SUMMARY
TRF2 is a telomere-binding protein involved in the protection of chromosome ends. Interestingly, TRF2 is overexpressed in a number of human cancers. Mice with increased TRF2 expression (K5TRF2 mice) display a severe skin phenotype including an increase in skin cancer and premature skin degeneration, which includes increased skin hyperpigmentation and skin dryness; these pathologies are concomitant with dramatic telomere shortening and increased chromosomal instability. Here, we show that K5TRF2 mice have a severe epidermal stem cell (ESC) dysfunction, which is reversed by abrogation of p53 in the absence of rescue of telomere length. Importantly, p53 deletion also rescues severe skin hyperpigmentation in these mice through regulation of alpha-melanocyte-stimulating hormone (α-MSH). In addition, skin carcinogenesis is accelerated in K5TRF2/p53−/−mice owing to attenuated p21 induction, which enables cell proliferation to resume. Altogether, these results reveal the existence of a DNA damage-dependent checkpoint that acts on ESCs with critically short telomeres and restricts skin proliferation, thereby increasing protection against skin cancer; however, the checkpoint also leads to premature skin aging phenotypes. Finally, the results described here are relevant to our understanding of the pathobiology of those human diseases that are characterized by the presence of critically short telomeres (hereafter referred to as ‘telopathies’), such as dyskeratosis congenita which causes severe skin phenotypes including skin hyperpigmentation and skin cancer.
INTRODUCTION
Telomeres are ribonucleoprotein heterochromatic structures at the ends of eukaryotic chromosomes that consist of tandem TTAGGG repeats bound by an array of associated proteins (Chan and Blackburn, 2002; de Lange, 2002; Garcia-Cao et al., 2004). In addition, telomeres contain long non-coding AAUCCC-containing telomeric RNAs (TelRNAs or TERRAs), which are stably associated with the telomeric chromatin (Azzalin et al., 2007; Schoeftner and Blasco, 2008). Telomeric chromatin protects the 3′-single-stranded overhang (G-strand overhang) at the chromosome ends from degradation and prevents telomeres from being recognized as double-strand breaks (DSBs), probably through the formation of a looped structure (T-loop) (Griffith et al., 1999; de Lange, 2002; de Lange, 2004). Shortening of telomeres below a threshold length and/or altered functioning of the telomere-binding proteins result in a loss of telomere protection that leads to chromosomal end-to-end fusions and cell cycle arrest and/or apoptosis, which in turn are proposed to impinge on the pathogenesis of cancer and aging (van Steensel et al., 1998; de Lange, 2005).
Telomere length is regulated by the activity of a cellular reverse transcriptase known as telomerase (telomerase reverse transcriptase, TERT), which generates de novo telomeric repeats using an associated RNA molecule (telomerase RNA component, TERC) as a template (Chan and Blackburn, 2002). Telomere repeats are bound by a six-protein complex known as shelterin, which includes the POT1-TTP1 heterodimer and the telomere-binding proteins TRF1 and TRF2, as well as their interacting factors RAP1 and TIN2 (reviewed by de Lange, 2005; Liu et al., 2004; Ye et al., 2004; Smith et al., 1998). TRF2, together with other shelterin components, plays a crucial role in the regulation of telomere length and in telomere protection (de Lange, 2002; Muñoz et al., 2005). Deletion of TRF2 (also known as Terf2) in mice causes both embryonic lethality and a massive induction of end-to-end chromosome fusions in the absence of detectable telomere shortening (Celli and de Lange, 2005). However, the dramatic effect of TRF2 deletion on chromosomal integrity has little effect on the regeneration ability of some adult tissues, such as the liver (Celli and de Lange, 2005; Lazzerini Denchi et al., 2006), suggesting that TRF2 is not required to maintain the regenerative capacity of some adult stem cell compartments. Interestingly, mice with a 2–3-fold increase in telomere-bound TRF2 at the stem cell compartment of different epithelia [K5TRF2 mice; PM mouse line described in Muñoz et al. (Muñoz et al., 2005)] show severe telomere shortening and increased chromosomal instability in the presence of normal telomerase activity. K5TRF2 mice also show premature skin degenerative pathologies (hyperpigmentation, skin dryness, alopecia) and an increase in skin cancer; the latter is accelerated further in the absence of telomerase (Muñoz et al., 2005; Blanco et al., 2007). Interestingly, these K5TRF2 phenotypes are not rescued by telomerase overexpression, indicating that the short telomeres produced by TRF2 overexpression are not susceptible to elongation by telomerase; this is in agreement with loss of the G-strand overhang in these mice (Muñoz et al., 2005). Instead, the short telomeres in K5TRF2 transgenic mice are rescued by deletion of XPF (ERCC4), a component of the XPF-ERCC1 heterodimer, a TRF2-interacting nuclease involved in the repair of ultraviolet light (UV)-induced DNA damage through the nucleotide excision repair (NER) pathway (de Laat et al., 1999; Hoeijmakers, 2001; Zhu et al., 2003; Tian et al., 2004; Muñoz et al., 2005; Lazzerini Denchi et al., 2006), indicating that TRF2 overexpression shortens telomeres through an XPF–ERCC1-dependent mechanism. Interestingly, the skin phenotypes of K5TRF2 mice recapitulate the skin pathologies associated with the human xeroderma pigmentosum (XP) syndrome, which is caused by germline mutations in NER pathway components including the XPF-ERCC1 nuclease (Muñoz et al., 2005; Andressoo et al., 2006; Blanco et al., 2007; Blasco, 2007), further supporting an interplay between TRF2 and NER. The skin phenotypes of K5TRF2 mice are also very similar to the skin pathologies present in patients with a defective telomerase pathway and the presence of short telomeres, such as in some cases of dyskerayosis congenita, aplastic anemia and pulmonary fibrosis (Mitchell et al., 1999; Vulliamy et al., 2001; Yamaguchi et al., 2005; Armanios et al., 2007; Tsakiri et al., 2007). In agreement with the short telomeres in these mice, a model has been proposed in which increased TRF2 expression results in higher XPF-ERCC1 activity at telomeres, leading to rapid XPF-dependent telomere degradation (Muñoz et al., 2005; Wu et al., 2007) and to the early onset of skin-aging phenotypes (Muñoz et al., 2005). Concomitantly, decreased XPF activity at UV-induced lesions might lead to increased DNA damage and skin cancer (Muñoz et al., 2005; Muñoz et al., 2006; Blanco et al., 2007). Remarkably, in the absence of telomerase activity, telomere shortening is accelerated further in K5TRF2 mice and UV-induced carcinogenesis is also increased (Blanco et al., 2007), indicating that the skin phenotypes associated with increased TRF2 expression are provoked by the presence of critically short telomeres and increased DNA damage in these mice. Importantly, the increase in skin tumorigenesis that is associated with K5TRF2 expression correlates positively with severe skin hyperpigmentation in both K5TRF2 mouse models, suggesting that the two parameters are related (Muñoz et al., 2005; Blanco et al., 2007).
Critically short telomeres resulting from a telomerase deficiency in Terc-deficient mice have been shown previously to severely impair the functionality of epidermal stem cells (ESCs) (Flores et al., 2005). In turn, transgenic expression of telomerase in ESC compartments leads to increased ESC activation and proliferation (Flores et al., 2005; Sarin et al., 2005). Interestingly, the effects of telomere length and telomerase activity on ESC functionality correlate with their known effects on cancer and aging phenotypes in Terc-deficient mice, suggesting that defects in ESC behavior are at the origin of these diseases (Blasco, 2007). The profound impact of increased TRF2 expression on skin cancer and aging suggests a putative impact of TRF2 overexpression on ESC functionality. Here, we set to address this by studying ESC behavior in K5TRF2 mice. We found that mobilization of K5TRF2 ESCs in response to mitogens was severely impaired, in agreement with the critically short telomeres of these mice. However, in contrast to ESC mobilization defects in Terc-deficient mice, K5TRF2 ESC defects could not be rescued by telomerase overexpression, in accordance with the fact that Tert overexpression did not rescue telomere length and premature skin aging phenotypes in K5TRF2 mice (Muñoz et al., 2005; Blanco et al., 2007). Further, we show that deletion of XPF, the nuclease responsible for telomere degradation in K5TRF2 mice, partially rescued ESC clonogenic activity in these mice concomitantly with a rescue of short telomeres. These results demonstrate that TRF2-associated stem cell defects are provoked by the presence of critically short telomeres, which can be rescued in a genetic context where telomeres are elongated (i.e. by XPF deletion but not by Tert overexpression).
It has been reported previously that a p53-dependent checkpoint is activated in response to critically short telomeres and results in cell death and/or inhibition of proliferation (Chin et al., 1999; Artandi et al., 2000; d’Adda di Fagagna et al., 2004; Rajaraman et al., 2007; Sharpless and DePinho, 2007). We addressed whether a p53-dependent checkpoint was also acting on K5TRF2 ESCs, by anticipating their impact on cancer and aging. Remarkably, abrogation of p53 restored ESC functionality (mobilization and clonogenic activity) in K5TRF2 mice in the absence of telomere elongation. Furthermore, this rescue of ESC defects occurred in parallel with the rescue of premature skin aging pathologies while simultaneously leading to augmented UV carcinogenesis. Altogether, these findings suggest that increased TRF2 expression, and the subsequent accumulation of dysfunctional telomeres in K5TRF2 mice, activates a telomere-based p53-dependent checkpoint in ESCs that impairs their ability to regenerate the skin, while at the same time exerting a potent tumor suppressor barrier that limits skin cancer development.
