Abstract
MicroRNAs (miRNAs) are present in all known plant and animal tissues and appear to be somewhat concentrated in the mammalian nervous system. Many different miRNA expression profiling platforms have been described. However, relatively little research has been published to establish the importance of ‘upstream’ variables in RNA isolation for neural miRNA expression profiling. We tested whether apparent changes in miRNA expression profiles may be associated with tissue processing, RNA isolation techniques, or different cell types in the sample. RNA isolation was performed on a single brain sample using eight different RNA isolation methods, and results were correlated using a conventional miRNA microarray and then cross-referenced to Northern blots. Differing results were seen between samples obtained using different RNA isolation techniques and between microarray and Northern blot results. Another complication of miRNA microarrays is tissue-level heterogeneity of cellular composition. To investigate this phenomenon, miRNA expression profiles were determined and compared between highly-purified primary cerebral cortical cell preparations of rat primary E15–E18 neurons versus rat primary E15–E18 astrocytes. Finally, to assess the importance of dissecting human brain gray matter from subjacent white matter in cerebral cortical studies, miRNA expression profiles were compared between gray matter and immediately contiguous white matter. The results suggest that for microarray studies, cellular composition is important, and dissecting white matter from gray matter improves the specificity of the results. Based on these data, recommendations for miRNA expression profiling in neural tissues, and considerations worthy of further study, are discussed.
Keywords: miRNAs, microRNAs, brain, neurons, astrocytes, microarray, Northern, human, Alzheimer’s disease, RNA isolation
Introduction
MicroRNAs (miRNAs) are short regulatory RNAs that play important biological roles in plants and animals alike [1]. In mammals, there appears to be a concentration of miRNAs in the brain [2–5]. MiRNAs in brain have been implicated in many fundamental functions including neurodevelopment, plasticity, and apoptosis (see reviews [6, 7]). Furthermore, miRNA dysfunction has been implicated in brain cancers, neurodegeneration, and schizophrenia [8–11]. These are among the reasons that researchers have been interested in obtaining miRNA profiling data from neural tissues.
Many miRNA profiling platforms and techniques have been used to assay the miRNA repertoire of biological samples. Platforms have incorporated cloning, microarrays, PCR, high-throughput Northern blots, nanoscale technologies, and other modalities [2–4, 12–29]. Each technique presumably entails specific strengths and weaknesses, including sensitivity, specificity, and cost-related considerations.
While the miRNA expression profiling platforms have received considerable interest, less attention has been focused on the ‘upstream’ steps of miRNA expression profiling: tissue selection, tissue dissection, and RNA isolation. Yet these pre-analytical steps are very important. When isolating RNA from cells or tissues, there are many potential opportunities for the introduction of systematic bias and/or experimental error. Some of these have been shown to be important in human studies regarding mRNA integrity (see for example [30–35]), however, systematic studies of pre-analytical variables for miRNA expression have not been published. For studies of human brain tissue, important variables include pre-mortem factors, as well as autopsy conditions, tissue dissection technique, and RNA isolation technique.
We assessed preliminarily the effects on apparent miRNA repertoires (as quantified by a conventional miRNA microarray and Northern blotting) that may be associated with tissue processing, RNA isolation techniques, and the identity of the neuroepithelial cell types in the sample. It should be stressed that these experiments are just an initial effort because much additional work needs to be performed to assess the nature and degree of changes in a miRNA-containing sample that are brought about merely by the techniques with which the RNA is isolated. Furthermore, each expression profiling platform will have unique issues. With those caveats, considerations worthy of further study are discussed.
