific expression in the renal collecting ducts would be expected to be expressed only in collecting duct cells (if they play an enhancer role) or would be expected to be uniquely non-expressed in collecting duct cells (if they play a repressor role). To further identify candidate TRs that may be involved in tissue-specific expression of AQP2 in collecting ducts, we carried out mRNA profiling in native rat renal medullary thick ascending limb (MTAL) cells and native rat renal proximal tubule (PT) cells using Affymetrix oligonucleotide microarrays (Rat 230 2.0 Expression Arrays, Affymetrix) and compared the results to our published mRNA profiling data for native IMCD cells isolated from rats (18) (Tables 2 and 3). The MTAL and PT cells were isolated using standard cell purification methods for these 2 cell types (see Methods). Table 2 lists the 14 TRs that correspond to the model shown in Fig. 1 and were found in IMCD and not in MTAL or PT. These TRs included members of the following TR families: AP2, CREB, EBOX, ETS, Forkhead (FKHD), GATA, Homeobox (HOX), and retinoic acid receptor (RXR). This list includes Ets1, found above to correlate with AQP2 mRNA abundance in cultured mpkCCD cells (Table 1). Table 3 lists the 5 TRs found in both MTAL and PT but not in IMCD including members of the FKHD, HOX and RXR TR families.
Table 2.
TRs in IMCD but not in MTAL or PT cells
| TR family | Gene symbol | IMCD signal |
|---|---|---|
| AP2 | Tcfap2a | 3.28 |
| CREB | Atf3 | 48.17 |
| CREB | Atf2 | 0.45 |
| EBOX | Myc | 3.09 |
| ETS | Ehf | 23.80 |
| ETS | Elf5 | 9.43 |
| ETS | Ets1 | 2.42 |
| ETS | Elk3 | 1.57 |
| FKHD | Foxq1 | 6.90 |
| FKHD | Foxi1 | 1.30 |
| GATA | Gata3 | 13.98 |
| GATA | Gata2 | 5.15 |
| HOX | Hoxd3 | 2.17 |
| RXR | Rara | 1.15 |
Signals are mean values (n = 3) of median-normalized fluorescence readings from Rat 230 2.0 Expression Arrays (Affymetrix). Data for all PT and MTAL transcripts including SE values are given in Tables S3 and S4. IMCD values are from Uawithya et al. (18).
Table 3.
TRs not in IMCD but in MTAL and PT cells
| Tr family | Gene symbol | MTAL signal | PT signal |
|---|---|---|---|
| FKHD | Foxk2 | 1.10 | 0.64 |
| HOX | Hoxa10 | 8.05 | 1.05 |
| HOX | Hoxa5 | 9.42 | 1.29 |
| HOX | Hoxa9 | 4.84 | 1.63 |
| RXR | Thrb | 0.88 | 0.47 |
Signals are mean values (n = 3) of median-normalized fluorescence readings from Rat 230 2.0 Expression Arrays (Affymetrix). Data for all PT and MTAL transcripts including SE values are given in Tables S3 and S4. IMCD values are from Uawithya et al. (18).
In native PT cells, 7,502 transcripts were expressed above background (Table S3), and 8,003 were expressed above background in MTAL cells (Table S4). To provide the full PT and MTAL transcriptomic data to the general community, we have created permanent online databases on the NHLBI Proteomics and Transcriptomics Online Database (http://dir.nhlbi.nih.gov/papers/lkem/pttr and http://dir.nhlbi.nih.gov/papers/lkem/mtaltr).
Based on the model shown in Fig. 1, we hypothesize that other collecting duct-specific genes gain their cell specificity via TRBEs shared with AQP2. To test this prediction, we analyzed 1,000 bp of the 5′-flanking region for 379 transcripts that are uniquely expressed in the rat IMCD, comparing them to findings for 155 unique MTAL transcripts (Table 5), 301 unique PT transcripts (Table 4) or 379 transcripts whose expression is common to IMCD, mTAL, and PT (Table S5). (See Table S6 for full list.) Interestingly, the ETS BE motif was found significantly more frequently in IMCD-specific genes than in all 3 control sets. The AP2, CREB, and HOX BEs were found significantly more frequently in IMCD-specific genes than in 2 of the controls.