RESULTS
Severe ESC dysfunction in K5TRF2 mice
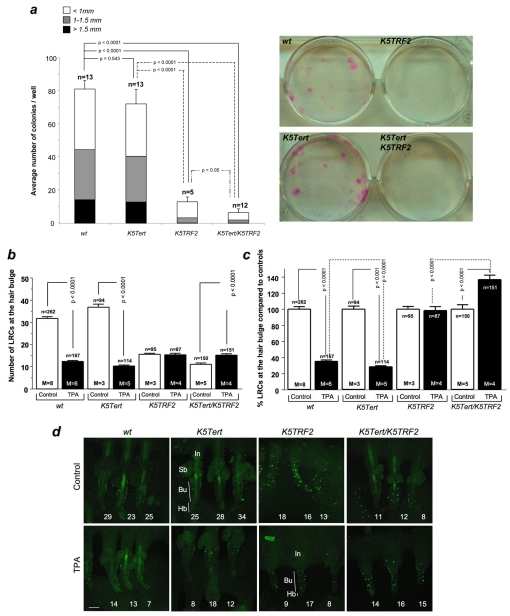
To study the impact of increased TRF2 expression (and the resulting 2–3-fold increase in telomere-bound TRF2) (Muñoz et al., 2005) on ESC behavior, we studied the number and functionality of ESCs by performing a label-retaining cell (LRC) assay in K5TRF2 mice and their corresponding wild-type controls (see Methods). In this assay, skin cells are pulse-labeled with bromodeoxyuridine (BrdU) and chased 2 months later to visualize slowly dividing LRCs, the putative ESCs (Flores et al., 2005). In wild-type skin, LRCs typically accumulate at the hair bulge area of the hair follicle where the putative stem cell niche is located (Flores et al., 2005) (see representative image in Fig. 1a). Interestingly, in control resting skin conditions, the skin of K5TRF2 mice showed significantly lower LRC numbers at the hair bulge than wild-type skin (P<0.0001) (Fig. 1b), which may suggest a lower number of ESCs or, alternatively, a lower labeling efficiency of these cells in K5TRF2 mice. To discern between these two possibilities, we determined the number of BrdU-positive cells present at the hair bulge 3 days after BrdU pulse labeling (see Methods). As shown in the supplementary material Fig. S1, K5TRF2 mice showed significantly fewer BrdU-positive cells at the hair bulge at 3 days post-injection than control wild-type mice, indicating a decrease in BrdU labeling efficiency. Furthermore, K5TRF2 mice showed lower numbers of proliferating cells (with Ki67-positive staining) in all skin compartments compared with wild-type controls (supplementary material Fig. S2), which might explain their lower BrdU labeling efficiency. A similar decrease in Ki67-positive cells was observed in the ventral skin (which has minimal exposure to UV light) of K5TRF2 mice compared with wild-type controls (supplementary material Fig. S1c and S3). Of note, we did not detect significant differences in active caspase-3 staining in the ventral skin of K5TRF2 and wild-type mice (supplementary material Fig. S4). In addition, we observed a significant decrease in the percentage of hair follicles that were positive for the K15 hair bulge stem cell marker in K5TRF2 mice compared with in wild-type controls (Fig. 1e), suggesting decreased ESC numbers in these mice.
Fig. 1.
Severe ESC mobilization defects in K5TRF2 mice. (a) Representative image of wild-type skin whole mounts showing LRCs (green cells) enriched at the hair bulge area of the mouse hair follicle. Bar, 20 μm. (b-d) Quantification of the absolute number of LRCs per hair follicle (b) and normalization to the corresponding untreated wild-type controls, 100% (c). The percentage of hair follicles with sebaceous glands was determined in confocal images of skin whole mounts (d). (e) Representative confocal images of cytokeratin 15 (K15) staining (red) in hair follicles of wild-type (wt, left panels) and K5TRF2 (right panels) mouse skin whole mounts (the blue staining is DAPI; Bu, bulge area; Hb, hair bulb). Bar, 80 μm. The percentage of hair follicles showing K15 staining in the bulge area is quantified (upper bar chart), as well as the percentage of K15-positive hair follicles relative to wild type (100%) (lower bar chart). Numbers indicate the number of positive follicles out of the number of follicles scored. (b-e) Bars represent mean±s.e.m., n=number of follicles scored/analyzed, M=number of mice per genotype. Statistical significance was calculated using unpaired two-tailed Student’s t-tests with Welch’s correction.
Topical skin treatment with the mitogen 12-O-tetradecanoylphorbol-13-acetate (TPA) stimulates ESCs to migrate out of the stem cell niche and to proliferate in the transient amplifying compartments, resulting in skin hyperplasia and hair growth (Flores et al., 2005). In agreement with this hypothesis, studies have shown that following TPA treatment, a 63% decrease in LRCs was detected at the hair bulge of wild-type skin compared with untreated skin, indicating ESC activation and mobilization (Flores et al., 2005) (Fig. 1b). Interestingly, similarly treated K5TRF2 mice did not show any detectable LRC mobilization in response to TPA (Fig. 1b,c), suggesting a severe defect in ESC activation in these mice. An in-depth look at the hair follicle morphology revealed that sebaceous glands were absent in around 60% of the hair follicles in K5TRF2 mice (Fig. 1d). Together, these results indicate that lower LRC numbers as well as a severe impairment of ESC mobilization (proliferation and migration) in K5TRF2 mice, which occur simultaneously with defects in hair follicle morphology, might explain the premature skin aging phenotypes observed in these mice, including hair loss, severe skin hyperpigmentation and severe skin dryness (Muñoz et al., 2005).
Next, we assessed the ex vivo proliferative potential of ESCs by performing in vitro clonogenic assays with newborn keratinocytes derived from wild-type and K5TRF2 mice. In this assay, individual colonies are supposedly derived from single ESCs (Barrandon and Green, 1987) and the size of the colonies is proposed to reflect their proliferative capacity (Barrandon and Green, 1987). In agreement with the in vivo LRC results, K5TRF2 mice formed significantly fewer and smaller colonies than wild-type controls (P<0.0001) (Fig. 2a), indicating that transgenic TRF2 expression decreases the ex vivo proliferative capacity of ESCs.
Fig. 2.
ESC defects in K5TRF2 mice are not rescued by telomerase overexpression. (a) The ex vivo proliferative potential of ESCs was determined by quantification of macroscopic clones obtained from newborn keratinocytes derived from mice of the indicated genotypes. Representative images are shown in the right panels. Quantification was performed according to number and size of clones per well (mean±s.e.m.). n=number of independent newborns per genotype used for the analysis. For statistical comparison of the total number of observed colonies per group we performed an unpaired two-tailed Student’s t-test with Welch’s correction. (b-c) Quantification of the absolute number of LRCs per hair follicle (b) and normalization to corresponding untreated wild-type controls, 100% (c). Values are mean±s.e.m.; n=number of analyzed follicles from a total of 3-8 mice per genotype and condition; M=number of mice per genotype and condition. Statistical significance was calculated using an unpaired two-tailed Student’s t-test with Welch’s correction. (d) Representative confocal images of skin whole mounts showing the tail hair follicles from mice of the indicated genotypes, stained with BrdU (LRC labeling), with and without TPA treatment. Tail hair follicles are grouped in sets of three. LRCs accumulate in the bulge area (Bu) of the hair follicle in all genotypes. The numbers in white identify results for each individual hair follicle. Bar, 80 μm.
Telomerase overexpression cannot rescue ESC defects in K5Tert/K5TRF2 mice
Poor ESC mobilization and decreased ESC clonogenic activity in K5TRF2 mice are likely to be the direct consequence of critically short telomeres and severe telomere dysfunction in these mice (Muñoz et al., 2005). To study whether telomerase was able to rescue ESC defects in these mice, we generated K5TRF2 mice that simultaneously overexpress Tert in the same skin compartment (K5Tert/K5TRF2 mice; see Methods). Again, untreated control K5TRF2 mice showed significantly decreased LRC numbers compared with wild-type mice, and these LRCs were not mobilized in response to TPA (Fig. 2b–d). In contrast, the TPA-treated wild-type and K5Tert control mice showed ESC mobilization rates of 61% and 72%, respectively (P≤0.0001) (Fig. 2b–d). Of note, the higher ESC mobilization in K5Tert mice is in agreement with previous findings showing that Tert augments the ESC mobilization response (Flores et al., 2005; Sarin et al., 2005). Strikingly, double transgenic K5Tert/K5TRF2 mice showed an even greater increase in the number of LRCs at the hair bulge upon TPA treatment (P≤0.0001) (Fig. 2b–d), suggesting that TPA treatment provoked proliferation of hair bulge LRCs but that migration of these cells out of their niche was severely impaired. These results indicate that ESC defects provoked by increased TRF2 expression cannot be rescued by telomerase overexpression.