Methods
RNA isolation from a human cerebral cortex using different methods
All RNA isolation methods were used on powdered brain (91 y.o.non-demented female, post-mortem interval 1.75 hours; neuropathology from subjacent tissue showed very minimal cortical Alzheimer’s-type pathology which is usual for the patient’s age) from superior and middle temporal cortical tissue which was prepared thusly: Tissue (6 gms) that had been snap-frozen in liquid nitrogen (LN) and then transferred to a −80C freezer was placed in an RNAse-free pestle and bathed in LN, and then ground to fine powder in LN for ten minutes with multiple changes of LN. Homogeneously powdered tissue in LN was then transferred to a −80C freezer without ever having thawed. Tissue for RNA isolation was directly placed in lysis buffers (see below) for the subsequent RNA isolation. Trizol LS (Invitrogen) was the only technique that did not specify or provide a specific lysis buffer. We performed one set of experiments with the exact protocol given in the Trizol LS instructions, and one experiment with an additional overnight −20C precipitation step. MiRVANA kit: after homogenization as described above, we isolated small, large, and total RNA exactly as described in the manufacturer’s protocol. Stratagene total RNA and microRNA: the same as the protocol, except for the homogenization caveat above. Performing the Qiagen RNeasy was unsuccessful when we used the needle homogenization technique as described in the manufacturer’s protocol, so a dounce-type homogenizer was employed instead with success.
RNA isolation from tissue cultured cells
RNA was isolated from primary rat cultures (embryonic day 15–18) that were highly purified (>98%) for either hippocampal neurons, or cerebral cortical astrocytes. Cells were purified and cultured using conventional methods as described previously [36–38]. Cells were washed in PBS, scraped in PBS, and then total RNA was isolated using Trizol LS as described above according to manufacturer’s protocol. For the studies involving H4 glioneuronal tumor-derived cell line [39, 40], the cells were cultured in conditions as per their vendor (ATCC, Inc.) and harvested at ~80% confluencein a 100cm culture dish. Cells were washed in PBS, collected in PBS, and RNA was isolated using Trizol LS according to manufacturer’s recommended protocol.
RNA isolation from gray matter and subjacent white matter
RNA was isolated from gray matter and white matter immediately subjacent under RNAse-free conditions. For this experiment, the superior and mid-temporal isocortex was obtained after having been snap-frozen in LN from three different patients with similar clinical histories from the University of Kentucky ADC (Alzheimer’s Disease Center) Brain Bank: a 93 year old female (Patient 1); an 84-year old female (Patient 2); and a 90 year old female (Patient 3). A razor blade was used to quickly separate the two different tissues, (<500mg each) which were then each homogenized using a Dounce-type glass and Teflon homogenizer on ice. Total RNA was isolated from the homogenate using Trizol LS as described above according to the manufacturer’s protocol. RNA was analyzed using the Agilent Bioanalyzer.
Microarray methods and analyses
The mercury Labeling Kit (Exiqon) was used for target labeling and hybridization to a conventional miRNA microarray. The oligonucleotide probe arrays were printed on Codelink substrates, processed and scanned at the University of Pennyslvania Microarray Core Facility. The protocols used are indicated in Supplementary File #1. R-squared values were calculated (and charted) using Log(10) values of the raw microarray data.
Northern blots
Northern blot analysis
10 µg of total RNA, and the equivalent total small RNA isolated from the same quantity of tissue was separated on a denaturing 15% polyacrylamide gel and transferred electrophoretically to a Hybond-N-Nylon filter membrane. After transfer, the membrane was soaked briefly in 2XSSPE, and was cross-linked using a UV cross-linker. Specific miRNA probes using their antisense DNA oligonucleotides were labeled by T4 polynucleotide kinase with 30 µCi gamma-32P-dATP (6000 Ci/mmole). The membrane was first prehybridized in a hybridization buffer (50% formamide, 5X SSPE, 5X Denhardt’s, 0.5% SDS, 80 µg/ml shark sperm DNA)for at least 2 hr at 37 °C, follow by hybridization with a specific probe for 6 hr to overnight. Afterwards, the membrane was washed 2–3 times with 2X SSC containing 0.1% SDS for 20 min at the same temperature. The membranes were wrapped and exposed to a phosphor-storage screen. The signals were detected by scanning the phosphor-storage screen using a phosphorimager scanner (Typhoon 9400, Molecular Dynamics, GE Healthcare Bio-Sciences, Piscataway, NJ). The detected signals were quantified using ImageQuant TL software (Amersham Biosciences, Piscataway, NJ), and normalized against the signal intensity of 5SrRNA by ethidium bromide gel staining. The membrane was stripped and subsequently hybridized with other miRNA probes.