Table 5.
Overrepresented TRBEs in IMCD-specific genes vs. genes expressed in MTAL
| Genomatix TRBE | Name | BioBase TRBE | fIMCD | fmTAL | Ratio | P |
|---|---|---|---|---|---|---|
| ETS | Human and murine ETS1 factors | V$ETS_Q6 | 3.65 | 3.16 | 1.16 | *** |
| FKHD | Forkhead domain factors | V$HNF3B_01 | 1.94 | 1.68 | 1.16 | * |
| GATA | GATA-binding factors | V$GATA4_Q3 | 3.75 | 3.36 | 1.11 | * |
| HOX | Homeobox-binding factor | V$NCX_01 | 8.67 | 8.02 | 1.08 | * |
BioBase ExPlain software suite was used to locate TRBEs in 5′-flanking regions of 379 IMCD-specific transcripts (signal above median and no signal in MTAL or PT), and 155 MTAL-specific transcripts (signal above median and no signal in IMCD or PT). Significant overrepresentation of TRBEs in IMCD-specific transcripts versus in MTAL was tested by χ2 analysis. fmTAL, frequency of TRBE in MTAL.
Significance: *, P < 0.05;
**, P < 0.01;
***, P < 0.005.
Table 4.
Overrepresented TRBEs in IMCD-specific genes vs. genes expressed in proximal tubule
| Genomatix TRBE | Name | BioBase TRBE | fIMCD | fPT | Ratio | P |
|---|---|---|---|---|---|---|
| AP2 | Activator protein 2 | V$AP2_Q6 | 4.33 | 3.33 | 1.30 | *** |
| CREB | cAMP-responsive element-binding proteins | V$CREBATF_Q6 | 0.82 | 0.64 | 1.29 | *** |
| ETS | Human and murine ETS1 factors | V$CETS1P54_03 | 6.29 | 5.93 | 1.06 | * |
| RXR | RXR heterodimer-binding sites | V$VDR_Q3 | 5.64 | 4.95 | 1.14 | *** |
| SRF | Serum response element-binding factor | V$SRF_Q6 | 0.07 | 0.03 | 2.02 | * |
BioBase ExPlain software suite was used to locate TRBEs in 5′-flanking regions of 379 IMCD-specific transcripts (signal above median and no signal in MTAL or PT), and 301 PT-specific transcripts (signal above median and no signal in IMCD or MTAL). Significant overrepresentation of TRBEs in IMCD-specific transcripts versus PT-specific transcripts was tested by χ2 analysis. fIMCD and fPT, frequency of TRBE in IMCD, and PT transcripts.
*, P < 0.05;
**, P < 0.01;
***, P < 0.005.
A common feature of all 4 parts of the foregoing systems-level analysis of TRBEs and TRs in collecting duct cells was the ETS-binding element and its associated TRs (Fig. 1, Tables 1–5). To address in greater detail the possible role of ETS family TRs in the regulation of AQP2 gene transcription, we cloned 1,124 bp of the mouse AQP2 5′-flanking region into a luciferase reporter construct and coexpressed it with each of 3 ETS family TRs in vasopressin-responsive LLC-PK1 cells. The 3 TRs chosen were exceptionally highly expressed in the IMCD: Elf3 (signal 23.5 above median on microarray), Elf5 (9.4 above median), and Ehf (23.8 above median). All 3 of the TRs transactivated the AQP2 gene (Fig. 4). Elf3 did so without addition of factors to increase intracellular cAMP levels, whereas Elf5 and Ehf required addition of the vasopressin analog dDAVP and the cyclic nucleotide phosphodiesterase inhibitor IBMX.