Next we addressed whether Tert overexpression was able to rescue ESC clonogenic activity in K5Tert/K5TRF2 mice. In agreement with the in vivo results, expression of transgenic Tert was not able to rescue the proliferation defects of K5TRF2 cells in vitro. In fact, cells from K5Tert/K5TRF2 mice showed even fewer and smaller colonies than those from K5TRF2 mice (P<0.05) (Fig. 2a).
Finally, we addressed whether persisting ESC defects in K5Tert/K5TRF2 mice were caused by the presence of critically short telomeres in the skin of newborn mice. We performed quantitative fluorescent in situ hybridization (Q-FISH) on newborn skin from 1–3-day-old wild-type, K5Tert, K5TRF2 and K5Tert/K5TRF2 mice (supplementary material Fig. S5 and S7a). Interestingly, the newborn keratinocytes [day 1-3 postpartum (pp)] in K5TRF2 mice showed a dramatic reduction in average telomere length compared with wild-type littermates; 34.9% of telomere signals were below 200 units of telomere fluorescence compared with 21.6% in wild-type controls (supplementary material Fig. S5). The skin of K5Tert mice showed slightly elongated telomeres compared with wild-type controls, as only 11.5% of the telomere signals were below 200 units of telomere fluorescence (supplementary material Fig. S5), indicating that overexpressed Tert resulted in elongated telomeres in newborn keratinocytes that are wild type for TRF2. However, overexpressed Tert in K5Tert/K5TRF2 mice failed to rescue either the average telomere length or the percentage of short telomeres to wild-type levels (supplementary material Fig. S5 and S7a). This finding indicates that telomerase overexpression is not sufficient to elongate the critically short telomeres generated in the context of TRF2 overexpression, as we have described previously for adult skin (Muñoz et al., 2005). Altogether, these results indicate that persisting ESC defects in K5Tert/K5TRF2 mice are the consequence of a failure to rescue critically short telomeres by telomerase overexpression in these mice.
XPF deficiency rescues ESC proliferation defects in K5TRF2 mice
TRF2 interacts with the XPF-ERCC1 nuclease (Zhu et al., 2003), which rapidly degrades telomeres in the context of increased TRF2 expression (Muñoz et al., 2005). We set out to determine a putative role for XPF in the behavior of K5TRF2 ESCs. Xpf-knockout mice die before weaning (Tian et al., 2004); therefore, we could not address the impact of XPF deficiency on in vivo LRC assays.
To circumvent this, we performed in vitro clonogenic assays with newborn keratinocytes derived from wild-type, K5TRF2, Xpf+/−K5TRF2 and Xpf−/−K5TRF2 mice, as well as from the corresponding Xpf single-mutant controls (see Methods). We noticed that XPF abrogation resulted in significantly lower ESC clonogenic activity compared with in wild-type controls (P<0.05) (Fig. 3a); however, this was still higher than that observed in K5TRF2 mice (Fig. 3a). Importantly, XPF abrogation elevated Xpf−/−K5TRF2 clonogenic activity to the levels observed in control Xpf−/− mice (Fig. 3a). Although the difference did not reach statistical significance when compared with control K5TRF2 mice (Fig. 3a), these results suggest that XPF abrogation alleviates the severe ESC proliferative defect observed in K5TRF2 mice.
Fig. 3.
XPF deletion rescues the clonogenic activity of ESCs in K5TRF2 mice. The number and size of macroscopic colonies obtained from newborn keratinocytes of the indicated genotypes. n=number of independent newborns per genotype used for the analysis. A two-tailed Student’s t-test with Welch’s correction was performed for statistical significance comparisons. Representative images are shown in the panels below the bar chart.
We addressed whether the rescue of ESC clonogenic activity by XPF deficiency was the result of longer telomeres in the Xpf−/−K5TRF2 mice (Muñoz et al., 2005) by performing Q-FISH analyses directly on skin sections from newborn mice (day 1–3 pp) of the different genotypes (supplementary material Fig. S6 and S7b).
In agreement with previous findings in adult skin (Muñoz et al., 2005), the abrogation of XPF in K5TRF2 newborn skin resulted in significantly longer telomeres when compared with K5TRF2 mice (supplementary material Fig. S6 and S7b), which explains the increased ESC clonogenic activity of these mice. These results suggest that XPF depletion rescues ESC defects in K5TRF2 mice by preventing telomere degradation. Interestingly, heterozygous XPF levels resulted in significant rescue of both telomere length and ESC defects in Xpf+/−K5TRF2 mice (Fig. 3; supplementary material Fig. S6 and S7b), suggesting that XPF levels are rate limiting in mediating TRF2-induced telomere degradation.
A p53-dependent checkpoint limits the contribution of K5TRF2 ESCs to skin regeneration
We reported previously that the skin of K5TRF2 mice shows a higher frequency of cells containing telomere-associated DNA damage, as detected by co-localization of γ-H2AX and telomeres (Muñoz et al., 2005; Blanco et al., 2007). Given the known role of p53 in signaling different types of DNA damage (Sharpless and DePinho, 2007), including telomere damage, we investigated a potential role for p53 in mediating ESC defects in K5TRF2 mice.
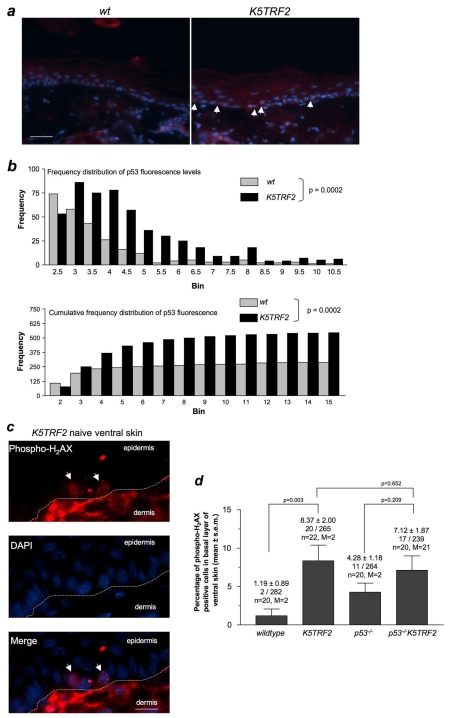
We studied whether p53 protein levels were increased in the skin of K5TRF2 mice, compared with in wild-type skin, by using immunofluorescence with anti-p53 antibodies (Fig. 4a). In particular, p53 fluorescence was quantified on a single-cell basis using Metamorph analysis (see Methods). Skin sections from p53-null mice were used as a threshold for p53 detection. We found significantly increased p53 protein levels in the basal layer of K5TRF2 epidermis compared with in wild-type skin (Fig. 4b), suggesting that there were increased levels of p53 in the skin of K5TRF2 mice.
Fig. 4.
Increased p53 expression and DNA damage in the skin of K5TRF2 mice. (a) Representative images of p53 immunofluorescence (white arrows indicate p53-positive cells) in basal layer skin keratinocytes from wild-type and K5TRF2 mice. Bar, 20 μm. (b) The level of p53 expression is depicted both in frequency distribution (upper panel) and cumulative frequency distribution (bottom panel) diagrams. Statistical differences were determined by unpaired two-tailed Student’s t-test with Welch’s correction. (c) Representative image of a K5TRF2 ventral skin section stained for phospho-H2AX (red) and DAPI (blue). White arrows indicate cells in the basal layer of the epidermis that are positive for phospho-H2AX. Bar, 10 μm. (d) Quantification of phospho-H2AX-positive basal cells in ventral (non-UV exposed) skin of the indicated genotypes. All groups included two mice (M) and at least 10 images (n) were analyzed from every mouse; in total, over 200 basal cells were analyzed per genotype. The percentage of phospho-H2AX-positive cells is shown (mean±s.e.m.), as well as the absolute number of phospho-H2AX-positive cells per total number of basal cells analyzed. Statistical analysis was performed by using the Student’s t-test with Welch’s correction.
It has been reported previously that TRF2 can prevent a DNA damage response when overexpressed in cells (Karlseder et al., 2004). To investigate whether p53 accumulation in K5TRF2 skin could be the consequence of increased DNA damage, we used immunofluorescence to detect γ-H2AX foci, a well-known marker of double-strand DNA breaks and of critically short telomeres (Takai et al., 2003; d’Adda di Fagagna et al., 2004). Interestingly, the ventral skin of K5TRF2 mice (which receives minimal exposure to UV light) showed higher numbers of γ-H2AX-positive cells compared with wild-type skin (Fig. 4c,d), indicating that increased DNA damage is associated with K5TRF2 overexpression.
