Abstract
An accepted prerequisite for clinical trials of a compound in humans is the successful alleviation of the disease in animal models. For some diseases, however, successful translation of drug effects from mouse models to the bedside has been limited. One question is whether the current models accurately reproduce the human disease. Here, we examine the mouse models that are available for therapeutic testing in Huntington disease (HD), a late-onset neurodegenerative disorder for which there is no effective treatment. The current mouse models show different degrees of similarity to the human condition. Significant phenotypic differences are seen in mouse models that express either truncated or full-length human, or full-length mouse, mutant huntingtin (mHTT). These differences in phenotypic expression may be attributable to the influences of protein context, mouse strain and a difference in regulatory sequences between the mouse Htt and human HTT genes.
The vision: from mechanisms in mice to therapies in humans
The most important rationale for modelling human disorders in a non-human organism is the identification of fundamental pathogenic mechanisms that lead to novel therapeutic targets, and the evaluation of the efficacy and safety of potential new drugs. Historically, mice have been used to model human disease because of their physiological, anatomical and genomic similarities to humans. For neurodegenerative diseases, the mammalian neuronal physiology and the human-like brain anatomy makes mouse models especially useful. Nevertheless, their short life span of approximately 2 years is a serious limitation for late-onset neurodegenerative disease and the nervous system of humans is significantly more similar to that of primates. However, the costs and logistics of performing large-scale therapeutic trials in non-human primates are prohibitive. The question therefore remains about whether any positive outcome from therapeutic trials in mice is sufficient evidence to proceed with studies in humans. An alternative question is whether human trials should commence when a drug against a compelling target has been identified that did not show efficacy in mice.
The reality: the predictive value of therapeutic trials in mice
For amyotrophic lateral sclerosis (ALS), the most widely used mouse model has proven to be of poor predictive value: approximately 3% of all cases of ALS are caused by point mutations in the superoxide dismutase (SOD1) gene, and the most commonly used ALS mouse model expresses human mutant SOD1 (G93A) as a transgene (Gurney et al., 1994). A recent review summarized 78 individual and 12 combination compound trials that have been conducted using this mouse model (Benatar, 2007), which subsequently led to 11 double-blind, placebo-controlled clinical trials in humans, all of which failed (Vincent et al., 2008). Even compounds such as creatine, minocycline or cyclooxygenase-2 inhibitors that were most promising in mouse trials (Klivenyi et al., 1999; Drachman et al., 2002; Zhu et al., 2002) have not yet been shown to be beneficial in human clinical trials (Groeneveld et al., 2003; Shefner et al., 2004; Cudkowicz et al., 2006; Gordon et al., 2007). Riluzole remains the only FDA approved drug for ALS (Miller et al., 2003).
The SOD1 mouse model more closely resembles familial than sporadic ALS and the possibility of biologically relevant differences between these forms of the disorder might limit the usefulness of the model. In addition, treatment in mice is commonly started before the onset of symptoms; this cannot be replicated in predominantly sporadic human diseases, such as ALS. These serious limitations apply to all diseases where familial and sporadic forms exist, such as Alzheimer’s and Parkinson’s diseases.
Therapeutic trials for Huntington disease (HD), however, have significant advantages: the expansion of a polyglutamine tract in the huntingtin HTT protein has been determined as the single cause of the disease (The Huntington’s Disease Collaborative Research Group, 1993). Therefore, it should be easier to generate an accurate mouse model of HD than for other conditions with multiple, often unknown, causes. Furthermore, predictive testing is available to accurately predict whether, and when, a person will be affected (Tibben, 2007), making presymptomatic treatment in humans a feasible option. To assess the efficacy of potential therapeutics in the HD mouse model, multiple endpoints have been established that allow the monitoring of neuropathological and behavioural changes in the YAC128 HD mouse model (Table 1) (Slow et al., 2003; Van Raamsdonk et al., 2005a; Van Raamsdonk et al., 2005b).
Table 1.
Neuropathological and behavioural endpoints for evaluation of therapeutic candidates in the YAC128 HD mouse model
| Primary endpoints
| |||
|---|---|---|---|
| Neuropathological | Behavioural Motor function | ||
| Striatal volume loss | Rotarod abnormalities* | ||
| Striatal neuronal loss | |||
| Secondary endpoints
| |||
|---|---|---|---|
| Pathological | Behavioural | ||
| Neuropathology | Peripheral pathology | Motor function | Cognitive function |
| Reduction in average volume of striatal neurons | Testicular degeneration | Open-field activity | Pre-pulse inhibition |
| Brain weight loss | |||
| Cortical volume loss | |||
| Nuclear translocation of mutant HTT | |||
| Caspase-6 activation | |||
Deficits on the fixed and accelerating rotarod. References: Slow et al., 2003; Van Raamsdonk et al., 2005a; Van Raamsdonk et al., 2005b.
HD mouse models: too much choice?
