Abstract
Single gold-tagged epidermal growth factor (EGF) molecules bound to cellular EGF receptors of fixed fibroblast cells were imaged in liquid with a scanning transmission electron microscope (STEM). The cells were placed in buffer solution in a microfluidic device with electron transparent windows inside the vacuum of the electron microscope. A spatial resolution of 4 nm and a pixel dwell time of 20 μs were obtained. The liquid layer was sufficiently thick to contain the cells with a thickness of 7 ± 1 μm. The experimental findings are consistent with a theoretical calculation. Liquid STEM is a unique approach for imaging single molecules in whole cells with significantly improved resolution and imaging speed over existing methods.
Keywords: cellular imaging, molecular labels
Understanding cellular function at a molecular level requires imaging techniques capable of imaging whole cells with a resolution sufficient to image individually tagged proteins. Electron microscopy and X-ray diffraction are traditionally used to resolve the structures of individual proteins and to image proteins distributions in cells (1). Imaging with these techniques demands extensive sample preparation to obtain, e.g., proteins crystals, stained thin sections, or frozen samples. The cells are thus not in their native liquid state. Light microscopy is used to image protein distributions via fluorescent labels on fixed cells in liquid and in live cells to investigate cellular function (2). Superresolution techniques surpass the diffraction limit in optical microscopy (3–6), but despite recent advances, these methods are still restricted to spatial resolutions >10–20 nm. Further, their optimal performance requires extended imaging times, and significant data postprocessing. The speed can only be increased at the cost of resolution.
Here, we describe a direct technique for imaging whole cells in liquid that offers nanometer spatial resolution and a high imaging speed. The principle is explained in Fig. 1. The eukaryotic cells in liquid are placed in a microfluidic flow cell with a thickness of up to 10 μm contained between 2 ultrathin electron transparent windows. This flow cell is placed in the vacuum of a STEM, using a fluid specimen holder. The annular dark field (ADF) detector in the STEM is sensitive to scattered electrons, which are generated in proportion to the atomic number (Z) of the atoms in the specimen (7, 8), so-called Z contrast, where the contrast varies with ≈Z2. It is thus possible to image specific high-Z atoms, such as gold, inside a thick (several micrometer) layer of low-Z material, such as water, protein, or the embedding medium of a thin section (9). We used this approach to raster image single gold-tagged epidermal growth factor (EGF) molecules bound to cellular EGF receptors on fibroblast cells with a spatial resolution of 4 nm and a pixel dwell time of 20 μs.
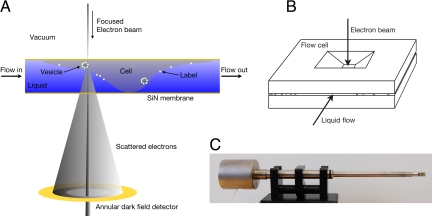
Fig. 1.
The principle of liquid scanning transmission electron microscopy (STEM). (A) A cell in liquid is enclosed between 2 electron-transparent windows of silicon nitride. Images are obtained by scanning a focused electron beam over the sample and detecting the elastically scattered electrons with an annular dark field detector. Labels made of a high atomic number material can be distinguished. (B) A microfluidic flow cell is formed from 2 silicon chips with silicon nitride windows spaced by microspheres. (C) The flow cell is placed in the vacuum of the microscope, using a fluid holder. The dimensions are not in proportion; in a typical experiment, the flow cell has a thickness of 10 μm, and the distance between the flow cell and the detector is 84 mm.
Results
COS7 fibroblast cells were labeled with 10-nm gold nanoparticles conjugated with epidermal growth factor (EGF-Au). The cells were grown, labeled, and fixed directly on the silicon nitride windows. Fig. 2A shows the edge of a cell that was incubated for 5 min with EGF-Au. Gold labels are visible as bright spots and the cellular material as light-gray matter over the dark gray background. Bright spots vary in size from 2 to 5 pixels. The broader spots are generally dimmer, thus suggesting that these spots represent labels that were not at the vertical position of the focus in the sample. The density of the gold labels at the edge of the cell is a factor of 10 larger than in the dark background region, indicating specific labeling. Individual labels are scattered over the cell apart from several clusters containing 2 or 3 labels and up to a maximum of 9 labels. Images were recorded at 11 different positions at cell edges and data were also recorded for two other flow cells, showing similar results (data not shown). The localization of the labels at the cell edges is consistent with EGF receptors dispersed along the cell surface after 5 min of label incubation (10). A fraction of 0.42 of the electron beam was scattered by an angle larger than 70 mrad into the ADF detector. The time needed to image Fig. 2A was 21 s for a 1,024 × 1,024 pixel image with a pixel dwell time of 20 μs.