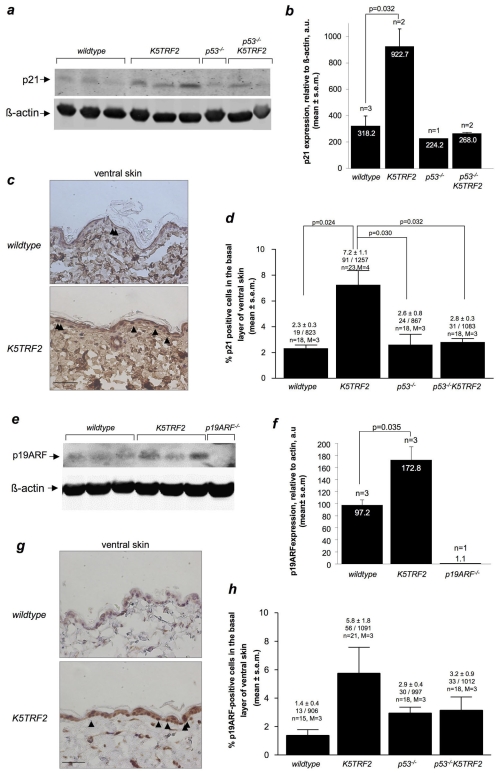
Among many other targets, p53 transactivates the well-known cell cycle inhibitor p21, thus exerting its activity as a potent inducer of cell cycle arrest in response to various types of DNA damage, including the presence of critically short telomeres (Choudhury et al., 2007). Therefore, to address the functional significance of p53 accumulation in K5TRF2 skin, we investigated p21 accumulation as an indicator of p53 transcriptional activity. Western blotting analysis of tail skin showed a significant increase in p21 protein in K5TRF2 mice compared with in wild-type mice (Fig. 5a,b), which would correlate with an increase in p53 activation. These results were confirmed by immunohistochemistry using an anti-p21 antibody on ventral skin sections from wild-type and K5TRF2 mice (Fig. 5c,d). We conclude that p53 deletion in p53−/−K5TRF2 mice attenuates p21 induction in the skin of these mice (Fig. 5a–d), demonstrating that p53 activity is responsible for p21 accumulation. Furthermore, decreased p21 levels in the skin of p53−/−K5TRF2 mice were also accompanied by increased proliferation compared with the K5TRF2 controls, as shown by Ki67 staining (supplementary material Fig. S3). Interestingly, the number of γ-H2AX-positive cells, as well as the number of apoptotic cells in ventral skin (supplementary material Fig. S4), were not significantly decreased in p53−/−K5TRF2 mice compared with in the K5TRF2 controls (Fig. 4c,d), suggesting that DNA-damaged cells are not eliminated in this context.
Fig. 5.
Accumulation of p21 and p19ARF in K5TRF2 skin. (a) Representative image of a western blot analysis of p21 protein expression in wild-type and K5TRF2 ventral skin. (b) Quantification of p21 expression relative to β-actin expression is expressed as mean (values indicated) ± s.e.m. n=number of samples analyzed per group. Statistical analysis was performed by using the Student’s t-test with Welch’s correction for unequal variance. (c) Representative images of wild-type and K5TRF2 ventral skin stained for p21 (brown) and hematoxylin (blue). Black arrowheads indicate basal cells that are positive for p21 expression. Bar, 20 μm. (d) Percentage of p21-positive basal cells observed in ventral skin sections. All groups include at least three mice (M) and at least six microscopic fields (n) were analyzed per mouse; in total, over 800 basal cells were analyzed. The percentage of p21-positive cells is shown, as well as the number of p21-positive cells per total number of basal cells analyzed. (e) Representative image of western blot analysis of p19ARF protein expression in wild-type and K5TRF2 ventral skin. (f) Quantification of p19ARF expression relative to β-actin expression. (g) Representative image of ventral skin from wild-type and K5TRF2 mice stained for p19ARF (brown) and hematoxylin (blue). Black arrowheads indicate the p19ARF-positive basal cells. Bar, 20 μm. (h) Percentage of p19ARF-positive basal cells detected in the ventral skin. All groups include at least three mice (M) and at least six microscopic fields (n) were analyzed per mouse; in total, over 900 basal cells were analyzed. The percentage of p19ARF-positive cells is shown, as well as the number of p19ARF-positive cells per total number of basal cells analyzed. (b,d,f,h) All data expressed as mean (values indicated) ± s.e.m. n=number of samples analyzed per group. Statistical analysis was performed by using the Student’s t-test with Welch’s correction for unequal variance.
Finally, as an additional indication of p53 accumulation in the skin of K5TRF2 mice, we studied the levels of p19ARF, an important tumor suppressor that exerts its tumor suppressing activity by stabilizing p53 protein levels (Collado et al., 2007). As shown in Fig. 5e,f, there was an accumulation of p19ARF in K5TRF2 keratinocytes. These results were confirmed by immunohistochemistry with anti-p19ARF antibodies on non-UV-exposed ventral skin sections (Fig. 5g,h).
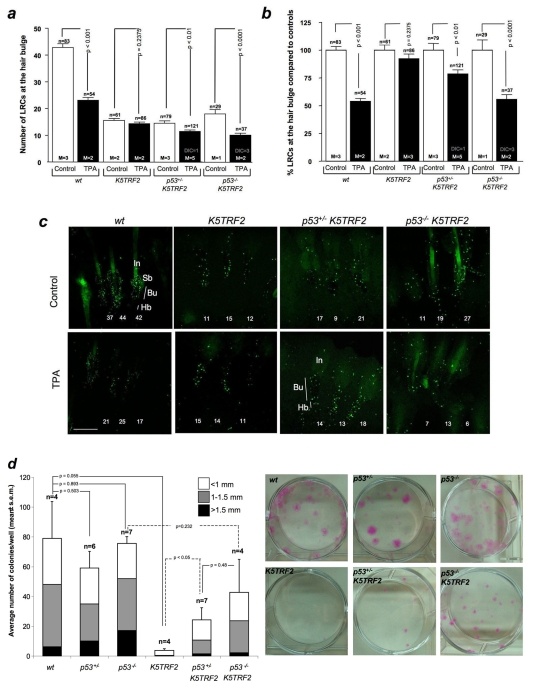
We addressed whether induction of p53 and increased DNA damage were important in mediating the ESC phenotypes of K5TRF2 mice by generating K5TRF2 mice that were deficient for p53 (p53−/−K5TRF2 mice) (see Methods). Again, LRC experiments indicated that K5TRF2 ESCs do not mobilize out of the hair bulge niche in response to TPA treatment (Fig. 6a–c). Remarkably, deletion of one p53 allele (p53+/−K5TRF2) resulted in an increase in LRC mobilization of up to 21%, which was further increased to 44% when both p53 alleles were deleted in the p53−/−K5TRF2 mice (Fig. 6a–c). These findings indicate that p53 mediates the mobilization defect of ESCs in K5TRF2 mice in an allele-dose-dependent manner, suggesting that there is a p53-dependent checkpoint that specifically acts on ESCs with critically short/dysfunctional telomeres.
Fig. 6.
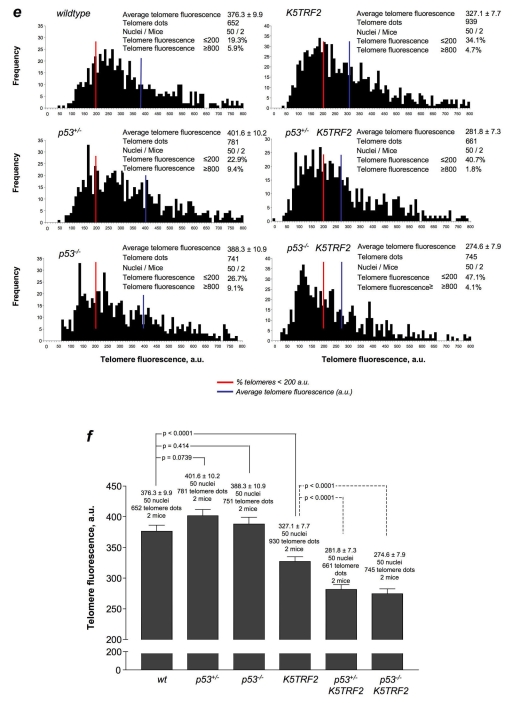
p53 deletion rescues ESC defects in K5TRF2 skin keratinocytes in the absence of telomere elongation. (a) Quantification of absolute number of LRCs per hair follicle before (control) and after TPA treatment (TPA). (b) Normalization of LRC numbers with respect to the corresponding untreated controls (100%). Values are mean±s.e.m. n=number of analyzed follicles per genotype and condition; M=number of mice used for the analysis per genotype and condition; DIC=number of animals that died during the course of the experiment. (c) Representative confocal images of hair follicles from the tails of mice with the indicated genotypes after LRC labeling and TPA treatment. Hair follicles in the tail are grouped in sets of three and LRCs accumulate at the bulge area (Bu) of the hair follicle in all genotypes. The numbers in white identify the results obtained from each individual hair follicle. Bar, 80 μm. (d) Ex vivo proliferative potential of ESCs from mice of the indicated genotypes. Quantification was performed according to the number and size of the clones. n=number of independent newborns from each genotype that were used for the analysis. (e) Telomere fluorescence, as determined by Q-FISH on neonatal skin sections from the indicated genotypes of mice. Histograms represent the frequency of telomere fluorescence in arbitrary units (a.u.) per telomere dot from a total of 50 nuclei, representing at least 650 telomeric dots per genotype. The red line indicates telomeres showing <200 a.u. of fluorescence. The blue line indicates average telomere length. (f) Comparisons of average telomere length. (a,b,d,f) An unpaired two-tailed Student’s t-test with Welch’s correction was used for all statistical comparisons.