HD is characterised by the progressive deterioration of cognitive and motor functions, and the selective loss of GABAergic medium spiny striatal neurons, as well as glutamatergic cortical neurons that project to the striatum (Albin et al., 1990). Aberrant proteolysis of mutant HTT (mHTT) has been proposed to play a role in HD (Wellington et al., 2000; Gafni et al., 2004; Graham et al., 2006), leading to the accumulation of N-terminal mHTT fragments in intracellular inclusions (DiFiglia et al., 1997).
Huntingtin fragment mouse models
HD is characterised by its late onset, therefore reproducing the human HD phenotype within the life span of a mouse represents a major challenge. Different approaches have been taken to increase the severity of the disease phenotype in various models. The R6/2 and N-171 mouse models use the fact that the expression of N-terminal mHTT fragments causes acute neuronal toxicity, by expressing truncated human mHTT consisting of either the first of 67 exons (Mangiarini et al., 1996) or the N-terminal 171 amino acids (Schilling et al., 1999). The drawbacks of these models are the loss of the natural genomic and protein context of the polyglutamine expansion, which could lead to altered regulation and a loss of potential disease-modifying post-translational modifications and protein interactions. Additionally, it has been shown that mHTT fragments generated by intracellular cleavage have a different subcellular localisation than truncated mHTT (Warby et al., 2008). It is therefore possible that the process of mHTT cleavage is in itself relevant to the pathogenesis of HD. In other words, although certain truncated fragments may cause neuronal toxicity, this might not necessarily recapitulate the disease.
The R6/2 fragment mouse model (Mangiarini et al., 1996) has been used in a number of therapeutic trials, and many compounds that have been effective in R6/2 mice have proceeded to clinical trials, with mixed results. Riluzole, remacemide and CoQ10 have successfully ameliorated the disease phenotype in R6/2 mice (Schilling et al., 2001; Ferrante et al., 2002; Schiefer et al., 2002), but were not beneficial for patients in the corresponding clinical trials (Huntington Study Group, 2001; Landwehrmeyer et al., 2007). The clinical trial of CoQ10 showed a trend towards a slowing in the decline of functional capacity, although it was not statistically significant (Huntington Study Group, 2001).
Full-length huntingtin mouse models
Murine models expressing full-length human mHTT were first reported in 1999 with the generation of the YAC46 and YAC72 models (Hodgson et al., 1999). In an effort to create a mouse model with more robust and earlier behavioural abnormalities that would be better suited for therapeutic trials, an additional YAC transgenic mouse was established in which the mHTT contained 128 glutamines (Slow et al., 2003). These mice show a uniform phenotype with age-dependent striatal and subsequent cortical neurodegeneration (Van Raamsdonk et al., 2005a), and have progressive motor and cognitive deficits (Van Raamsdonk et al., 2005b). A progressive deficit on the rotarod is observed in this line starting at 2–4 months of age, which correlates highly with neuronal loss in the striatum (Slow et al., 2003).
Trials of several candidate therapeutic compounds have been performed in the YAC128 mice: ethyl-EPA showed a modest rescue of the behavioural deficits (Van Raamsdonk et al., 2005d), whereas a trial of cystamine demonstrated protection from neuronal loss but no effect on the motor phenotype (Van Raamsdonk et al., 2005c). In human patients, ethyl-EPA led to a slight improvement of motor dysfunction as well as a reduction in cerebral atrophy in the caudate and thalamus (Puri et al., 2005), which was not seen in the YAC128 mice. Tetrabenazine had positive effects on the behavioural phenotype in mice (Tang et al., 2007), which is in agreement with the results from a clinical trial (Huntington Study Group, 2006). However, the true predictive value of this mouse model will only be established when additional compounds are tested and proceed to clinical trials.
Recently, a bacterial artificial chromosome (BAC) mouse expressing full-length human mHTT was established (BACHD mice) (Gray et al., 2008). The BACHD mice exhibit progressive motor deficits and late-onset selective neurodegeneration in the cortex and striatum, consistent with the findings in the YAC128 animals. Similar to the YAC model, the BACHD mice are also inherently well suited for therapeutic trials.
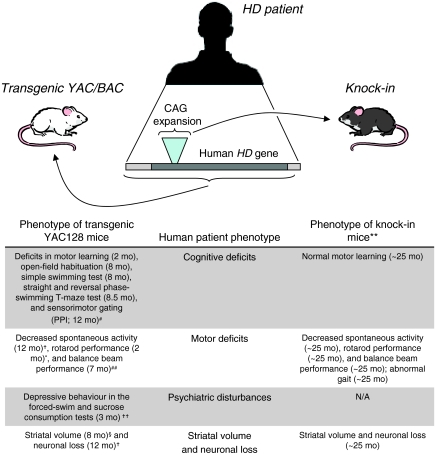
The knock-in mouse model of HD expresses mHTT in the most appropriate genomic and protein context, since it has a polyQ sequence inserted into the endogenous mouse Htt gene (Shelbourne et al., 1999). These mice display a very late-onset phenotype, which is strongest in the 150 CAG homozygotes (Heng et al., 2007). A rotarod deficit was reported at ~25 months of age and occurs in parallel with a reduction in striatal neuron counts and striatal volume (Heng et al., 2007), representing a protracted time course compared with the behavioural phenotypes observed in the YAC128 and BACHD animals (Fig. 1). mHTT expression levels cannot explain this striking difference in age of onset since the YAC128 mice express mHTT at similar levels to the endogenous mouse protein (Slow et al., 2003). However, a patch/matrix pattern of HTT immunoreactivity, as seen in the human striatum (Ferrante et al., 1997), has not been observed in the mouse (Bhide et al., 1996), which could point to differences in expression patterns between the human and mouse proteins. A lingering question is whether inherent differences between the human and mouse HTT protein-coding or regulatory sequences could result in the earlier and more profound changes in transgenic HD mouse models compared with the knock-in HD mouse models.