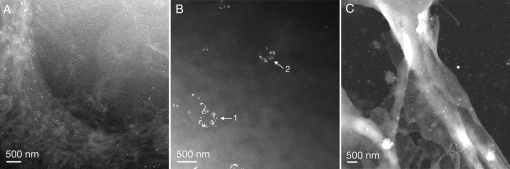
Fig. 2.
Liquid STEM images of COS7 fibroblast cells labeled EGF-Au. (A) Image of the edge of a fixed COS7 cell after 5-min incubation with EGF-Au. The labels are visible as bright spots and the cellular material is shown as light-gray matter on a dark-gray background. The pixel size was 5.7 nm. (B) Image of a COS7 cell incubated with EGF-Au for 10 min and incubated in buffer (without EGF-Au) for an additional 15 min. The pixel size was 4.4 nm. (C) Image of the sample used in A recorded after the flow cell was opened and the sample was dried in air. The pixel size was 8.9 nm. Note that the salt of this sample was not removed.
To observe molecular rearrangements in the COS7 cells, the cells were incubated for 10 min with EGF-Au followed by additional 15-min incubation in buffer. Liquid STEM images of these cells are shown in Fig. 2B. Circular clusters of labeled EGF receptors are visible. Imaging of a second flow cell with cells treated with the same protocol provided a similar result (data not shown). Sharp-edged gold labels are visible in the cluster at arrow 1, whereas the labels in the cluster at arrow 2 appear blurred and cannot be distinguished as individual labels, indicating that cluster 2 is out of the vertical plane of focus. The observation of circular clusters of labels at different vertical positions in the cell indicates that the labels were internalized. Circular clusters were not found in cells that were fixed after 5 min of incubation, where labels occurred individually, or in small groups of 2 - 9 labels (see Fig. 2A and Figs. S1–S4). The clustering of the EGF receptors and internalization of the labels is consistent with the known behavior of EGF-activation of EGF receptors, which cluster as internalized endosomes upon receptor activation (10). These images showed less contrast of the gold labels than in Fig. 2A. The detector semiangle was increased to 94 mrad to optimize the visibility of the labels. In this case, a fraction of 0.37 of the electron beam was scattered into the detector.
After STEM imaging, the flow cell used for Fig. 2A was opened and inspected for the presence of liquid, which was verified visually. The sample was then dried in air and imaged in the STEM (Fig. 2C). The fraction of electrons scattered into the detector was <3%, which is much less than for the liquid samples and is expected for a dry sample in the absence of a liquid layer. A further difference between the dry sample and the liquid sample is the much stronger contrast on the cellular material in the dry sample. The white shades in the image show the edge of the dried cell, and the gold labels are still visible as brighter spots. Several other features are visible on the silicon nitride membrane, such as salt crystals and other debris.
To measure the resolution of liquid STEM imaging, we examined several EGF-Au nanoparticles (Fig. 3 A and B). The line-profile over an individual gold nanoparticle is shown in Fig. 3C. As measure of the resolution, we determined the width in which the signal rose from 20% to 80%, i.e., the 20–80% edge width. Averaged over 5 gold nanoparticles, this value was determined to be 3.9 ± 0.4 nm, using both the left and right sides of the peak. The diameter of the electron probe in vacuum for these images is 1 nm, but the probe beam is broadened by interactions of the electron beam with the liquid. In addition, the scattering function depends on both the shape and electron density of the individual gold nanoparticle. The shape of the electron probe beam and the electron scattering probability affects the line-profile. Because the measured value contains both of these contributions, it can be concluded that the resolution of STEM with our liquid flow cell is 4 nm or better.
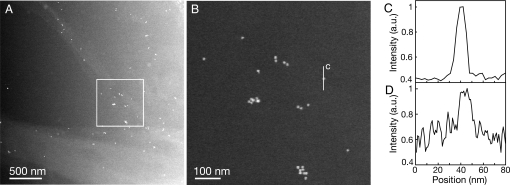
Fig. 3.
Resolution in Liquid STEM. (A) Image of a COS7 cell edge labeled with EGF-Au. The pixel size was 2.9 nm. (B) Magnified image of area in A showing the individual labels. (C) Line-scan of the detector signal as function of the position over a single gold nanoparticle indicated by the line “c” in B. (D) Line-scan over a gold nanoparticle of 10 nm diameter at the bottom of 1.3 μm liquid (10% PBS buffer in water) in a test sample without cells. The pixel size was 0.91 nm. See also Fig. S5.