Next, we focused on a potential role of p53 in limiting the ex vivo clonogenic activity of K5TRF2 ESCs (see Methods). Again, K5TRF2 cells showed severely reduced clonogenic activity (Fig. 6d). Deletion of one or both p53 alleles in control p53+/− and p53−/−mice, respectively, had no significant effect on ESC clonogenic activity compared with in wild-type controls (Fig. 6d). Notably, the decreased clonogenic activity associated with K5TRF2 expression was largely abrogated in p53−/−K5TRF2 mice (Fig. 6d) suggesting that p53 is actively limiting the clonogenic potential of K5TRF2 cells.
Finally, to address whether the impact of p53 status on ESC behavior was mediated by putative effects of p53 on telomere length, we performed Q-FISH analysis directly on skin sections from newborn mice (Fig. 6e,f; supplementary material Fig. S7c). We observed normal telomere lengths in the skin cells of single mutant p53+/− and p53−/− mice, which were indistinguishable from those of wild-type skin (Fig. 6e,f). Again, the keratinocytes from the skin of newborn K5TRF2 mice showed a decrease in average telomere length and a significant increase in the percentage of short telomeres (telomere signals with less than 200 units of telomere fluorescence) (Fig. 6e,f; supplementary material Fig. S7c). Interestingly, deletion of p53 in p53−/−K5TRF2 mice resulted in further shortening of telomeres in newborn skin keratinocytes compared with those in p53+/+K5TRF2 mice (Fig. 6e; supplementary material Fig. S7c); this was probably the consequence of increased proliferative rates in these cells owing to p53 deletion and decreased p21 levels (see Fig. 5). In support of this finding, proliferation was increased in the skin of p53−/−K5TRF2 mice compared with K5TRF2 mice, as determined by Ki67-positive staining (supplementary material Fig. S3), whereas no differences in apoptosis were detected by active caspase-3 staining (supplementary material Fig. S4). These findings demonstrate that the recovery of ESC mobilization and the proliferation defects in p53−/−K5TRF2 mice occur independently of telomere length, indicating a role for p53 in signaling dysfunctional telomeres in ESCs thereby limiting their contribution to tissue regeneration.
p53 deletion accelerates UV-induced skin cancer in K5TRF2 mice
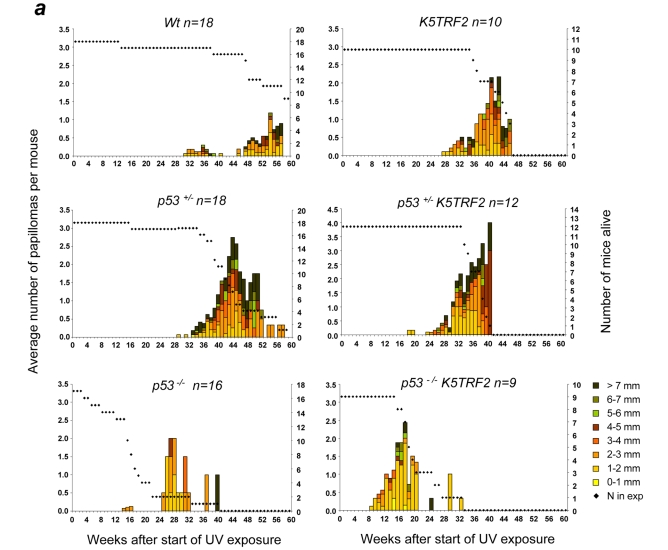
p53 acts as a potent tumor suppressor in the context of the organism by forcing cells with chromosomal damage, or with activated oncogenes, to undergo cellular arrest or apoptosis (Sharpless and DePinho, 2007). Because K5TRF2 mice show increased p53 levels and DNA damage in the skin, as well as decreased ESC clonogenic activity, and because both ESC mobilization and clonogenic defects can be rescued in these mice by p53 ablation, we decided to investigate the potential role of p53 in K5TRF2 skin tumorigenesis. We subjected mice with different genotypes to chronic, mild UV exposure, which does not lead to increased skin cancer in wild-type controls (see Fig. 7 and Methods). As expected, wild-type and heterozygous p53 mutant mice showed very few skin lesions by 40 weeks after the start of treatment (Fig. 7a). In contrast, p53−/− mice developed papillomas after 25 weeks of UV treatment (Fig. 7a). K5TRF2 mice also developed papillomas at around week 26 of treatment (Fig. 7a). Interestingly, p53−/−K5TRF2 mice showed a dramatically accelerated onset of papillomas (the first ones appeared at week 8), which were more abundant and larger when compared with K5TRF2 and p53−/− control mice (Fig. 7a). In addition to papilloma formation, we observed the appearance of both squamous cell carcinomas (SCC) and basal cell carcinomas (BCC) in both K5TRF2 and p53+/−K5TRF2 mice, beginning at around 28 weeks after the start of treatment (Fig. 7b). Control p53−/−mice also developed SSCs by 27 weeks after the start of UV treatment (Fig. 7b). Complete p53 abrogation in p53−/−K5TRF2 mice decreased the time to SCC and BCC onset to only 12 weeks, and resulted in larger and more severe lesions when compared with the other genotypes (Fig. 7b). These results indicate that p53 deletion dramatically accelerates the UV-induced skin carcinogenesis that is associated with increased TRF2 expression. This combined effect of p53 deletion and TRF2 overexpression on skin carcinogenesis was also observed when considering the total number of skin lesions in the different genotypes of mice at their time of death (Fig. 7b). p53−/−K5TRF2 mice showed more pre-neoplastic skin lesions (hyperplasia, dysplasia, hyperkeratosis and actinic keratosis) and more neoplastic skin lesions (spindle cell carcinomas; BCC; and well differentiated, poorly differentiated and in situ types of SSC) when compared with p53−/− mice. Altogether, these findings suggest that p53 acts as a potent tumor suppressor in the context of K5TRF2 skin, by decreasing the incidence and delaying the onset of TRF2-induced skin cancers. This tumor suppressor effect of p53 occurs in combination with increased levels of the tumor suppressors p21 and p19ARF (Fig. 5a–d) and with decreased proliferation rates in the skin of K5TRF2 mice compared with p53−/−K5TRF2 mice (supplementary material Fig. S3).
Fig. 7.
p53 abrogation accelerates UV-induced skin carcinogenesis in K5TRF2 mice. (a,b) Total number and size of papillomas (a) and carcinomas (b) observed in dorsal mouse skin following chronic UV treatment (shown in weeks after protocol initiation) in mice of the indicated genotypes. The dotted line represents the number of alive mice at each time point. (c) The total number of skin lesions per genotype in UV-exposed areas (dorsal skin, tail and ear) at the time of death is depicted. Representative images of the skin lesions are shown in the right panel. (i) Normal skin. (ii) Dorsal mouse skin with hyperkeratosis, squamous cell hyperplasia and cellular dysplasia, showing diffuse chronic inflammatory reactions in the skin. (iii) In situ carcinoma in dorsal mouse skin with severe cellular dysplasia and hyperchromatic nuclei – note that tumoral cells invade the basement membrane. (iv) Spindle cell carcinoma in dorsal mouse skin showing closely packed spindle-shaped epithelial cells with numerous mitotic figures. The intensely stained cells are slightly acidophilic and have dark blue nuclei. (v) SCC in dorsal mouse skin showing a nest of squamous cells with an invasive growth pattern into the dermis, as well as numerous atypical nuclei and mitotic figures. (vi) BCC in dorsal mouse skin with foci of closely packed cells with numerous mitotic figures. Intensely stained cells are slightly basophilic and have dark blue nuclei. Bar, 100 μm.
p53 mediates skin hyperpigmentation in K5TRF2 mice
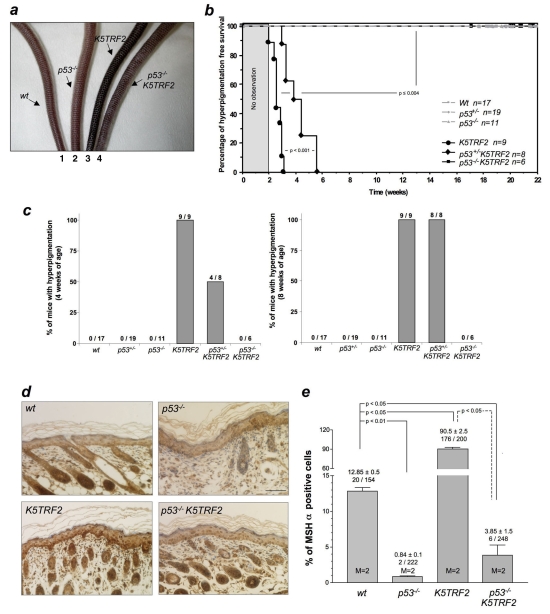
K5TRF2 mice show accelerated skin aging including development of severe hyperpigmentation in areas of skin that are exposed to light, such as the ear, paws and tail (Muñoz et al., 2005). Interestingly, it has been shown recently that p53 mediates the secretion of alpha-melanocyte-stimulating hormone [α-MSH, a cleavage product of proopiomelanocortin (POMC)] through transcriptional regulation of the POMC promoter in response to UV light, leading to skin hyperpigmentation (Pathak and Fanselow, 1983; Riley, 1997; Brash, 2006; Cui et al., 2007). This prompted us to investigate whether p53 could be responsible for the severe hyperpigmentation phenotype observed in K5TRF2 mice. As a result, we studied skin hyperpigmentation in K5TRF2, p53+/−K5TRF2 and p53−/−K5TRF2 mice, as well as in the corresponding p53 single mutant genotypes.