Fig. 1.
Comparison of phenotypes between transgenic and knock-in mouse models of HD and human patients. Transgenic YAC mice express human mHTT with its regulatory sequences and exhibit an earlier and more severe phenotype than knock-in mice, which express the CAG expansion in the context of the mouse Htt gene. References: *Graham et al., 2006; †Slow et al., 2003; #Van Raamsdonk et al., 2005b; §Lerch et al., 2008; **Heng et al., 2007; ††Pouladi et al., 2009; ##Tang et al., 2007.
Can we attribute the phenotypic differences in full-length transgenic and knock-in models to species-specific sequence differences in huntingtin between mice and humans?
The human HTT protein (NP_002102) is 3144 amino acids long and the mouse protein (NP_034544) contains 3120 amino acids. Excluding the differences in their polyglutamine and polyproline tracts, the human and mouse proteins are 91% identical and 95% similar.
The majority of the functional elements in human and mouse HTT do not differ or have only the most conservative amino acid substitutions and are thus unlikely to account for differences in the disease phenotypes of YAC128 and knock-in mouse models. Conserved functional elements include caspase, calpain and aspartyl protease cleavage sites; palmitoylation, phosphorylation, sumoylation and ubiquitylation sites; nucleocytoplasmic shuttling signals; the HTT membrane-associated domain; and a proteasome-targeting sequence (Table 2). One apparent exception is the calpain cleavage site at residues 468–470 that contributes to the toxicity of mHTT when cleaved (Gafni et al., 2004). The human sequence at this site is Leu-Thr-Ala; in the mouse it is Phe-Ala-Ala. This is consistent with the human site being a more favourable cleavage site than that in the mouse (Tompa et al., 2004), which might be significant in light of the increased calpain activity in caudate tissue from human HD patients (Gafni and Ellerby, 2002).
Table 2.
Protein sequence elements that do not differ between human and mouse HTT
| Functional site | Location* | References |
|---|---|---|
| Caspase-6 cleavage | IVLD 586 | Wellington et al., 2000 |
| Caspase-2/3 cleavage | DLND 552 | Hermel et al., 2004 |
| Caspase-3 cleavage | DVSD 513 | Wellington et al., 2000 |
| Palmitoylation | C214 | Yanai et al., 2006 |
| Phosphorylation | S421 | Warby et al., 2005 |
| Calpain cleavage | VLS 438 | Gafni and Ellerby, 2002 |
| SSS 537 | ||
| Aspartyl protease cleavage | HCLT…NIVA 104–114 | Lunkes et al., 2002 |
| RMVA…LLPC 146–214 | ||
| Sumoylation, ubiquitylation | K6, K9, R15 | Steffan et al., 2004 |
| Proteasome-targeting sequences | FQKLL 127 | Chandra et al., 2008 |
| Nuclear export signal | IIIS…RLPL 2397–2406† | Xia et al., 2003 |
| Putative nuclear localisation signal | PIRR…KEKE 1182–1191 | Bessert et al., 1995 |
| Membrane-associated domain/cytoplasmic retention signal | MATL…LKSF 1–17 | Steffan et al., 2004; Truant et al., 2006 |
Motif sequence and position of the last residue in the motif in human HTT NP_002102, or sequence range for motifs >6 residues.
The nuclear export signal has two conservative Ile to Val changes between the human and mouse.
The genomic organisation of the human HTT gene is well conserved in the mouse. This large gene consists of 67 exons that comprise about 170 kb of the human genome and 150 kb of the mouse genome. Both organisms have two mRNAs that are generated by alternative cleavage and polyadenylation of the primary transcript, producing a long and a short 3′ untranslated region (UTR) that differ by 3 kb (Lin et al., 1993; Lin et al., 1994). In both organisms, the long-UTR transcript is predominantly expressed in the brain, whereas the short-UTR transcript is more widely expressed (Lin et al., 1993). Separate from the alternative 3′UTRs, two HTT/Htt transcripts that differ by ~1.4 kb are seen in both the human and mouse brain (Lin et al., 1994). Although there is experimental evidence for this second pair of transcripts in both the mouse and human, Ensembl predictions only support their existence in the mouse genome, with one transcript missing ~480 amino acids from exons 36 to 43 (see ENSMUSG00000029104 at http://www.ensembl.org) (Hubbard et al., 2007). The corresponding human genomic data do not support this alternative splicing event (ENSG00000197386), which could influence phenotypes in mouse models.