The contrast obtained with liquid STEM on labels is reduced when the vertical position of the label is further down in the liquid because of electron-sample interactions leading to beam blurring. A test sample with gold nanoparticles on the bottom silicon nitride window was prepared to measure the effect of beam blurring (see Fig. S5). It was found that 10-nm diameter gold nanoparticles could still be imaged with sufficient contrast below 1.3 μm of water. A line-scan of a 10-nm gold nanoparticle (Fig. 3D) revealed a decreased signal-to-noise ratio. However, the 20–80% edge resolution amounted to 4.5 ± 0.5 nm, which is equal within the error of the measurement to the resolution of 4 nm measured in upper regions of the liquid.
For comparison of the resolution obtained with liquid STEM with the resolution of confocal laser microscopy COS7 cells were incubated for 5 min with quantum-dot-labeled epidermal growth factor. The optical resolution of the smallest single points (consistent with the brightness of single quantum dots in our instrument) was ≈200 nm as expected for the 40 × 1.3 NA objective lens and 1 airy unit confocal pinhole diameter (11). Fig. 4A shows the combined images obtained with fluorescence signals and with differential interference contrast (DIC). Quantum dots are visible at the edges of the cells. Fig. 4B shows a close-up of the fluorescent labels and demonstrates that it is not possible to elucidate whether the bright spots are generated by individual labels or by clusters with the available resolution. For comparison between confocal laser microscopy and liquid STEM a second close-up is shown in Fig. 4C, that is the same image size as Fig. 3A. The resolution of liquid STEM is superior to that of confocal microscopy by more than a factor of 50.
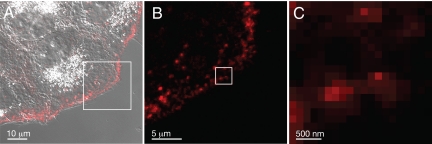
Fig. 4.
Confocal laser microscope images of fixed COS7 cells incubated for 5 min with EGF- Qdot 655. The pixel size was 140 nm. (A) Overlay of the fluorescent image and the image obtained with differential interference contrast (DIC). (B) Magnified image of the same group of cells as in A indicating EGF binding at the surface of the cell and showing the maximal useful resolution of the confocal microscope. (C) Further magnified image of B, resulting in an image of the same size as Fig. 3A obtained with liquid STEM, demonstrating the difference in resolution between confocal laser microscopy and liquid STEM.
Theory of Liquid STEM Imaging.
The STEM images of the annular dark field (ADF) detector are primarily formed by electrons that are elastically scattered on the atoms in the specimen. The number N of electrons scattered into the ADF detector with semiangle β can be calculated using the partial cross section for elastic scattering σ(β) (12). For a certain thickness of the material T, this number is given by:
with N0 being the number of incident electrons, mean-free-path length for elastic scattering l, mass density ρ, the atomic weight W, and Avogadro's number NA. The partial cross section for elastic scattering can be estimated by integration of the differential cross section dσ/dΩ assuming a simple screened Rutherford scattering model based on a Wentzel potential (12):
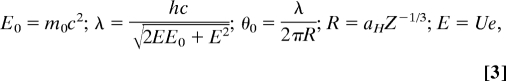 |
with electron accelerating voltage U (in V), atomic number Z, aH the Bohr radius, m0 the rest mass of the electron, c the speed of light, h Planck's constant, and e the electron charge.
The images Figs. 2A and 3A were recorded with β = 70 mrad and U = 200 kV. For these instrument settings, we find that lgold = 73 nm. For the water medium it follows that lwater = 10.5 μm, using the average Z number of water of 4.7 (13). The fraction of N/N0 = 0.42 was measured (ratio of detector currents for wet and dry sample). Using Eq. 1, the fraction can be translated into a thickness T = 5.7 μm of the water layer. For Fig. 2B the settings were different and β = 94 mrad. The fraction of N/N0 was 0.37, corresponding with a water layer of T = 8.6 μm. This calculation provides values of the thickness of the sample that agree with the thicknesses of the cells measured with the confocal microscope of 7 μm within 20%.
The calculated thickness of T = 5.7 μm obtained from images at the edges of cells is smaller than the thickness that would be expected on the basis of the spacer between the 2 silicon chips formed by polystyrene microspheres of 10 μm at the 4 corners of the chips. A possible explanation of this discrepancy is a compression of the microspheres and a minor deformation of the silicon chips as a result of the pressure applied by the lid of the fluid specimen holder to maintain a vacuum seal by the O-rings (note that the silicon nitride windows bulge outside into the vacuum). The salt concentration was neglected in this calculation, because 10% PBS buffer would increase l by <1% and thus the calculation of the liquid surrounding the cells, using water only is of sufficient precision.