Macroscopic skin inspection revealed that all K5TRF2 tails showed hyperpigmentation as early as 4 weeks of age, whereas none of the wild-type or single mutant p53 genotypes showed this phenotype (Fig. 8a–c) (Muñoz et al., 2005). Remarkably, the absence of one p53 allele in p53+/−K5TRF2 mice significantly delayed the onset of hyperpigmentation by approximately 2 weeks (Fig. 8b,c). Strikingly, p53 ablation completely eliminated the hyperpigmentation phenotype in p53−/−K5TRF2 mice, which did not develop any signs of skin hyperpigmentation during the time course of the experiment (Fig. 8a–c) – even in response to UV radiation (not shown).
Fig. 8.
p53 deletion rescues K5TRF2 skin hyperpigmentation. (a) Representative images of tail skin pigmentation in wild-type (1), p53−/− (2), K5TRF2 (3) and p53−/−K5TRF2 (4) mice at 8 weeks of age. (b) Hyperpigmentation-free survival was generated in mice of the indicated genotypes. n=number of mice analyzed. A Kruskal-Wallis statistical test was used to determine statistical differences in the development of tail hyperpigmentation over time. (c) Percentage of mice of the indicated genotypes that showed tail skin hyperpigmentation at 4 weeks (left panel) or 8 weeks (right panel) of age. The total numbers of mice used per genotype are indicated. (d) Representative images of immunohistochemical staining for α-MSH. Bar, 50 μm. (e) Quantification of α-MSH-positive cells (mean±s.e.m.). An unpaired Student’s t-test with Welch’s correction was performed to compare the differences between the groups. M=number of mice analyzed per genotype. The number of α-MSH-positive cells out of the total number of cells scored is also indicated.
We studied the expression of α-MSH by performing immunohistochemistry directly on to skin sections from mice of the different genotypes (see Methods) (Fig. 8d). Interestingly, K5TRF2 mice showed significantly higher numbers of α-MSH-positive cells in the basal layer of the skin epidermis compared with wild-type mice (Fig. 8e), which was in agreement with the higher p53 levels in the K5TRF2 mice (Fig. 4a,b). This increase was completely abolished by p53 deletion in p53−/−K5TRF2 mice (Fig. 8e) indicating that p53-dependent regulation of α-MSH expression is responsible for the skin hyperpigmentation phenotype of K5TRF2 mice. Taken together these results suggest that increased telomere damage results in p53-mediated signaling of the hyperpigmentation response.
Finally, it is worth noting that decreased skin hyperpigmentation in the absence of p53 is unlikely to be responsible for the increased tumorigenesis observed in p53−/−K5TRF2 mice, as we have previously reported a positive correlation between skin hyperpigmentation and increased skin cancer in K5TRF2 mice (Muñoz et al., 2005). We have also reported the same positive correlation in compound Terc−/−K5TRF2 telomerase-deficient mice, which are heavily hyperpigmented but show a dramatically accelerated skin carcinogenesis (Blanco et al., 2007). In agreement with the notion that rescue of hyperpigmentation is unlikely to be the cause of increased skin cancer in the K5TRF2 model, K5TRF2 mice were also reported to be more susceptible to skin tumors when using non-UV-based skin carcinogenesis protocols, such as the DMBA/TPA protocol (Blanco et al., 2007).
DISCUSSION
TRF2 is a central component of the shelterin complex that binds to telomeres, and has essential roles in the protection of chromosome ends and regulation of telomere length. TRF2 has been found to be upregulated in human cancers, including in SCC and BCC in the skin (Hsu et al., 2005; Oh et al., 2005; Muñoz et al., 2005). In this regard, mice with increased TRF2 expression show critically short telomeres, increased telomere damage and an increased incidence of cancer (Muñoz et al., 2005), suggesting that deregulated TRF2 expression might favor a pro-tumorigenic phenotype (Muñoz et al., 2006). Here, we describe the impact of increased TRF2 levels on ESC behavior and, using mouse genetics, we further dissect the pathways involved in this process. In particular, we show that increased TRF2 expression in the stem cell compartment of hair follicles in the mouse skin (K5TRF2 mice) results in impaired ESC mobilization in response to mitogenic stimuli and decreased ESC proliferation when using in vitro clonogenic assays. Importantly, we show that these ESC defects are produced by the presence of critically short telomeres in K5TRF2 mice, as they can be largely rescued by deleting the TRF2-interacting XPF/ERCC1 nuclease, which is associated with significant telomere elongation in these mice. These results suggest that decreased XPF activity (i.e. arising from the XFP mutations that cause xeroderma pigmentosum) would favor the growth of cells with critically short/dysfunctional telomeres because of TRF2 overexpression (i.e. human tumors with increased TRF2 expression).
Although re-introduction of telomerase was shown recently to rescue the ESC defects associated with critically short telomeres in the telomerase-deficient mouse model (Siegl-Cachedenier et al., 2007), increased expression of telomerase in K5Tert/K5TRF2 double transgenic mice failed to rescue ESC function in these mice. In addition, telomerase was not able to rescue the short telomeres in these mice. These results suggest that the short telomeres produced by TRF2 overexpression are not susceptible to elongation by telomerase, in agreement with loss of the G-strand overhang in these mice (Muñoz et al., 2005). Alternatively, overexpression of K5Tert might fail to elongate short telomeres in K5TRF2 mice, partly owing to the severe proliferative defects in the skin of these mice.
Finally, we describe the existence of a p53-dependent checkpoint acting on ESCs with critically short telomeres, which prevents them from contributing to skin regeneration and skin cancer. In particular, in the absence of changes in telomere length, we show that deletion of p53 rescues ESC proliferation and migration defects in the skin of K5TRF2 mice. Together with rescue of ESC functionality, p53 deletion rescued premature skin aging phenotypes, such as severe skin hyperpigmentation, and led to dramatically accelerated skin carcinogenesis in K5TRF2 mice. The rescue of skin hyperpigmentation in the presence of critically short telomeres by p53 abrogation is of interest because skin hyperpigmentation is a landmark phenotype associated with human diseases caused by germline mutations in telomerase or telomere-binding proteins (e.g. Tin2), including dyskeratosis congenita, some cases of aplastic anemia and idiopathic pulmonary fibrosis (Mitchell et al., 1999; Vulliamy et al., 2001; Yamaguchi et al., 2005; Armanios et al., 2007; Tsakiri et al., 2007; Savage et al., 2008). These findings highlight the important role of ESC defects in the pathobiology of cancer and aging, and suggest a model in which p53 limits the contribution of ESCs with dysfunctional telomeres to tissue regeneration, thus providing cancer protection and ensuring healthy tissue condition. This role of p53 in stem cell function represents a novel p53-dependent damage control mechanism by which the amount of p53 in ESC populations functionally selects the most appropriate ESCs (i.e. those with sufficiently long telomeres) to contribute to tissue regeneration. Eventually, the pool of ESCs with sufficiently long telomeres may become exhausted, for instance owing to the aging process, and could result in severe stem cell dysfunction, leading to decreased tissue regeneration and to premature aging. In this situation, fortuitous mutations in the p53 pathway might however lead to aberrant restoration of stem cell mobilization and to increased stem cell proliferation, thereby prolonging the healthy state of the tissues (Choudhury et al., 2007); although, the mutations might also increase the risk of cancer (Chin et al., 1999; Artandi et al., 2000; Artandi and DePinho, 2000). Overall, these results indicate an important impact for altered TRF2 expression in stem cell behavior, which explains the impact of TRF2 overexpression in cancer and aging phenotypes.
METHODS
Mice
K5Tert/K5TRF2 double mutant mice and Xpf (Ercc4)/K5TRF2 mice were generated as described previously (Muñoz et al., 2005). To generate the p53/K5TRF2 mice, we crossed K5TRF2 males from the previously described PM founder transgenic line (Muñoz et al., 2005) with p53-knockout females and then crossbred the offspring. Wild-type and mutated p53 alleles were detected as described previously (Jacks et al., 1994). Cross-breeding was always performed in p53 and Xpf heterozygous backgrounds to avoid any possible genomic effects arising from XPF or p53 deficiencies.
Mice were kept with standard mouse chow and water, which were available ad libitum, and maintained under specific pathogen-free conditions in accordance with the recommendations of the Federation of European Laboratory Animal Science Associations. All mice were exposed to day-night cycles (12 hours each of light and dark) at the Spanish National Cancer Research Centre mouse facility. The light source was white fluorescent lamps (TLD36W/840 and TLD58W/840, Phillips), which are known to emit low doses of ultraviolet radiation at between 250 nm and 400 nm.