Differences in human and mouse huntingtin regulatory sequences
Transcription factor binding sites
YAC128 mice contain ~308 kb of the human HTT region including ~24 kb upstream of the coding sequence (M.R.H. and R. Devon, unpublished) (Bates et al., 1992; Hodgson et al., 1996; Slow et al., 2003). Human-mouse conservation of this region is characterised by a few segments of high nucleotide sequence identity (>80%) interrupted by long regions of very dissimilar sequences, precluding alignment. Since HTT in YAC mice includes all the local conserved regions and CpG islands that are expected to influence HTT regulation (based on ‘Mammal Cons’ and ‘CpG islands’ tracks in the UCSC genome browser, http://genome.ucsc.edu/, March 2006 genome assembly) (Kent et al., 2002), we would expect that it is regulated in the mouse as it is in humans, defined by its native promoter and internal regulatory elements. The inclusion of ~24 kb upstream of the HTT transcription start site plus ~116 kb downstream of the gene makes it highly likely that even distant regulatory elements have been included in the YAC mouse model. BACHD mice are expected to retain a similar range of crucial elements since their human transgene includes ~20 kb upstream and ~50 kb downstream of the HTT gene (Gray et al., 2008). Regulatory elements that are farther than 20 kb from a gene’s transcription start site are less common. However, they cannot be excluded as a possibility for the HD gene and would not be present in either the BACHD or YAC mouse models.
Some experimentally validated sites of transcription factor binding show differences between the mouse and human. For example, human HTT is bound by the transcription factor NRSF/REST (neuron-restrictive silencer factor/repressor element 1 silencing transcription factor) (Johnson et al., 2008), which represses the expression of neuronal genes in non-neuronal cells (Schoenherr and Anderson, 1995). This site is not conserved in the mouse and the relevant Htt region shares no alignable sequence with human HTT in this intronic region. Differences between mouse Htt and human HTT targeting by NRSF might influence pathology.
Putative microRNA targets
Human HTT and mouse Htt may also be differentially regulated by microRNA (miRNA) families that function in both humans and mice, which could provide another basis for the differences between mice expressing the mouse gene versus the human gene. There is a precedent for miRNA modulation of cytotoxicity in the polyglutamine-expansion disorder spinocerebellar ataxia 1: several miRNAs co-regulate ataxin 1 at both the RNA and protein levels and influence cyotoxicity of the polyglutamine-expanded protein (Lee et al., 2008). Using TargetScan 4.2, we identified sites that, owing to sequence differences, would be targeted exclusively in the long 3′ UTRs in either the human or mouse. Context scores in TargetScan can be used to predict site performance (Lewis et al., 2003; Grimson et al., 2007), and we selected only miRNA targets that are likely to be significantly correlated with protein downregulation (Baek et al., 2008). Human HTT is predicted to be targeted by ten miRNAs that are conserved in humans and mice, but whose seed sequences are not conserved in the homologous site in the mouse 3′UTR (Table 3). Conversely, TargetScan predicts that mouse Htt is targeted by 12 miRNAs that are conserved in humans and mice but whose seed sequences are not conserved in the homologous site in the human 3′UTR (Table 3). miRNAs from both columns of Table 3, miR-133, miR-204, miR-30-5p and miR-195, have been implicated in HD pathology and related processes (Kim et al., 2007; Johnson et al., 2008; Mellios et al., 2008), pointing to a complex network of dysregulation. The binding of miRNAs can be strongly influenced by single nucleotide differences in their seed regions (Grimson et al., 2007), so unidentified differences between the HTT/Htt 3′UTRs in different mouse models could exacerbate phenotypic differences. Although YAC and knock-in mouse models express mHTT proteins at similar levels in different tissues (Slow et al., 2003), the potential for differential miRNA regulation suggests that there could be important differences in cell type-specific regulation of HTT. Clearly, further studies are needed to explore whether these might form the basis for different phenotypic manifestations of the mouse Htt and the human HTT gene.
Table 3.
Human HTT or mouse Htt sequence-specific targeting by miRNAs*
| Human sequence-specific miRNAs | Mouse sequence-specific miRNAs |
|---|---|
| miR-23 | miR-15/miR-16/miR-195/miR-424/miR-497† |
| miR-24 | miR-22 |
| miR-30-5p† | miR-24 |
| miR-103/miR-107 | miR-30-3p |
| miR-128 | miR-31 |
| miR-148/miR-152 | miR-133† |
| miR-181 | miR-223 |
| miR-204†/miR-211 | miR-224 |
| miR-214 | miR-331 |
| miR-370 | miR-342 |
| miR-378 | |
| miR-491 |
These miRNAs function in both humans and mice but are strongly predicted to target either the human HTT or mouse Htt 3′UTR, where that site sequence is not conserved in both organisms.
miRNAs implicated in HD pathology.
Strain differences between knock-in and YAC or BAC mouse models
Another important issue when comparing different mouse models is the respective background strain of mouse used. Breeding of a mouse model onto another strain can lead to changes in phenotypic severity. An Alzheimer’s disease mouse model expressing mutant amyloid precursor protein is lethal on pure FVB/N or C57BL/6J backgrounds but has a milder phenotype on outbred mouse strains (Carlson et al., 1997). A similar phenomenon was observed in caspase-3 knockout mice, which show perinatal lethality on a 129/B6F1 background, whereas backcrossing to the strain C57BL/6J dramatically improved the survival rate (Zheng et al., 1999; Houde et al., 2004).