We will now calculate the resolving power of liquid STEM, which can be considered as the sample-limited resolution. When the electron beam is scanned over a gold nanoparticle with thickness z in a water layer with thickness T, the ADF detector receives both the electrons scattered by the particle and those scattered from the water, resulting in Nsignal electrons (12).
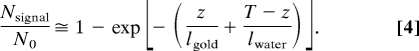 |
For our experiment, z amounts to 10 nm and T varies from 5–10 μm; therefore, we can assume that T − z ≈ T and that both z/lgold and T/lwater are small numbers. Using the first-order Taylor expansion of the exponential function, we can thus write:
When the beam is shifted just from the particle, the detector receives only Nbkg background electrons:
The key issue in the detection of a gold particle is to “see” it with sufficient electrons such that it “lights-up” above the noise in the background signal produced by the water. In other words, it has to be detected at a sufficient confidence level. Assuming 100% detection efficiency and assuming Poisson statistics, the noise in the detection is given by and the signal-to-noise ratio (SNR) becomes (14):
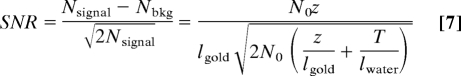 |
If we then use the relation z/lgold ≪ T/lwater, the SNR becomes:
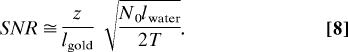 |
According to the Rose criterion the signal should be at least a factor χ = 5 larger than the noise to be able to detect 1 pixel (15–17). Thus, we can calculate the minimum particle height z that can be detected within a water layer of thickness T given a certain amount of electrons:
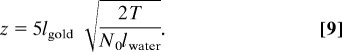 |
The value of z can be considered as the (sample-related) resolution of liquid STEM imaging of a nanoparticle in a liquid layer. For the experimental conditions of Figs. 2A and 3A it follows that z = 1.3 nm for T = 5 μm and z = 1.9 nm for T = 10 μm. This calculation theoretically confirms our observation that gold nanoparticles of a size of 10 nm can be detected in water layers of up to 10-μm thickness.
Discussion
We have demonstrated that 10-nm gold labels can be detected in a thick liquid sample by STEM with excellent contrast, and this experimental finding was verified by a theoretical calculation. The high signal-to-noise ratio in our figures also suggests that smaller gold nanoparticles will be detectable as well. The theoretical resolution limit calculated in the above is 1.9 nm for a water thickness of 10 μm. We expect that this limit can be achieved at similar and at lower electron doses as used here, by further optimizing the detector efficiency in the presence of a high background signal, and applying noise reduction techniques. The electron dose used for 1 image was estimated to be 740 electrons/Å2 or less, which is an order of magnitude larger than the maximum dose of 80 electrons/Å2 for imaging frozen samples in tilt-series TEM (18). The electron dose increases with the square of the resolution (14, 16), so imaging with a lower resolution can be used to reduce the electron dose. The continuous exchange of the liquid in the flow cell may have been beneficial by removing free electrons, radicals and heat generated by the impact of the electron beam, thereby limiting beam damage. The resolution of 4 nm also opens the possibility to measure the distance between labels that are in close proximity. Labels of different sizes and materials could be used to perform experiments in which different proteins or other biomolecules are labeled with specific and distinguishable labels. Imaging may also be conducted with an aberration corrected STEM, to provide the ability to image in 3 dimensions with an axial resolution of several nanometers (19). The axial resolution obtained with our noncorrected microscope was calculated to be <80 nm.
The goal of nanometer resolution on whole cells in their native liquid environment dates back to the early days of electron microscopy, see (20) and references therein. The high spatial resolution of liquid STEM obtained on sample volumes compatible with whole eukaryotic cells is not achievable with a liquid cell for TEM imaging (21), because the TEM contrast mechanism is limited to sample thicknesses typically <1 μm. In the case of such thin and weakly scattering samples, TEM yields better resolution and signal-to-noise ratio than STEM (22), but in our case, STEM offers a particular advantage based on the imaging of high-Z labels. A liquid compartment (23) placed in a scanning electron microscope (SEM) provides a resolution of ≈20 nm on gold labels, but this is a surface technique that images the top 100 nm of the sample. The resolution we achieved through a thick liquid layer also surpasses alternative approaches, such as imaging of cooled cells in water vapor, using an environmental TEM (24, 25), environmental SEM with a STEM detector (26), X-ray microscopy (27), or atomic force microscopy (28).