Generation of primary keratinocytes
Newborn male mice (1-3 days old) were sacrificed by CO2 asphyxiation and soaked in Betadine (for 5 minutes) followed by two washes in 70% ethanol (5 minutes) and then PBS. Skin detachment and isolation of the epidermis was performed essentially as described previously (Dlugosz et al., 1995; Flores et al., 2005). In brief, the limbs and tail were amputated and, after a longitudinal incision from tail to snout, the skin was peeled from the newborn mouse using forceps. A 0.5×0.5 cm piece of this skin was isolated and fixed overnight in 10% formalin and then embedded in paraffin (see Q-FISH methodology). Epidermises were stretched and flattened, then floated overnight on 1×trypsin (Sigma) at 4°C. The epidermis was separated from the dermis, then minced and stirred at 37°C for 30 minutes in serum-free CnT-02 medium (CELLnTEC advanced cell systems AG) supplemented with 1.4 mM of CaCl2. Subsequently, the cell suspension was filtered through a sterile teflon mesh (70 μm cell strainer, Falcon) to eliminate cornified cell material. Keratinocytes were collected by centrifugation.
Colony formation assay
Ten thousand primary mouse keratinocytes per newborn mouse were seeded in triplicate onto mitomycin C-treated (10 μg/ml, 2 hours) J2-3T3 fibroblasts (105 cells per well, six-well plates) and cultured at 37°C/5% CO2 in CnT-02 medium. After 10 days of culture, dishes were rinsed with PBS, and fixed in formaldehyde-PBS and rhodamine B to visualize colony formation. Colony size and number were measured. Statistical analysis of the average total number of colonies was performed using the Student’s t-test with Welch’s correction.
Q-FISH telomere length analysis
Perpendicular sections of neonatal mouse skin were cut, deparaffinized and incubated with a Cy3-labeled telomeric peptide nucleic acid (PNA) probe (Gonzalez-Suarez et al., 2000; Gonzalez-Suarez et al., 2001). Telomere fluorescence in basal cells of the epidermis (interphase nuclei) was determined as described previously by our group (Gonzalez-Suarez et al., 2000; Gonzalez-Suarez et al., 2001; Muñoz et al., 2005; Blanco et al., 2007). In brief, more then 50 nuclei per genotype were captured at a 100× magnification using a Leica CTR MIC microscope and COHU high performance CCD camera. We integrated telomere fluorescence signals using the TFL-TELO program (provided by P. Lansdorp, Vancouver, Canada). Telomere fluorescence frequencies were represented as histograms. Statistical analysis of the average telomere length was performed by using the Student’s t-test with Welch’s correction.
Label-retaining cell (LRC) assay
LRC assays were carried out as described previously (Bickenbach et al., 1986; Braun et al., 2003; Flores et al., 2005). Neonatal mice were injected with 50 mg/kg bodyweight of BrdU (Sigma B-5002) diluted in PBS. Each animal received two sets of daily injections starting on day 4 pp, with each set of injections lasting for 5 days. After the labeling period, mice were either sacrificed after only 3 days in order to study the amount of ESCs at the start of the experiment or they were kept for 60 days (wash-out period) before the initiation of any treatment.
LRC mobilization, hyperplasia of the interfollicular epidermis and anagen entry were induced by topical treatment of the tail skin with the potent mitogen, TPA. Tails were treated with four doses of 20 nmol TPA in acetone at 48-hour intervals. Mice were sacrificed 24 hours after the last TPA treatment and tails were amputated for preparation of whole mounts, as described previously (Braun et al., 2003). Briefly, the skin was peeled from the tail bone, stretched on 3MM paper and incubated (floating) on 5 mM EDTA/PBS for 4 hours at 37°C. Using forceps, the epidermis was separated from the dermis in intact sheets, which were subsequently fixed in neutral-buffered formalin for 30 minutes at room temperature (RT). After rinsing in PBS, whole mounts were stored in PBS/0.2% sodium azide at 4°C until use.
For the detection of LRCs, 0.5×0.5 cm pieces of fixed epidermal sheets were blocked and permeabilized by incubations in TBS containing 0.5% BSA and 0.5% Triton for 30 minutes. The whole mounts were then denatured in 2 M HCl for 2 hours followed by neutralization in 1 M Tris. After washing in TBS, the whole mounts were incubated overnight with FITC-conjugated anti-BrdU antibody (Roche) diluted 1:50 in TBS-BSA-Triton. The next day, whole mounts were washed extensively in TBS with 0.2% Tween20 and mounted in Vectashield (Vector Labs).
Imaging of whole mounts was performed with laser scanning confocal microscopy (LEICA SP5). Image stacks of 25-60 μm in the z-dimension were obtained at 2 μm intervals using a water immersion 20×/0.7 NA (numerical aperture) lens. Image stacks were converted to maximum intensity projections using Leica LAS AF v.1.6.3 software.
LRCs were identified as cells that retained the BrdU label at the end of treatment. The number of LRCs in treated and untreated whole mounts was determined per hair follicle [as CD34+, K15+ cells in the bulge area (Flores et al., 2005)] in 10 representative projections per mouse. The average number of LRCs and the number of LRCs normalized to a corresponding untreated control (mean±s.e.m.; controls were set to 100%) were generated to determine the stem cell mobilization. Statistical analysis was performed by using the Student’s t-test with Welch’s correction.
Confocal microscopy of Ki67 and K15
Whole mounts were permeabilized by incubation in TBS with 0.5% BSA and 0.5% Triton for 30 minutes. After washing in TBS, the whole mounts were incubated overnight with cytokeratin 15 Ab1 (1:50; Neomarkers, Clone LHK15) and Ki67 (1:400; Master Diagnostica), which were diluted in TBS-BSA-Triton. The next day, whole-mounts were washed extensively in TBS with 0.2% Tween20 and mounted in Vectashield with DAPI (Vector Labs). Imaging of whole mounts was performed as described for the LRC assay.
Chronic UV experiments
The skin from the backs of 6–8-week-old male mice (at least 7 mice per genotype) was shaved with electric clippers and irradiated chronically, three times per week, with UVB at a dose of 1.3 kJ/m2 per exposure. Tumors were scored once a week. Mice were sacrificed at human endpoints (when signs of poor health developed). Different parts of the skin were harvested, fixed in 10% buffered formalin, embedded in paraffin and subjected to full histopathological analysis.
Western blot analysis
A lysis buffer (20 mM HEPES pH 7.9, 25% glycerol, 0.4 M NaCl, 1 mM EGTA, 1 mM EDTA, 1% NP40) supplemented with a protease inhibitor cocktail was used to extract the total protein from skin samples (dermis and epidermis) obtained from littermate male mice at 9–10 weeks of age. After repetitive cycles of freezing and thawing, supernatants were sonicated and collected to determine the protein concentration. 40 μg of protein was loaded onto a 1 mm pre-casted 4–12% acrylamide gradient gel (NuPage). After transfer, nitrocellulose membranes were incubated overnight at 4°C with monoclonal antibodies against p21 (1:500; Santa Cruz C-19-G) and MDM2 (1:200; Abcam AB16895-50), and with a polyclonal antibody against p19 (1:200, Abcam Ab80), all of which were diluted in PBS-BSA (3%). After incubation (at RT for 1 hour) with respective Alexa Fluor 680-conjugated secondary antibodies (1:5000; Molecular Probes A21057 and A21076), fluorescent signals were detected by infrared scanning (LICOR Odessey) and quantified by ImageJ. The intensity of the bands was represented by the area under the plot and normalized for β-actin. Protein expression levels are represented and statistical analysis was performed by using the Student’s t-test with Welch’s correction.
Immunofluorescence
Skin sections of 4 μm were deparaffinized and rehydrated, and the antigens were subsequently retrieved with citric acid. After permeabilization in PBS with 0.1% Triton X-100, sections were blocked in 2% BSA for 1 hour then incubated with primary antibody.
p53
Permeabilized sections were incubated with rabbit anti-p53 (1:1400; CM5) and Cy3-conjugated goat anti-rabbit (1:400). Slides were washed in PBS with 0.1% Triton X-100 and then mounted in Vectashield with DAPI (Vector). Immunofluorescent images were analyzed using the Metamorph platform (version 6.3r6, Molecular Devices) to quantify the p53 levels. A threshold was set for the DAPI images, which were then segmented and converted to 1-bit binary images. This binary DAPI mask was applied to the matching Cy3 image in order to quantify nuclei for the level of p53. Cy3 fluorescence intensity was measured in units of ‘average gray values’. Fluorescence values for the basal layer were exported to Microsoft Excel where frequency histograms were generated.
Phospho-H2AX
Permeabilized sections were incubated with phospho-H2AXser139 (1:200, Upstate Biotechnologies 06–636) diluted in PBS with 2% BSA. After extensive washing, slides were incubated with a Cy3-conjugated secondary antibody (1:400, Jackson 115-166-071) and mounted in Vectashield with DAPI. The number of positive cells per total number observed basal cells was scored in at least 10 randomly chosen images.