Fig. 4.
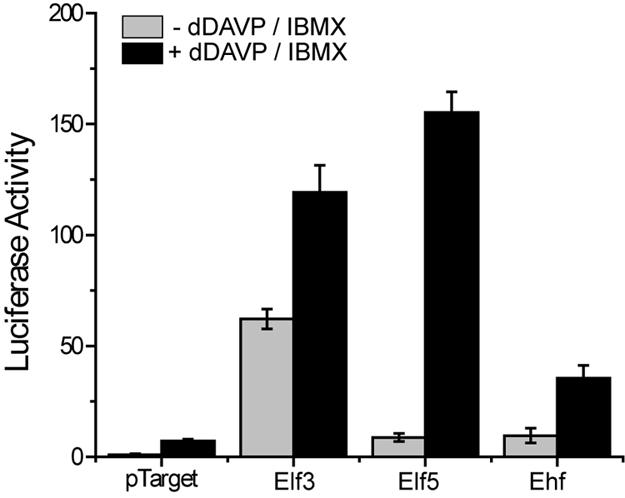
Ets family transcriptional regulators transactivate the mouse proximal AQP2 promoter. Luciferase activity was measured in LLC-PK1 cells cotransfected with the AQP2-Luc reporter and either pTarget (empty vector) or Elf3, Elf5, or Ehf and stimulated with 10−7 M dDAVP and 400 μM IBMX (+dDAVP/IBMX) or vehicle (-dDAVP/IBMX). Data represent mean ± SE. (n = 12 per condition).
Discussion
We have used computational techniques to identify conserved BE motifs in the 5′-flanking region of the AQP2 gene and have used oligonucleotide microarrays in cultured collecting duct cells and kidney cells from the proximal tubule, thick ascending limb of Henle and inner medullary collecting duct to identify candidate TR proteins involved in cell-specific expression of the AQP2 gene. The broad view afforded by these approaches identified several BEs and TR families that play likely roles in the regulation of AQP2 gene expression. Among the TRs likely to be involved in cell-specific regulation of AQP2 gene expression are TRs that bind to ETS, HOX, RXR, CREB and GATA BEs of the AQP2 gene.
The conserved ETS-binding element, located ≈500 bp upstream from the transcription start site in mouse (Fig. 1), stands out in this study. Four separate approaches all pointed to this binding element or to the ETS family TRs as a likely determinant of cell-specific AQP2 expression in collecting duct cells: (i) the identification of a conserved ETS BE in the AQP2 gene via computational methods (Fig. 1); (ii) the identification of the Ets1 transcript in cultured mouse mpkCCD cells and the finding that its level is strongly correlated with the AQP2 transcript level among mpkCCD clonal cell lines (Table 1); (iii) the selective expression of the Ets1 transcript in native IMCD cells but not in native proximal tubule cells or medullary thick ascending limb cells (Table 2); and (iv) the overrepresentation of putative ETS BE motifs in the 5′-flanking regions of IMCD-specific transcripts other than AQP2 (Tables 4 and 5). Based on the consilient findings from multiple approaches, we conclude that ETS family TRs are likely to be determinants of cell-specific gene expression in the collecting duct. Accordingly, we tested the ability of selected ETS family TRs to enhance AQP2 transcription in a promotor-reporter assay. Indeed, all 3 TRs (Elf3, Elf5, and Ehf) increased reporter activity either alone or with measures to increase intracellular cAMP (Fig. 4).
The ETS family consists of ≈29 members in mouse and human and belongs to the winged helix-turn-helix super family (20). ETS-family TRs are expressed in a wide variety of tissues and play a central role in development, differentiation and physiology. In situ hybridization analysis of a number of ETS factors in the developing mouse kidney (www.gudmap.org) shows that ETS factors Pea3, Erm, Elf4, Elk1, Elk4, Tel, Elf3, Elf5, and Ehf are highly expressed in the ureteric bud, the embryonic precursor of the renal collecting duct. ETS family TRs have been implicated in urea-mediated regulation of Egr-1 transcription in cultured IMCD cells (21).