For fixed samples, liquid STEM presents an alternative to fluorescence. The resolution of liquid STEM is a factor of 50 higher than that of confocal microscopy, as illustrated in Fig. 4. Current ultrahigh-resolution optical methods such as stimulated emission depletion (STED) (3, 4), photo-activated localization microscopy (PALM) (5) and stochastical optical reconstruction microscopy (STORM) (6) offer subdiffraction limit images. Those techniques can obtain fluorescence images of fixed cells, with lateral resolutions reaching 10–20 nm and sub-100-nm vertical resolution (29). However, this resolution is still not sufficient to image individually tagged proteins as needed to understand cellular function on a molecular level. Another disadvantage of ultrahigh-resolution optical methods is the limited imaging speed (e.g., images can tens of minutes to obtain the highest resolution). In our initial work, we have already achieved a resolution of 4 nm, while providing imaging at 20 μs pixel dwell time (20 s for a 1,024 × 1,024 pixel image, 5 s for 512 × 512). Thus, liquid STEM presents a unique approach for imaging single molecules in whole cells, which is significantly improved over existing methods in spatial resolution and imaging speed. In addition, liquid STEM is a real-time technique that does not require any data postprocessing, which is needed for all of the current high-resolution optical methods. It is noteworthy that a STEM and the fluid holder are straightforward to operate.
A major advantage of optical microscopy is the capability to image time-evolving cellular processes (2), but improving temporal resolution comes at the expense of spatial resolution. PALM imaging on live cells has been shown a temporal resolution of 300 s with a spatial resolution of ≈40 nm (30), or down to 25 s with a resolution of 60 nm (31). Likewise, live-cell STED imaging was accomplished at video frequency (a pixel dwell time of 4 μs), but at a reduced resolution of 60 nm (32). It has already been shown that cells can survive and grow in microfluidic chambers such as the ones we are using (33). Although electron microscopy is not similarly compatible with live cell imaging, one can imagine uses for liquid STEM to acquire a nanometer resolution “snapshot” of molecular localizations. Because of the relatively short imaging times needed in liquid STEM, such a snapshot would allow elucidation of specific molecular patterns, which could be usefully interpreted in combination with time-lapse live cell fluorescence data.
Finally, the capabilities of liquid STEM will also be important for chemistry and materials science by allowing direct imaging of liquid:solid interfaces. The liquid thickness of up to 10 μm allows the imaging of micrometer-sized objects and even functional units, such as a micrometer-sized battery, to be probed with nanometer resolution.
Methods
Construction and Preparation of the Silicon Chips.
Custom designed silicon chips with silicon nitride windows (Protochips Inc, NC) were used to form a liquid enclosure. They were fabricated using low stress silicon nitride of 50-nm thickness deposited with a low-pressure chemical vapor deposition process onto 300-μm-thick silicon wafers. Openings in the silicon wafer were obtained by anisotropic etching in a heated bath of KOH, leaving open silicon nitride membrane windows with an area of 0.2 × 0.05 mm2. One set of chips was used for cell growth. A chip was cleaned with acetone and ethanol. It was then plasma cleaned and coated with a thin layer of polyL-lysine applied to make the window hydrophilic and to enhance the attachment of the cells. Another set of chips was used as covers of the liquid enclosure. For spacing between a chip with cells and a cover chip, a solution of polystyrene microspheres of 10-μm thickness was placed in droplets at the 4 corners of the chip and the solution was dried.
Placement of a Liquid Sample in the Electron Microscope.
A STEM/TEM specimen holder (Hummingbird Scientific) was designed for placing a liquid sample in the vacuum of the electron microscope. The specimen holder fitted 2 chips in a slot with their silicon nitride surfaces facing, such that the silicon nitride windows in the middle of both chips overlapped. The holder contained 2 fluid lines (one input and one output) connected to the chips. The fluid lines were fed to plastic tubing for microfluidics (Upchurch Scientific). One tube was connected to a syringe pump (Harvard Scientific, MA). Liquid flow occurred between the 2 chips and through a bypass channel. For loading of the sample in the fluid holder, a cover chip was first placed in the slot of the holder with its silicon nitride surface facing up. A droplet of 50/50 glycerol/10% PBS buffer was placed on the cover chip. The chip with cells was transferred from its storage in liquid to the fluid holder and placed on top of the cover chip with the cells facing down. The silicon nitride surfaces of both chips were kept under liquid continuously. The use of 50% glycerol prevented rapid evaporation of the liquid during loading. The tip of the fluid holder was closed by a coverlid and a vacuum seal was obtained using O-rings above and below the silicon chips. A flow of 10% PBS buffer of 2 μL/min was initiated at the input tube of the fluid holder until liquid appeared at the exit tube typically a few minutes later. The holder was then inspected for leaks visually in a vacuum test chamber. After this inspection the flow rate was maintained at 2 μL/min and the holder was transferred to the electron microscope.