Results
RNA isolation – effect on RNA quality parameters
The same starting material was used for each isolation technique (see above for detailed description). The different RNA isolation techniques (Table 1) were chosen to include widely-used methods, associated with manufacturers’ supplied protocols, that incorporated different one-step biochemical principles (e.g., phenol/chloroform and resin-based columns, total RNA versus large or small RNA). PAGE-based RNA isolation techniques were not used because they employ a two-stage isolation strategy. The results of these experiments can be described in terms of RNA quality parameters; miRNA expression profiling results; and the relationship between the microarray results and Northern blots results (See Table 1, Figure 1–Figure 2, and Supplementary File 1–Supplementary File 5). In terms of RNA quality, Agilent Bioanalyzer results are shown in detail in Supplementary File 2–Supplementary File 4. The Agilent Bioanalyzer “RIN” (RNA integrity number) is measurement for degree of degradation in a total RNA sample based on integrated Bioanalyzer signal amounts under the expected ribosomal peaks or detected in smaller degradation products, and can be used to compare the quality of different RNA samples. Small RNA is not correctly assessed by the RIN algorithm and the resulting RINs should not be compared to those from total RNA.
Table 1. Quality and Quantity parameters of Isolated RNA using different methods.
RNA parameters using different RNA isolation techniques on human brain tissue. Note, the ng/µl results are only presented in order to give an idea of the consistency of the yield within a given method, because the inputted amounts were different for each method. Also, there were apparent differences in the A260/A230 ratios for which we do not have a ready explanation.
| Sample ID--all replicates | Method Type* | RNA type** | Yield | RIN*** | |||
|---|---|---|---|---|---|---|---|
| ng/uL | A260 | 260/280 | 260/230 | Readout | |||
| Stratagene Total RNA Rep 1 | P/C | Tot | 359.3 | 9.0 | 2.1 | 1.7 | 7.2 |
| Stratagene Total RNA Rep 2 | 476.1 | 11.9 | 2.1 | 1.9 | 7.6 | ||
| Stratagene Total RNA Rep 3 | 463.5 | 11.6 | 2.0 | 1.8 | 7.5 | ||