Based on extensive prior study of transcriptional networks (17), we would not expect that ETS family TRs alone or any other TR to be the sole determinant of cell-specific gene expression in the collecting duct. Rather, AQP2 transcriptional regulation is likely to be combinatorial, integrating the effects of multiple TRs. Based on the systems level analysis presented in this article, it appears that the HOX, RXR, CREB and GATA binding elements are also excellent candidates to play roles in collecting-duct-specific expression of AQP2 and other genes.
Homeobox or HOX transcriptional regulator proteins have long been supposed to be a major determinant of renal tubule segmentation and collecting duct specific gene expression (22). These TRs are responsible for diverse developmental processes. Hox genes are organized in 4 clusters (A–D) derived from gene duplication events. Each cluster is made up of multiple genes, each with paralogs in the other clusters. Here, using Affymetrix microarrays, we found only genes from cluster A expressed in proximal tubule (HoxA10, A9, A5, and A4). In contrast, we found Hox genes from all 4 clusters expressed in thick ascending limb. Finally, the IMCD expressed only genes from cluster B (HoxB8, B7, B5, B4, and B3) and cluster D (HoxD10, D9, and D3). The HoxB7 5′-flanking region has been used to target gene expression to the ureteric bud and mature collecting duct (23, 24).
RXR-binding elements bind dimers of ligand activated transcription factors, most commonly RAR/RXR TR heterodimers. Besides RAR/RXR heterodimerization, RXR also heterodimerizes with thyroid receptors, vitamin D receptors, peroxisome proliferator-activator receptors and other ligand-activated nuclear receptors. RXR and RAR are recognized to play important roles in renal development especially in structures derived from the ureteric bud (25). RXR-binding motifs were substantially and significantly more common in 5′-flanking regions of IMCD-specific genes than in other genes expressed in PT, MTAL or IMCD (Table S5) consistent with a role as a determinant of cell-specific gene expression in the collecting duct.
The presence of a functional CRE element in the 5′-flanking region of the AQP2 gene has been documented (10–12). This site has been supposed to be responsible for cAMP-mediated regulation of AQP2 gene transcription by binding the CREB1 transcriptional regulator after phosphorylation at Ser-133 by protein kinase A. Although CREB1 is expressed in mpkCCD cells, it is expressed at an extremely low level in native rat IMCD cells (18), and it seems possible that other CREB family proteins bind to this site to regulate AQP2 gene expression. In mpkCCD cells, CREB family TRs Atf1, Atf4 and CREB3-like 2 (Creb3l2) are all expressed at levels above the median signal and Creb3l2 mRNA abundance shows a correlation with AQP2 mRNA levels (Table 1). Furthermore, both Atf2 and Atf3 are selectively expressed in IMCD (Table 2). Thus, the specific TRs that bind to the CRE site in the AQP2 gene remain to be discovered.
Consistent with our computational analysis (Fig. 1), Rai et al. (9) found a DNase I-protected GATA-binding element just downstream of the CREB binding element in the AQP2 5′-flanking region. They showed that deletion and mutation of this cis-element abolished protein-DNA binding and increased promoter activity in hetaptocyte Ac2F cells. Furthermore, they found that deletion of a portion of the 5′ flanking region containing the GATA element led to an increased reporter activity in mouse outer medullary collecting duct cells, suggesting a repressor role of the GATA binding element (13). However, in a later study using the same mouse outer medullary collecting duct cells, over-expression of the GATA-3 TR increased AQP2 transcription (26). These seemingly conflicting findings can be resolved in light of the finding that the GATA BE is part of a cluster of binding elements that together may constitute a cis-regulatory module (Fig. 1). We propose that the GATA element plays an enhancer role, but that mutations introduced into promoter-reporter constructs may have disrupted binding of other TRs involved in this cis-regulatory module and that at least 1 of these TRBEs plays a repressor role. Hence, we suggest that the HOX, CREB or SRF BEs may mediate the repressor activity attributed to GATA.