Imaging and Analysis.
Electron microscope images were recorded with a CM200 TEM/STEM (Philips) at 200 kV, using the ADF detector. The microscope was set for an electron probe semiangle of 11 mrad, a probe current of 0.59 nA (measured on the phosphor screen), a detector semiangle of 70 mrad, and a pixel dwell time of 20 μs. The probe diameter obtained with these settings was 1 nm. The images on liquid biological specimen were recorded at various magnifications and with pixel sizes between 2.9 and 5.7 nm. A conservative estimate of the electron dose is thus 7.4 × 102 electrons/Å2. Liquid STEM imaging of the COS7 cells took place in 10% PBS buffer in water at magnifications varying between 8,000 and 48,000, while the flow rate in the fluid system was 0.1 μL/min. Regions of interest were selected from the recorded images of a size of 1024 × 1024 pixels. The contrast and brightness were adjusted for maximal visibility of the labels and the cells. The liquid STEM images presented throughout this article represent the original data and no procedures were applied to reduce the noise. STEM images of dried samples in Figs. S1–S4 were enhanced by applying a despeckle procedure to reduce the noise.
Confocal imaging was performed on a Zeiss LSM510 microscope equipped with a 40 × 1.3 NA oil-immersion objective. A 488-nm argon laser was used for excitation of the quantum dots and fluorescence was detected through a 650- to 710-nm band-pass emission filter. The image size was 1,024 × 1,024 pixels. Images at a single focal plane were selected. The thickness of the fixed COS7 cells was determined by recording z stacks and determining the distance between the lowest and the highest focal plane still containing fluorescent signals. A total of 44 cells were imaged and the average thickness was 7.0 ± 0.7 μm.
Labeling of COS7 Cells with EGF-Gold Nanoparticles.
We optimized existing procedures (34, 35) to gold-label EGF receptors on living COS7 cells. Gold-labeled streptavidin (KPL) was diluted in PBS containing 0.5% BSA (PBS-BSA). The gold particles were washed twice by centrifugation and resuspension of the gold pellet. A 16 nM gold nanoparticle solution in PBS-BSA was incubated with 0.4 μM Biotin-EGF (Invitrogen) for 1 h at 35 °C. Unbound biotin-EGF was removed using a size exclusion column. The filtrate, containing EGF-gold nanoparticles (EGF-Au) was diluted with Tyrode's buffer, supplemented with 50 mM glucose and 0.5% BSA (Tyrode's-BSA), washed once and resuspended to yield 5 nM EGF-Au in Tyrode's-BSA.
COS7 cells (African Green Monkey fibroblast) were cultured in DMEM (ATCC), supplemented with 10% FBS, in a 5% CO2 atmosphere, at 37 °C. Confluent COS7 cells were harvested by rinsing in Dulbecco's PBS and dissociating the adherent layer with CellStripper (Mediatech), followed by a quench in supplemented media. Poly-l-lysine coated silicon chips were placed at the bottom of the wells of a 96-well plate, filled with 200 μL of supplemented media, 20 μL of harvested cells in suspension were added per chip. The chips with the cells were incubated for at least 4 h or overnight, in a 5% CO2 atmosphere, at 37 °C.
For EGF-Au labeling, the medium in the wells was exchanged by serum free DMEM. After 4 h of incubation in serum free medium, cells were washed once with Tyrode's-BSA. Eleven-microliter droplets of EGF-gold nanoparticle solution were placed in plastic wells and 1 silicon chip per droplet was placed, inclined upside down on the droplet. The setup with droplets and chips was then stored in a closed box with a 100% humidity environment. The chips remained in this environment for either 5 or 10 min at room temperature with slight agitation. The 5-min samples were washed 3 times with PBS and fixed for 15 min in 4% glutaraldehyde diluted in PBS. The 10-min samples were placed in a new well, filled with Tyrode's-BSA (without EGF-gold nanoparticles) and incubated for an additional 15 min, before washing and fixation. The chips were then washed 3 times with PBS, once with 10% PBS in water, incubated for 5 min in 100 mM Glycine to quench un-reacted aldehyde groups after fixation, washed twice with 10% PBS and left in this solution at 4 °C until imaging.