The transgenic full-length YAC128 and BACHD models are on a pure FVB/N background, whereas most mHTT fragment and knock-in models are on a C57BL/6 mixed background (Table 4) (Mangiarini et al., 1996; Shelbourne et al., 1999; Slow et al., 2003; Gray et al., 2008). Neuronal toxicity mediated by the activation of glutamate receptors (excitotoxicity) contributes to the selective neurodegeneration observed in HD (Pouladi et al., 2006). Mounting evidence reveals that there is a strong genetic component influencing the susceptibility to excitotoxicity; the C57BL/6 strain background confers more resistance to most of the excitotoxins examined when compared with the FVB/N background (Schauwecker, 2005). This difference in susceptibility to excitotoxic neuronal death might underlie the differences in severity of the HD phenotype that were observed between the full-length YAC and BAC HD models compared with the knock-in mouse models.
Table 4.
Strain differences between HD mouse models
| Mouse HD model | CAG size | Strain | References |
|---|---|---|---|
| R6/2 model | 115–150 CAG | CBA C57BL/6 mixed background | Mangiarini et al., 1996 |
| N-171 model | 18, 44, 82 CAG | C3H/HEJ C57BL/6JF1 mixed background | Schilling et al., 1999 |
| YAC models | 18, 46, 72, 128 CAG | FVB/N | Hodgson et al., 1999; Slow et al., 2003 |
| BACHD model | 97 CAG | FVB/NJ | Gray et al., 2008 |
| Knock-in models | 72–80 CAG | 129/Svx C57BL/6 mixed background | Shelbourne et al., 1999 |
| 129/Svx FVB/N mixed background | |||
| 50, 92, 111 CAG | 129/CD1 mixed background | Wheeler et al., 2000 | |
| 94, 140 CAG | 129/Sv C57BL/6 mixed background | Menalled et al., 2002; Menalled et al., 2003 | |
| 150 CAG | 129/Ola C57BL/6J mixed background | Lin et al., 2001 |
Conclusion
The use of genetically modified mice has led to great strides in advancing our understanding of disease owing to their physiological similarity to humans. However, over the last decade, the use of murine models of neurodegenerative diseases in preclinical testing shows that therapeutic success in animal models is not always paralleled by clinical success in patients. The experience with HD mouse models is instructive in this regard, showing that introduction of the polyQ expansion sequence into the mouse Htt gene leads to a less severe phenotype and a more protracted time course when compared with that seen with transgenic expression of the full-length mutant human HTT gene. Furthermore, mouse models expressing a truncated form of mHTT that have been used extensively for preclinical testing had very limited predictive value for therapeutic success in patients, suggesting that success in more than one mouse model should be a prerequisite for embarking on clinical trials. However, for those drugs with appropriate ADME (adsorption, distribution, metabolism, excretion) and safety profiles against a compelling disease-relevant target, human trials could commence as soon as the compound becomes available. This approach could lead to expeditious and parallel preclinical trials that target verified disease-relevant pathways, and could provide hope for HD patients who currently have no therapy to influence the course of the illness, while raising the bar for drugs with less well-defined mechanisms of action.
Footnotes
COMPETING INTERESTS
The authors declare no competing financial interests.
REFERENCES
- Albin RL, Young AB, Penney JB, Handelin B, Balfour R, Anderson KD, Markel DS, Tourtellotte WW, Reiner A. (1990). Abnormalities of striatal projection neurons and N-methyl-D-aspartate receptors in presymptomatic Huntington’s disease. N. Engl. J. Med. 322, 1293–1298 [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. (2008). The impact of microRNAs on protein output. Nature 455, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates GP, Valdes J, Hummerich H, Baxendale S, Le Paslier DL, Monaco AP, Tagle D, MacDonald ME, Altherr M, Ross M, et al. (1992). Characterization of a yeast artificial chromosome contig spanning the Huntington’s disease gene candidate region. Nat. Genet. 1, 180–187 [DOI] [PubMed] [Google Scholar]
- Benatar M. (2007). Lost in translation: treatment trials in the SOD1 mouse and in human ALS. Neurobiol. Dis. 26, 1–13 [DOI] [PubMed] [Google Scholar]
- Bessert DA, Gutridge KL, Dunbar JC, Carlock LR. (1995). The identification of a functional nuclear localization signal in the Huntington disease protein. Brain Res. Mol. Brain Res. 33, 165–173 [DOI] [PubMed] [Google Scholar]
- Bhide PG, Day M, Sapp E, Schwarz C, Sheth A, Kim J, Young AB, Penney J, Golden J, Aronin N, et al. (1996). Expression of normal and mutant huntingtin in the developing brain. J. Neurosci. 16, 5523–5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson GA, Borchelt DR, Dake A, Turner S, Danielson V, Coffin JD, Eckman C, Meiners J, Nilsen SP, Younkin SG, et al. (1997). Genetic modification of the phenotypes produced by amyloid precursor protein overexpression in transgenic mice. Hum. Mol. Genet. 6, 1951–1959 [DOI] [PubMed] [Google Scholar]