Conclusion
The results implicate ETS family TRs in cell-specific expression of AQP2 and point to HOX, RXR, CREB, and GATA family TRs as playing likely additional roles. Aside from the knowledge gained about transcriptional regulators and cell-specific regulation of AQP2 gene expression, the work described in this article has created several valuable byproducts. First, we have provided internet-accessible comprehensive mRNA profiling databases for native rat proximal tubule cells and native thick ascending limb cells. (See Results for URLs). Second, we have produced a clonal cell line derived from the original mpkCCDc14 cells of Vandewalle and coworkers (27) that express AQP2 protein at a level equivalent to native collecting duct cells and exhibit vasopressin-dependent AQP2 phosphorylation similar to that seen in native cells. Finally, we have provided an internet-accessible comprehensive mRNA profiling database listing ≈8,000 transcripts expressed in this clonal line (mpkCCD clone 11) (see Results for URL).
Methods
Computational Methods.
The BLAT function in the University of California, Santa Cruz, Genome Browser (http://genome.ucsc.edu) was used to identify evolutionarily conserved regions in the 5′-flanking region of the AQP2 gene. To identify phylogenetically conserved TRBEs, 1,000 bp of the AQP2 5′-flanking regions from 5 species (human, cow, dog, rat, and mouse) were analyzed using Gene2Promoter and Frameworker software from the Genomatix database and software suite. In addition, the Biobase ExPlain software suite was used to analyze TRBEs overrepresented in selected transcript sets.
Cell Culture and Cloning.
Cell culture conditions for the mouse kidney cortical collecting duct cell line (mpkCCDc14) are described in ref. 14. Cells from the original mpkCCDc14 were sorted using a MoFlo XDP Cell Sorter (Beckman Coulter) into 10 96-well plates and grown into colonies. Cells were grown on membrane supports (24-mm Transwell, Corning) until polarization (transepithelial resistance >5 kΩ.cm2) and exposed to 0.1 nM vasopressin analog (1-desamino-8-D-arginine vasopressin, dDAVP) added to the basolateral medium (serum- and hormone-free) for 5 days. Media were changed daily.
Immunoblotting.
Immunoblotting was carried out as described (28). Protein was quantified using the BCA method (Thermo Scientific). Protein amounts between 10 and 25 μg were separated on 4–15% gradient polyacrylamide gels, and transferred to nitrocellulose membranes. Secondary antibodies conjugated to infrared fluorescent dyes were from LI-COR. Protein bands were visualized and quantified with an infrared fluorescence scanner using Odyssey software (LI-COR).
Primary Antibodies.
Primary antibodies for AQP2 included K5007 detecting the COOH terminus (29) and N-20 detecting the NH2 terminus (SC-9880, Santa Cruz Biotechnology). Phospho-specific antibodies against AQP2 were generated in our laboratory (29, 30).
Immunofluorescence Confocal Microscopy; Laser-Scanning Cytometry.
Immunofluorescence labeling was done as described in ref. 28. Confocal fluorescence micrographs were obtained using a Zeiss LSM 510 microscope and software (Carl Zeiss MicroImaging; NHLBI Light Microscopy Core Facility). Some slides were analyzed by laser-scanning cytometery (CompuCyte) to determine the distribution of AQP2 protein expression among cells.
Isolation of Native Renal Proximal Tubules and Medullary Thick Ascending Limbs.
Animal experiments followed animal protocol H-0110 (NHLBI Animal Care and Use Committee). Proximal tubules were isolated from rat renal cortex, as described (31). Thick ascending limbs were isolated from rat outer medulla as described (32) with minor modifications.