STEM images were recorded of 3 dried samples with (i) 5-min incubation with EGF-Au, (ii) 10-min incubation with EGF-Au and an additional 15-min incubation in buffer, and (iii) a control with 10-min incubation with streptavidin-Au in buffer and 15-min incubation in buffer. Figs. S1–S4 demonstrate that specific labeling of the EGF receptor was obtained. The existence of circular vesicles after 10 + 15 min incubation indicates that internalization of the receptor took place (10). The size of the gold labels was determined from Fig. S3 and amounted to 10 nm (full width at half maximum of a line scan over the image of a gold nanoparticle).
Labeling of COS7 Cells with EGF-QD.
EGF-Qdot655 (6:1) labels were prepared by mixing 10 μL of 6 μM EGF-biotin (Invitrogen) and 10 μL of 1 μM Qdot655-streptavidin (Invitrogen) in 80 μL of 50 mM sodiumborate pH 8.3. After 2 h shaking at room temperature, unbound EGF-biotin was removed using a Microcon YM-100 column (Millipore). The EGF-Qdot655 pellet was resuspended in 100 μL of sodium borate buffer to make a 100 mM stock solution. COS7 cells were grown in 35-mm dishes containing No. 1 glass coverslips (MatTek) in standard DMEM (Invitrogen) supplemented with 10% FBS and were incubated in serum-free medium for 4 h before the labeling. Cells were labeled with 5 nM EGF-Qdot655 in BMHH buffer [125 mM NaCl, 5.7 mM KCl, 2.5 mM CaCl2, 1.2 mM MgCl2, 0.1% BSA, 10 mM glucose, 10 mM Hepes (pH 7.4)] for 5 min at 37 °C. The cells were then washed 3 times with BMHH buffer and either fixed immediately with 2% formaldehyde or incubated another 25 min at 37 °C before fixation.
Supplementary Material
Acknowledgments.
We thank D. C. Joy and S. J. Pennycook for discussions and J. Bentley, W. Bigelow, C. Chisholm, M. Cole, R. Dona, M. J. Dukes, C. J. Easley, W. S. Head, P. S. Herrell, T. E. McKnight, K. L. More, E. A. Ring, and G. M. Veith for discussions and help with the experiments. We thank Hummingbird Scientific for providing the fluid holder and Protochips Inc. for providing the silicon chips. This work was supported by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory, managed by UT-Battelle, LLC for the U.S. Department of Energy under Contract DE-AC05-00OR22725; Shared Research Equipment (SHARE); Vanderbilt University Medical Center; and National Institutes of Health Grants R01-RR018470 (to P. Mazur for D.B.P.) and P20-GM072048 (to D.W.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809567106/DCSupplemental.
References
- 1.Sali A, Glaeser R, Earnest T, Baumeister W. From words to literature in structural proteomics. Nature. 2003;422:216–225. doi: 10.1038/nature01513. [DOI] [PubMed] [Google Scholar]
- 2.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 3.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 4.Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW. STED microscopy reveals that synapthotagmin remains clustered after synaptic vesicle exocytosis. Nature. 2006;440:935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- 5.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 6.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crewe AV, Wall J, Langmore J. Visibility of single atoms. Science. 1970;168:1338–1340. doi: 10.1126/science.168.3937.1338. [DOI] [PubMed] [Google Scholar]
- 8.Mueller SA, Engel A. Biological scanning transmission electron microscopy: Imaging and single molecule mass determination. Chimia. 2006;60:749–753. [Google Scholar]
- 9.Sousa AA, Hohmann-Marriott M, Aronova MA, Zhang G, Leapman RD. Determination of quantitative distributions of heavy-metal stain in biological specimens by annular dark-field STEM. J Struct Biol. 2008;162:14–28. doi: 10.1016/j.jsb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lidke DS, et al. Quantum dot ligands provide new insights into erB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 11.Sandison DR, Piston DW, Williams RM, Web WW. Quantitative comparison of background rejection, signal-to-noise ratio, and resolution in confocal and full-field laser-scanning microscopes. Applied Optics. 1995;34:3576–3588. doi: 10.1364/AO.34.003576. [DOI] [PubMed] [Google Scholar]
- 12.Reimer L. Transmission Electron Microscopy. Heidelberg, Germany: Springer; 1984. [Google Scholar]
- 13.Joy DC, Joy CS. Scanning electron microscope imaging in liquids – some data on electron interactions in water. J Microscopy. 2005;221:84–99. doi: 10.1111/j.1365-2818.2006.01548.x. [DOI] [PubMed] [Google Scholar]