- Chandra S, Shao J, Li JX, Li M, Longo FM, Diamond MI. (2008). A common motif targets huntingtin and the androgen receptor to the proteasome. J. Biol. Chem. 283, 23950–23955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudkowicz ME, Shefner JM, Schoenfeld DA, Zhang H, Andreasson KI, Rothstein JD, Drachman DB. (2006). Trial of celecoxib in amyotrophic lateral sclerosis. Ann. Neurol. 60, 22–31 [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. (1997). Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- Drachman DB, Frank K, Dykes-Hoberg M, Teismann P, Almer G, Przedborski S, Rothstein JD. (2002). Cyclooxygenase 2 inhibition protects motor neurons and prolongs survival in a transgenic mouse model of ALS. Ann. Neurol. 52, 771–778 [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Gutekunst CA, Persichetti F, McNeil SM, Kowall NW, Gusella JF, MacDonald ME, Beal MF, Hersch SM. (1997). Heterogeneous topographic and cellular distribution of huntingtin expression in the normal human neostriatum. J. Neurosci. 17, 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Andreassen OA, Dedeoglu A, Ferrante KL, Jenkins BG, Hersch SM, Beal MF. (2002). Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington’s disease. J. Neurosci. 22, 1592–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni J, Ellerby LM. (2002). Calpain activation in Huntington’s disease. J. Neurosci. 22, 4842–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni J, Hermel E, Young JE, Wellington CL, Hayden MR, Ellerby LM. (2004). Inhibition of calpain cleavage of huntingtin reduces toxicity: accumulation of calpain/caspase fragments in the nucleus. J. Biol. Chem. 279, 20211–20220 [DOI] [PubMed] [Google Scholar]
- Gordon PH, Moore DH, Miller RG, Florence JM, Verheijde JL, Doorish C, Hilton JF, Spitalny GM, MacArthur RB, Mitsumoto H, et al. (2007). Efficacy of minocycline in patients with amyotrophic lateral sclerosis: a phase III randomised trial. Lancet Neurol. 6, 1045–1053 [DOI] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. (2006). Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125, 1179–1191 [DOI] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, et al. (2008). Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci. 28, 6182–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. (2007). MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneveld GJ, Veldink JH, van der Tweel I, Kalmijn S, Beijer C, de Visser M, Wokke JH, Franssen H, van den Berg LH. (2003). A randomized sequential trial of creatine in amyotrophic lateral sclerosis. Ann. Neurol. 53, 437–445 [DOI] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. (1994). Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–1775 [DOI] [PubMed] [Google Scholar]
- Heng MY, Tallaksen-Greene SJ, Detloff PJ, Albin RL. (2007). Longitudinal evaluation of the Hdh(CAG)150 knock-in murine model of Huntington’s disease. J. Neurosci. 27, 8989–8998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermel E, Gafni J, Propp SS, Leavitt BR, Wellington CL, Young JE, Hackam AS, Logvinova AV, Peel AL, Chen SF, et al. (2004). Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington’s disease. Cell Death Differ. 11, 424–438 [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Smith DJ, McCutcheon K, Koide HB, Nishiyama K, Dinulos MB, Stevens ME, Bissada N, Nasir J, Kanazawa I, et al. (1996). Human huntingtin derived from YAC transgenes compensates for loss of murine huntingtin by rescue of the embryonic lethal phenotype. Hum. Mol. Genet. 5, 1875–1885 [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F, Singaraja R, Smith DJ, Bissada N, McCutcheon K, Nasir J, et al. (1999). A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron 23, 181–192 [DOI] [PubMed] [Google Scholar]
- Houde C, Banks KG, Coulombe N, Rasper D, Grimm E, Roy S, Simpson EM, Nicholson DW. (2004). Caspase-7 expanded function and intrinsic expression level underlies strain-specific brain phenotype of caspase-3-null mice. J. Neurosci. 24, 9977–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. (2007). Ensembl 2007. Nucleic Acids Res. 35, D610–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington Study Group (2001). A randomized, placebo-controlled trial of coenzyme Q10 and remacemide in Huntington’s disease. Neurology 57, 397–404 [DOI] [PubMed] [Google Scholar]
- Huntington Study Group (2006). Tetrabenazine as antichorea therapy in Huntington disease: a randomized controlled trial. Neurology 66, 366–372 [DOI] [PubMed] [Google Scholar]
- Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. (2008). A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol. Dis. 29, 438–445 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. (2007). A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317, 1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klivenyi P, Ferrante RJ, Matthews RT, Bogdanov MB, Klein AM, Andreassen OA, Mueller G, Wermer M, Kaddurah-Daouk R, Beal MF. (1999). Neuroprotective effects of creatine in a transgenic animal model of amyotrophic lateral sclerosis. Nat. Med. 5, 347–350 [DOI] [PubMed] [Google Scholar]
- Landwehrmeyer GB, Dubois B, de Yebenes JG, Kremer B, Gaus W, Kraus PH, Przuntek H, Dib M, Doble A, Fischer W, et al. (2007). Riluzole in Huntington’s disease: a 3-year, randomized controlled study. Ann. Neurol. 62, 262–272 [DOI] [PubMed] [Google Scholar]
- Lee Y, Samaco RC, Gatchel JR, Thaller C, Orr HT, Zoghbi HY. (2008). miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nat. Neurosci. 11, 1137–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Carroll JB, Dorr A, Spring S, Evans AC, Hayden MR, Sled JG, Henkelman RM. (2008). Cortical thickness measured from MRI in the YAC128 mouse model of Huntington’s disease. Neuroimage 41, 243–251 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. (2003). Prediction of mammalian microRNA targets. Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- Lin B, Rommens JM, Graham RK, Kalchman M, MacDonald H, Nasir J, Delaney A, Goldberg YP, Hayden MR. (1993). Differential 3′ polyadenylation of the Huntington disease gene results in two mRNA species with variable tissue expression. Hum. Mol. Genet. 2, 1541–1545 [DOI] [PubMed] [Google Scholar]
- Lin B, Nasir J, MacDonald H, Hutchinson G, Graham RK, Rommens JM, Hayden MR. (1994). Sequence of the murine Huntington disease gene: evidence for conservation, alternate splicing and polymorphism in a triplet (CCG) repeat [corrected]. Hum. Mol. Genet. 3, 85–92 [DOI] [PubMed] [Google Scholar]
- Lin CH, Tallaksen-Greene S, Chien WM, Cearley JA, Jackson WS, Crouse AB, Ren S, Li XJ, Albin RL, Detloff PJ. (2001). Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum. Mol. Genet. 10, 137–144 [DOI] [PubMed] [Google Scholar]
- Lunkes A, Lindenberg KS, Ben-Haiem L, Weber C, Devys D, Landwehrmeyer GB, Mandel JL, Trottier Y. (2002). Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol. Cell 10, 259–269 [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, et al. (1996). Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 [DOI] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. (2008). A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum. Mol. Genet. 17, 3030–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Wu Y, Olivieri M, Li XJ, Li H, Zeitlin S, Chesselet MF. (2002). Early motor dysfunction and striosomal distribution of huntingtin microaggregates in Huntington’s disease knock-in mice. J. Neurosci. 22, 8266–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. (2003). Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J. Comp. Neurol. 465, 11–26 [DOI] [PubMed] [Google Scholar]
- Miller RG, Mitchell JD, Lyon M, Moore DH. (2003). Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Amyotroph. Lateral Scler. Other Motor Neuron Disord. 4, 191–206 [PubMed] [Google Scholar]
- Pouladi MA, Bezprozvanny I, Raymond LA, Hayden MR. (2006). Molecular pathogenesis of Huntington’s disease: the role of excitotoxicity. In Genetic Instabilities and Neurological Diseases (ed. Wells RD, Ashizawa T. ), pp. 251 New York: Academic Press [Google Scholar]
- Pouladi MA, Graham RK, Karasinska J, Xie Y, Dar Santos R, Petersen A, Hayden MR. (2009). Prevention of depressive behaviour in the YAC128 mouse model of Huntington disease by mutation at residue 586 of huntingtin. Brain (in press) [doi:10.1093/brain/awp006]. [DOI] [PubMed] [Google Scholar]
- Puri BK, Leavitt BR, Hayden MR, Ross CA, Rosenblatt A, Greenamyre JT, Hersch S, Vaddadi KS, Sword A, Horrobin DF, et al. (2005). Ethyl-EPA in Huntington disease: a double-blind, randomized, placebo-controlled trial. Neurology 65, 286–292 [DOI] [PubMed] [Google Scholar]
- Schauwecker PE. (2005). Susceptibility to excitotoxic and metabolic striatal neurodegeneration in the mouse is genotype dependent. Brain Res. 1040, 112–120 [DOI] [PubMed] [Google Scholar]
- Schiefer J, Landwehrmeyer GB, Luesse HG, Sprunken A, Puls C, Milkereit A, Milkereit E, Kosinski CM. (2002). Riluzole prolongs survival time and alters nuclear inclusion formation in a transgenic mouse model of Huntington’s disease. Mov. Disord. 17, 748–757 [DOI] [PubMed] [Google Scholar]
- Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, et al. (1999). Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet. 8, 397–407 [DOI] [PubMed] [Google Scholar]
- Schilling G, Coonfield ML, Ross CA, Borchelt DR. (2001). Coenzyme Q10 and remacemide hydrochloride ameliorate motor deficits in a Huntington’s disease transgenic mouse model. Neurosci. Lett. 315, 149–153 [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. (1995). Silencing is golden: negative regulation in the control of neuronal gene transcription. Curr. Opin. Neurobiol. 5, 566–571 [DOI] [PubMed] [Google Scholar]