Transcriptome Analysis.
Total RNA was extracted using TriZOL reagent (15596–026, Invitrogen) following the manufacturer's protocol. For analysis of mouse mpkCCD clonal cell lines, 2 μg of total RNA was used for oligonucleotide microarray analysis using Affymetrix GeneChip Mouse Genome 430 2.0 Arrays (NHLBI Gene Expression Core Facility). For analysis of native rat proximal tubule or thick ascending limb cells, 2.5 μg of total RNA was used for oligonucleotide microarray analysis employing Rat 230 2.0 Expression Arrays from Affymetrix, Inc. Full details are as described (18). Microarray raw data were examined with Affymetrix GeneChip Operating System software version 1.4 and normalized based on MAS5 algorithm using Affymetrix Gene Console software version 1.1. The normalized data were subjected to principal component analysis to examine biological and technical variations before further statistical analysis and bioinformatics interpretation using the PANTHER Classification System (http://www.pantherdb.org).
Promotor-Reporter Assays.
A 1,511-bp fragment from the 5′-flanking region of the mouse AQP2 gene (−1,124 to + 386) was PCR amplified from mouse tail DNA and cloned into the pGEMT vector (Promega). The AQP2-pGEMT construct was cut with Xhol (−992) and Afel (−21) (New England BioLabs) and cloned into the Xhol and Hindlll sites of the pGL3 luciferase vector (Promega); the Afel site was filled using DNA polymerase I, large fragment (Klenow, New England Biolabs) so that it could be cloned into the HindIII site. Full length Elf3, Elf5 and Ehf cDNA were PCR amplified, sequence verified, and cloned into the pTarget vector downstream of the CMV promoter (Promega). LLCPK1 cells were transfected with 0.8 μg of total DNA: 0.4 μg of AQP2-pGL3 reporter and 0.4 μg of expression construct, i.e., pTarget (empty vector) or pTarget containing Elf3, Elf5, or Ehf using Lipofectamine 2000 (Invitrogen). Cells were grown to confluence and then stimulated with 10−7 M dDAVP and 400 μM IBMX or vehicle for 72 h before the luciferase readout. Cells were washed with PBS, lyzed, scraped, collected, freeze-thawed 3 times and centrifuged (10,000 × g). Luciferase activity was measured in 20 μL of supernatant using a luminometer.
Supplementary Material
Acknowledgments.
We thank Prof. M. Kretzler (University of Michigan) and Dr. C.-H. Tai (National Cancer Institute) for advice regarding Genomatix software. These studies used the Gene Expression Core Facility (Dr. N. Raghavachari, Director), Light Microscopy Core Facility (Dr. C. Combs, Director), and Flow Cytometry Core Facility (Dr. J. P. McCoy, Director) of the NHLBI. Support: Division of Intramural Research, NHLBI (project ZO1-HL001285, MAK) and National Institutes of Health Grant DK053990 (to R.D.N. and R.L.M.). M.M.R. was supported by the Braun Foundation (Melsungen, Germany) and the Biomedical Sciences Exchange Program (Hannover, Germany).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813002106/DCSupplemental.