- 14.Spence JCH. High-Resolution Electron Microscopy. Oxford: Oxford Univ Press; 2003. [Google Scholar]
- 15.Rose A. Television pickup tubes and the problem of noise. Adv Electron. 1948;1:131–166. [Google Scholar]
- 16.Colliex C, Jeanguillaume C, Mory C. Unconventional modes for STEM imaging of biological structures. J Ultrastruct Res. 1984;88:177–206. doi: 10.1016/s0022-5320(84)80007-6. [DOI] [PubMed] [Google Scholar]
- 17.Isaacson M, Johnson D, Crewe AV. Electron beam excitation and damage of biological molecules; its implications for specimen damage in electron microscopy. Rad Res. 1973;55:205–224. [PubMed] [Google Scholar]
- 18.Iancu CV, Wright ER, Heymann JB, Jensen GJ. A comparison of liquid nitrogen and liquid helium as cryogens for electron cryotomography. J Struct Biol. 2006;153:231–240. doi: 10.1016/j.jsb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 19.De Jonge N, Sougrat R, Peckys DB, Lupini AR, Pennycook SJ. In: Nanotechnology in Biology and Medicine-Methods, Devices and Applications. Vo-Dinh T, editor. Boca Raton, FL: CRC; 2007. pp. 13.11–13.27. [Google Scholar]
- 20.Parsons DF. Structure of wet specimens in electron microscopy. Science. 1974;186:407–414. doi: 10.1126/science.186.4162.407. [DOI] [PubMed] [Google Scholar]
- 21.Williamson MJ, Tromp RM, Vereecken PM, Hull R, Ross FM. Dynamic microscopy of nanoscale cluster growth at the solid-liquid interface. Nat Mater. 2003;2:532–536. doi: 10.1038/nmat944. [DOI] [PubMed] [Google Scholar]
- 22.Rez P. Comparison of phase contrast transmission electron microscopy with optimized scanning transmission annular dark field imaging for protein imaging. Ultramicroscopy. 2003;96:117–124. doi: 10.1016/S0304-3991(02)00436-9. [DOI] [PubMed] [Google Scholar]
- 23.Thiberge S, et al. Scanning electron microscopy of cells and tissues under fully hydrated conditions. Proc Natl Acad Sci USA. 2004;101:3346. doi: 10.1073/pnas.0400088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daulton TL, Little BJ, Lowe K, Jones-Meehan J. In situ environmental cell—transmission electron microscopy study of microbial reduction of chromium(VI) using electron energy loss spectroscopy. Microsc Microanal. 2001;7:470–485. doi: 10.1017.S1431927601010467. [DOI] [PubMed] [Google Scholar]
- 25.Gai PL. Development of wet environmental TEM (wet-ETEM) for in situ studies of liquid-catalyst reactions on the nanoscale. Microsc Microanal. 2002;8:21–28. doi: 10.1017/s143192760201005x. [DOI] [PubMed] [Google Scholar]
- 26.Bogner A, Thollet G, Basset D, Jouneau PH, Gauthier C. Wet STEM: A new development in environmental SEM for imaging nano-objects included in a liquid phase. Ultram. 2005;104:290–301. doi: 10.1016/j.ultramic.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Legros MA, McDermott G, Larabell CA. X-ray tomography of whole cells. Curr Opin Struct Biol. 2005;15:593–600. doi: 10.1016/j.sbi.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Chtcheglova LA, Waschke J, Wildling L, Drenckhahn D, Hinterdorfer P. Nano-scale dynamical recognition imaging on vascular endothelial cells. Biophys J. 2007;93:L11–13. doi: 10.1529/biophysj.107.109751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juette MF, et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 30.Hess ST, et al. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminating between raft theories. Proc Natl Acad Sci USA. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shroff H, Galbraith CG, Galbraith JA, Betzig E. LIve-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Westphal V, Rizzoli SO, Lauterbach MA, Kamin D, Jahn R, Hell SW. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- 33.Keymer JE, Galajda P, Muldoon C, Park SH, Austin RH. Bacterial metapopulations in nanofabricated landscapes. Proc Natl Acad Sci USA. 2006;103:17290–17295. doi: 10.1073/pnas.0607971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibaraki N, Lin LR, Reddy VN. A study of growth factor receptors in human lens epithelial cells and their relationship to fiber differentiation. Exp Eye Res. 1996;63:683–692. doi: 10.1006/exer.1996.0162. [DOI] [PubMed] [Google Scholar]
- 35.Driskell OJ, Mironov A, Alayan PM, Woodman PG. Dynein is required for receptor sorting and the morphogenesis of early endosomes. Nat Cell Biol. 2007;9:113–120. doi: 10.1038/ncb1525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.