- Shefner JM, Cudkowicz ME, Schoenfeld D, Conrad T, Taft J, Chilton M, Urbinelli L, Qureshi M, Zhang H, Pestronk A, et al. (2004). A clinical trial of creatine in ALS. Neurology 63, 1656–1661 [DOI] [PubMed] [Google Scholar]
- Shelbourne PF, Killeen N, Hevner RF, Johnston HM, Tecott L, Lewandoski M, Ennis M, Ramirez L, Li Z, Iannicola C, et al. (1999). A Huntington’s disease CAG expansion at the murine Hdh locus is unstable and associated with behavioural abnormalities in mice. Hum. Mol. Genet. 8, 763–774 [DOI] [PubMed] [Google Scholar]
- Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y, Oh R, Bissada N, Hossain SM, Yang YZ, et al. (2003). Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 12, 1555–1567 [DOI] [PubMed] [Google Scholar]
- Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, et al. (2004). SUMO modification of Huntingtin and Huntington’s disease pathology. Science 304, 100–104 [DOI] [PubMed] [Google Scholar]
- Tang TS, Chen X, Liu J, Bezprozvanny I. (2007). Dopaminergic signaling and striatal neurodegeneration in Huntington’s disease. J. Neurosci. 27, 7899–7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington’s Disease Collaborative Research Group (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- Tibben A. (2007). Predictive testing for Huntington’s disease. Brain Res. Bull 72, 165–171 [DOI] [PubMed] [Google Scholar]
- Tompa P, Buzder-Lantos P, Tantos A, Farkas A, Szilagyi A, Banoczi Z, Hudecz F, Friedrich P. (2004). On the sequential determinants of calpain cleavage. J. Biol. Chem. 279, 20775–20785 [DOI] [PubMed] [Google Scholar]
- Truant R, Atwal R, Burtnik A. (2006). Hypothesis: Huntingtin may function in membrane association and vesicular trafficking. Biochem. Cell Biol. 84, 912–917 [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Murphy Z, Slow EJ, Leavitt BR, Hayden MR. (2005a). Selective degeneration and nuclear localization of mutant huntingtin in the YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 14, 3823–3835 [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Pearson J, Slow EJ, Hossain SM, Leavitt BR, Hayden MR. (2005b). Cognitive dysfunction precedes neuropathology and motor abnormalities in the YAC128 mouse model of Huntington’s disease. J. Neurosci. 25, 4169–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Pearson J, Bailey CD, Rogers DA, Johnson GV, Hayden MR, Leavitt BR. (2005c). Cystamine treatment is neuroprotective in the YAC128 mouse model of Huntington disease. J. Neurochem. 95, 210–220 [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Pearson J, Rogers DA, Lu G, Barakauskas VE, Barr AM, Honer WG, Hayden MR, Leavitt BR. (2005d). Ethyl-EPA treatment improves motor dysfunction, but not neurodegeneration in the YAC128 mouse model of Huntington disease. Exp. Neurol. 196, 266–272 [DOI] [PubMed] [Google Scholar]
- Vincent AM, Sakowski SA, Schuyler A, Feldman EL. (2008). Strategic approaches to developing drug treatments for ALS. Drug Discov. Today 13, 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Chan EY, Metzler M, Gan L, Singaraja RR, Crocker SF, Robertson HA, Hayden MR. (2005). Huntingtin phosphorylation on serine 421 is significantly reduced in the striatum and by polyglutamine expansion in vivo.Hum. Mol. Genet. 14, 1569–1577 [DOI] [PubMed] [Google Scholar]
- Warby SC, Doty CN, Graham RK, Carroll JB, Yang YZ, Singaraja RR, Overall CM, Hayden MR. (2008). Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum. Mol. Genet. 17, 2390–2404 [DOI] [PubMed] [Google Scholar]
- Wellington CL, Singaraja R, Ellerby L, Savill J, Roy S, Leavitt B, Cattaneo E, Hackam A, Sharp A, Thornberry N, et al. (2000). Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J. Biol. Chem. 275, 19831–19838 [DOI] [PubMed] [Google Scholar]
- Wheeler VC, White JK, Gutekunst CA, Vrbanac V, Weaver M, Li XJ, Li SH, Yi H, Vonsattel JP, Gusella JF, et al. (2000). Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum. Mol. Genet. 9, 503–513 [DOI] [PubMed] [Google Scholar]
- Xia J, Lee DH, Taylor J, Vandelft M, Truant R. (2003). Huntingtin contains a highly conserved nuclear export signal. Hum. Mol. Genet. 12, 1393–1403 [DOI] [PubMed] [Google Scholar]
- Yanai A, Huang K, Kang R, Singaraja RR, Arstikaitis P, Gan L, Orban PC, Mullard A, Cowan CM, Raymond LA, et al. (2006). Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 9, 824–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng TS, Hunot S, Kuida K, Flavell RA. (1999). Caspase knockouts: matters of life and death. Cell Death Differ. 6, 1043–1053 [DOI] [PubMed] [Google Scholar]
- Zhu S, Stavrovskaya IG, Drozda M, Kim BY, Ona V, Li M, Sarang S, Liu AS, Hartley DM, Wu DC, et al. (2002). Minocycline inhibits cytochrome c release and delays progression of amyotrophic lateral sclerosis in mice. Nature 417, 74–78 [DOI] [PubMed] [Google Scholar]