References
- 1.Nielsen S, Frokiaer J, Marples D, Kwon TH, Agre P, Knepper MA. Aquaporins in the kidney: From molecules to medicine. Physiol Rev. 2002;82:205–244. doi: 10.1152/physrev.00024.2001. [DOI] [PubMed] [Google Scholar]
- 2.Fushimi K, et al. Cloning and expression of apical membrane water channel of rat kidney collecting tubule. Nature. 1993;361:549–552. doi: 10.1038/361549a0. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen S, DiGiovanni SR, Christensen EI, Knepper MA, Harris HW. Cellular and subcellular immunolocalization of vasopressin-regulated water channel in rat kidney. Proc Natl Acad Sci USA. 1993;90:11663–11667. doi: 10.1073/pnas.90.24.11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiGiovanni SR, Nielsen S, Christensen EI, Knepper MA. Regulation of collecting duct water channel expression by vasopressin in Brattleboro rat. Proc Natl Acad Sci USA. 1994;91:8984–8988. doi: 10.1073/pnas.91.19.8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi M, et al. Role of vasopressin V2 receptor in acute regulation of aquaporin-2. Kidney Blood Press Res. 1996;19:32–37. doi: 10.1159/000174043. [DOI] [PubMed] [Google Scholar]
- 6.Nelson RD, et al. Expression of an AQP2 Cre recombinase transgene in kidney and male reproductive system of transgenic mice. Am J Physiol. 1998;275:C216–C226. doi: 10.1152/ajpcell.1998.275.1.C216. [DOI] [PubMed] [Google Scholar]
- 7.Zharkikh L, et al. Renal principal cell-specific expression of green fluorescent protein in transgenic mice. Am J Physiol Renal Physiol. 2002;283:F1351–F1364. doi: 10.1152/ajprenal.0224.2001. [DOI] [PubMed] [Google Scholar]
- 8.Uchida S, Sasaki S, Fushimi K, Marumo F. Isolation of human Aquaporin-CD gene. J Biol Chem. 1994;269:23451–23455. [PubMed] [Google Scholar]
- 9.Rai T, Uchida S, Marumo F, Sasaki S. Cloning of rat and mouse aquaporin-2 gene promoters and identification of a negative cis-regulatory element. Am J Physiol. 1997;273:F264–F273. doi: 10.1152/ajprenal.1997.273.2.F264. [DOI] [PubMed] [Google Scholar]
- 10.Hozawa S, Holtzman EJ, Ausiello DA. cAMP motifs regulating transcription in the aquaporin-2 gene. Am J Physiol. 1996;270:C1695–C1702. doi: 10.1152/ajpcell.1996.270.6.C1695. [DOI] [PubMed] [Google Scholar]
- 11.Matsumura Y, Uchida S, Rai T, Sasaki S, Marumo F. Transcriptional regulation of aquaporin-2 water channel gene by cAMP. J Am Soc Nephrol. 1997;8:861–867. doi: 10.1681/ASN.V86861. [DOI] [PubMed] [Google Scholar]
- 12.Yasui M, Zelenin SM, Celsi G, Aperia A. Adenylate cyclase-coupled vasopressin receptor activates AQP2 promoter via a dual effect on CRE and AP1 elements. Am J Physiol. 1997;272:F443–F450. doi: 10.1152/ajprenal.1997.272.4.F443. [DOI] [PubMed] [Google Scholar]
- 13.Furuno M, Uchida S, Marumo F, Sasaki S. Repressive regulation of the aquaporin-2 gene. Am J Physiol. 1996;271:F854–F860. doi: 10.1152/ajprenal.1996.271.4.F854. [DOI] [PubMed] [Google Scholar]
- 14.Hasler U, et al. Long term regulation of aquaporin-2 expression in vasopressin-responsive renal collecting duct principal cells. J Biol Chem. 2002;277:10379–10386. doi: 10.1074/jbc.M111880200. [DOI] [PubMed] [Google Scholar]
- 15.Hasler U, et al. Tonicity-responsive enhancer binding protein is an essential regulator of aquaporin-2 expression in renal collecting duct principal cells. J Am Soc Nephrol. 2006;17:1521–1531. doi: 10.1681/ASN.2005121317. [DOI] [PubMed] [Google Scholar]
- 16.Li SZ, et al. Calcineurin-NFATc signaling pathway regulates AQP2 expression in response to calcium signals and osmotic stress. Am J Physiol. 2007;292:C1606–C1616. doi: 10.1152/ajpcell.00588.2005. [DOI] [PubMed] [Google Scholar]