Immunohistochemistry
Skin sections of 4 μm were deparaffinized and the antigens were subsequently retrieved with Tris-EDTA at pH 9 for α-MSH staining, citric acid for p19 staining, or with sodium citrate at pH6.5 for staining with p21, Ki67 and active caspase-3 (C3A). Endogenous peroxidase was blocked by incubation in H2O2 (DAKO) for 5 minutes. Sections were then blocked with fetal bovine serum (FBS) and incubated with rabbit anti-MSHalpha (1:1200; M0939, Sigma), goat polyclonal anti-p21 (1:75; Santa Cruz C-19-G, sc397-G), rat monoclonal antibody anti-p19arf (1:15; Santa Cruz Clone 5-C3-1), HRP-conjugated Ki67 (pre-diluted 1:200; Master Diagnostica 000310QD) and rabbit polyclonal anti-C3A (1:200; RyD AF835). Where necessary, sections were then incubated with HRP-conjugated secondary antibodies. Immunostaining was visualized with DAB and counterstained with hematoxylin.
Skin pigmentation
Pigmentation was studied macroscopically by weekly monitoring the tails of newborn animals from the age of 2 weeks. Hyperpigmentation-free survival curves, in which mice without development of pigmentation are considered to be survivors, were generated over the elapsed period of time.
Supplementary Material
Acknowledgments
We thank F. W. Alt for kind donation of the XPF-deficient mice; R. M. Serrano and M. E. Collado for mouse care and handling; E. Santos for genotyping; and C. J. McNees for critical reading of the manuscript. Thanks to the confocal microscopy and comparative pathology units at CNIO for excellent technical assistance. Juana Flores is acknowledged for pathological analysis of carcinogenesis experiments. G.J.S. is a Juan de la Cierva postdoctoral fellow funded by the Spanish Ministry of Health and Education. M.A.B.’s laboratory is funded by the MCyT (SAF2005-00277, GEN2001-4856-C13-08), the Regional Government of Madrid (GR/SAL/0597/2004), European Union (TELOSENS FIGH-CT-2002-00217, INTACT LSHC-CT-2003-506803, ZINCAGE FOOD-CT-2003-506850, RISC-RAD FI6R-CT-2003-508842, MOL CANCER MED LSHC-CT-2004-502943), the Spanish Association Against Cancer (AECC) and by the Korber European Science Award to M.A.B.
Footnotes
COMPETING INTERESTS
The authors declare no competing financial interests.
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://dmm.biologists.org/content/2/3-4/139/suppl/DC1
REFERENCES
- Andressoo JO, Hoeijmakers JH, Mitchell JR. (2006). Nucleotide excision repair disorders and the balance between cancer and aging. Cell Cycle 5, 2886–2888 [DOI] [PubMed] [Google Scholar]
- Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA, 3rd, et al. (2007). Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 356, 1317–1326 [DOI] [PubMed] [Google Scholar]
- Artandi SE, DePinho RA. (2000). A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 10, 39–46 [DOI] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. (2000). Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406, 641–645 [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 [DOI] [PubMed] [Google Scholar]
- Barrandon Y, Green H. (1987). Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA 84, 2302–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickenbach JR, McCutecheon J, Mackenzie IC. (1986). Rate of loss of tritiated thymidine label in basal cells in mouse epithelial tissues. Cell Tissue Kinet. 19, 325–333 [DOI] [PubMed] [Google Scholar]
- Blanco R, Muñoz P, Flores JM, Klatt P, Blasco MA. (2007). Telomerase abrogation dramatically accelerates TRF2-induced epithelial carcinogenesis. Genes Dev. 21, 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA. (2007). Telomere length, stem cells and aging. Nat. Chem. Biol. 3, 640–649 [DOI] [PubMed] [Google Scholar]
- Brash DE. (2006). Roles of the transcription factor p53 in keratinocyte carcinomas. Br. J. Dermatol. 154 Suppl. 1, 8–10 [DOI] [PubMed] [Google Scholar]
- Braun KM, Niemann C, Jensen UB, Sundberg JP, Silva-Vargas V, Watt FM. (2003). Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in wholemounts of mouse epidermis. Development 130, 5241–5255 [DOI] [PubMed] [Google Scholar]
- Celli GB, de Lange T. (2005). DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7, 712–718 [DOI] [PubMed] [Google Scholar]
- Chan SW, Blackburn EH. (2002). New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene 21, 553–563 [DOI] [PubMed] [Google Scholar]
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. (1999). p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97, 527–538 [DOI] [PubMed] [Google Scholar]
- Choudhury AR, Ju Z, Djojosubroto MW, Schienke A, Lechel A, Schaetzlein S, Jiang H, Stepczynska A, Wang C, Buer J, et al. (2007). Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat. Genet. 39, 99–105 [DOI] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M. (2007). Cellular senescence in cancer and aging. Cell 130, 223–233 [DOI] [PubMed] [Google Scholar]
- Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, D’Orazio J, Fung CY, Schanbacher CF, Granter SR, et al. (2007). Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell 128, 853–864 [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Teo SH, Jackson SP. (2004). Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 18, 1781–1799 [DOI] [PubMed] [Google Scholar]
- de Laat WL, Jaspers NG, Hoeijmakers JH. (1999). Molecular mechanism of nucleotide excision repair. Genes Dev. 13, 768–785 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2002). Protection of mammalian telomeres. Oncogene 21, 532–540 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2004). T-loops and the origin of telomeres. Nat. Rev. Mol. Cell. Biol. 5, 323–329 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- Dlugosz AA, Glick AB, Tennenbaum T, Weinberg WC, Yuspa SH. (1995). Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 254, 3–20 [DOI] [PubMed] [Google Scholar]
- Flores I, Cayuela ML, Blasco MA. (2005). Effects of telomerase and telomere length on epidermal stem cell behavior. Science 309, 1253–1256 [DOI] [PubMed] [Google Scholar]
- Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. (2004). Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 36, 94–99 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Samper E, Flores JM, Blasco MA. (2000). Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nat. Genet. 26, 114–117 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Samper E, Ramirez A, Flores JM, Martin-Caballero J, Jorcano JL, Blasco MA. (2001). Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 20, 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. (2001). Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 [DOI] [PubMed] [Google Scholar]
- Hsu CP, Lee LW, Shai SE, Chen CY. (2005). Clinical significance of telomerase and its associate genes expression in the maintenance of telomere length in squamous cell carcinoma of the esophagus. World J. Gastroenterol. 11, 6941–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. (1994). Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 [DOI] [PubMed] [Google Scholar]
- Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. (2004). The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2, E240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Denchi E, Celli G, de Lange T. (2006). Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes Dev. 20, 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, O’Connor MS, Qin J, Songyang Z. (2004). The telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 279, 51338–51342 [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. (1999). A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402, 551–555 [DOI] [PubMed] [Google Scholar]
- Muñoz P, Blanco R, Flores JM, Blasco MA. (2005). XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 37, 1063–1071 [DOI] [PubMed] [Google Scholar]
- Muñoz P, Blanco R, Blasco MA. (2006). Role of the TRF2 telomeric protein in cancer and ageing. Cell Cycle 5, 718–721 [DOI] [PubMed] [Google Scholar]
- Oh BK, Kim YJ, Park C, Park YN. (2005). Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am. J. Pathol. 166, 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak MA, Fanselow DL. (1983). Photobiology of melanin pigmentation: dose/response of skin to sunlight and its contents. J. Am. Acad. Dermatol. 9, 724–733 [DOI] [PubMed] [Google Scholar]
- Rajaraman S, Choi J, Cheung P, Beaudry V, Moore H, Artandi SE. (2007). Telomere uncapping in progenitor cells with critical telomere shortening is coupled to S-phase progression in vivo. Proc. Natl. Acad. Sci. USA 104, 17747–17752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley PA. (1997). Melanin. Int. J. Biochem. Cell Biol. 29, 1235–1239 [DOI] [PubMed] [Google Scholar]
- Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE. (2005). Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 436, 1048–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP. (2008). TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 82, 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. (2008). Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236 [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. (2007). How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell. Biol. 8, 703–713 [DOI] [PubMed] [Google Scholar]
- Siegl-Cachedenier I, Flores I, Klatt P, Blasco MA. (2007). Telomerase reverses epidermal hair follicle stem cell defects and loss of long-term survival associated with critically short telomeres. J. Cell Biol. 179, 277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T. (1998). Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487 [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. (2003). DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556 [DOI] [PubMed] [Google Scholar]
- Tian M, Shinkura R, Shinkura N, Alt FW. (2004). Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol. Cell. Biol. 24, 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK. (2007). Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc. Natl. Acad. Sci. USA 104, 7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. (1998). TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413 [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. (2001). The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413, 432–435 [DOI] [PubMed] [Google Scholar]
- Wu Y, Zacal NJ, Rainbow AJ, Zhu XD. (2007). XPF with mutations in its conserved nuclease domain is defective in DNA repair but functions in TRF2-mediated telomere shortening. DNA Repair (Amst.) 6, 157–166 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS. (2005). Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N. Engl. J. Med. 352, 1413–1424 [DOI] [PubMed] [Google Scholar]
- Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. (2004). TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279, 47264–47271 [DOI] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. (2003). ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12, 1489–1498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.