- 17.Busser BW, Bulyk ML, Michelson AM. Toward a systems-level understanding of developmental regulatory networks. Curr Opin Genet Dev. 2008;18:521–529. doi: 10.1016/j.gde.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uawithya P, Pisitkun T, Ruttenberg BE, Knepper MA. Transcriptional profiling of native inner medullary collecting duct cells from rat kidney. Physiol Genom. 2008;32:229–253. doi: 10.1152/physiolgenomics.00201.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert-Nicoud M, et al. Transcriptome of a mouse kidney cortical collecting duct cell line: Effects of aldosterone and vasopressin. Proc Natl Acad Sci USA. 2001;98:2712–2716. doi: 10.1073/pnas.051603198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharrocks AD, Brown AL, Ling Y, Yates PR. The ETS-domain transcription factor family. Int J Biochem Cell Biol. 1997;29:1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- 21.Cohen DM, Gullans SR, Chin WW. Urea inducibility of egr-1 in murine inner medullary collecting duct cells is mediated by the serum response element and adjacent Ets motifs. J Biol Chem. 1996;271:12903–12908. doi: 10.1074/jbc.271.22.12903. [DOI] [PubMed] [Google Scholar]
- 22.Patterson LT, Potter SS. Atlas of Hox gene expression in the developing kidney. Dev Dyn. 2004;229:771–779. doi: 10.1002/dvdy.10474. [DOI] [PubMed] [Google Scholar]
- 23.Srinivas S, et al. Expression of green fluorescent protein in the ureteric bud of transgenic mice: A new tool for the analysis of ureteric bud morphogenesis. Dev Genet. 1999;24:241–251. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<241::AID-DVG7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 24.Rubera I, et al. Collecting duct-specific gene inactivation of alphaENaC in the mouse kidney does not impair sodium and potassium balance. J Clin Invest. 2003;112:554–565. doi: 10.1172/JCI16956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burrow CR. Retinoids and renal development. Exp Nephrol. 2000;8:219–225. doi: 10.1159/000020672. [DOI] [PubMed] [Google Scholar]
- 26.Uchida S, Matsumura Y, Rai T, Sasaki S, Marumo F. Regulation of aquaporin-2 gene transcription by GATA-3. Biochem Biophys Res Commun. 1997;232:65–68. doi: 10.1006/bbrc.1997.6236. [DOI] [PubMed] [Google Scholar]
- 27.Bens M, et al. Corticosteroid-dependent sodium transport in a novel immortalized mouse collecting duct principal cell line. J Am Soc Nephrol. 1999;10:923–934. doi: 10.1681/ASN.V105923. [DOI] [PubMed] [Google Scholar]
- 28.Yu MJ, Pisitkun T, Wang G, Shen RF, Knepper MA. LC-MS/MS analysis of apical and basolateral plasma membranes of rat renal collecting duct cells. Mol Cell Proteom. 2006;5:2131–2145. doi: 10.1074/mcp.M600177-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffert JD, et al. Vasopressin-stimulated increase in phosphorylation at ser-269 potentiates plasma membrane retention of aquaporin-2. J Biol Chem. 2008;283:24617–24627. doi: 10.1074/jbc.M803074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sachs AN, Pisitkun T, Hoffert JD, Yu MJ, Knepper MA. LC-MS/MS analysis of differential centrifugation fractions from native inner medullary collecting duct of rat/ Am J Physiol. 2008;295:F1799–F1806. doi: 10.1152/ajprenal.90510.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curthoys NP, Taylor L, Hoffert JD, Knepper MA. Proteomic analysis of the adaptive response of rat renal proximal tubules to metabolic acidosis. Am J Physiol. 2007;292:F140–F147. doi: 10.1152/ajprenal.00217.2006. [DOI] [PubMed] [Google Scholar]
- 32.Eveloff J, Haase W, Kinne R. Separation of renal medullary cells: Isolation of cells from the thick ascending limb of Henle's loop. J Cell Biol. 1980;87:672–681. doi: 10.1083/jcb.87.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.