| Stratagene small RNA Rep 1 | P/C | Small | 183.7 | 4.6 | 1.9 | 1.6 | N/A |
| Stratagene small RNA Rep 2 | 339.7 | 8.5 | 2.0 | 1.7 | N/A | ||
| Stratagene small RNA Rep 3 | 95.0 | 2.4 | 1.9 | 1.1 | N/A | ||
| Qiagen RNEasy Rep 1 | Col | Large | 238.5 | 6.0 | 2.2 | 2.0 | 8 |
| Qiagen RNEasy Rep 2 | 222.4 | 5.6 | 2.2 | 2.1 | 8.4 | ||
| Qiagen RNEasy Rep 3 | 123.0 | 3.1 | 2.1 | 2.1 | 7.9 | ||
| Ambion Total RNA Rep 1 | Col | Tot | 181.7 | 4.5 | 2.1 | 1.8 | 8 |
| Ambion Total RNA Rep 2 | 172.1 | 4.3 | 2.1 | 1.6 | 7.4 | ||
| Ambion Total RNA Rep 3 | 183.7 | 4.6 | 2.1 | 1.7 | N/A | ||
| Ambion Large RNA Rep 1 | Col | Large | 164.6 | 4.1 | 2.1 | 0.5 | 5.9 |
| Ambion Large RNA Rep 2 | 118.4 | 3.0 | 2.1 | 0.5 | 7.8 | ||
| Ambion Large RNA Rep 3 | 160.3 | 4.0 | 2.1 | 0.8 | 7.4 | ||
| Ambion Small RNA Rep 1 | Col | Small | 21.4 | 0.5 | 1.9 | 0.6 | N/A |
| Ambion Small RNA Rep 2 | 27.8 | 0.7 | 1.9 | 0.7 | N/A | ||
| Ambion Small RNA Rep 3 | 21.6 | 0.5 | 1.9 | 0.5 | N/A | ||
| Invitrogen Trizol LS Conv Rep 1 | P/C | Tot | 318.5 | 8.0 | 2.0 | 1.9 | N/A |
| Invitrogen Trizol LS Conv Rep 2 | 354.6 | 8.9 | 2.1 | 1.7 | N/A | ||
| Invitrogen Trizol LS Conv Rep 3 | 311.9 | 7.8 | 2.1 | 2.0 | 7.6 | ||
| Trizol LS Add'l −20 Precip Rep 1 | P/C | Tot | 415.3 | 10.4 | 2.1 | 1.5 | 7.7 |
| Trizol LS Add'l −20 Precip Rep 2 | 311.9 | 7.8 | 2.1 | 1.7 | 8.1 | ||
| Trizol LS Add'l −20 Precip Rep 3 | 374.2 | 9.4 | 2.1 | 1.4 | 8.6 |
P/C-phenol chloroform
Col--Column
Tot-total
Agilent RNA Integrity Number
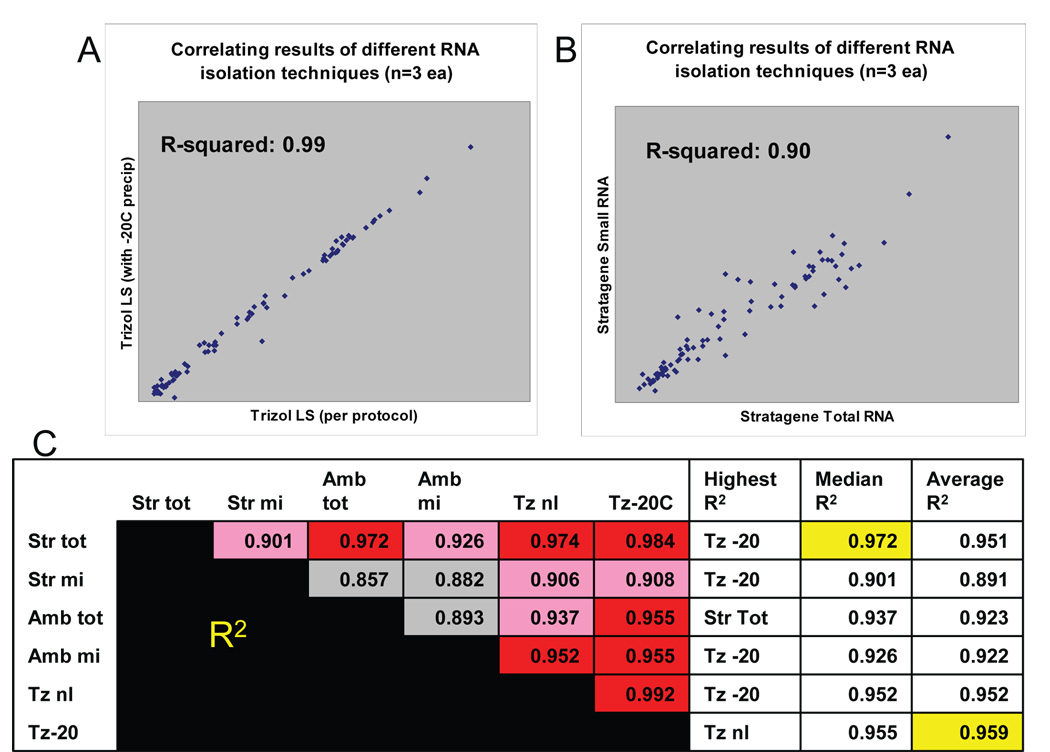
Figure 1.
Correlating the results between the six different RNA isolation techniques that are expected to include microRNAs. These comparisons show that distinct RNA isolation methods result in different miRNA profiling results. These data also may help to predict which of the techniques is most able to provide results that correlate with the outcomes of other methods. Correlations were performedusing linear regression in comparing the results of three different microarray experiments (biological replicates). Representative comparisons are depicted in Figs 1A and 1B. . The results of cross-comparisons between all the techniques is presented in Fig 1C. The RNA isolation methods shown here are, respectively, the Stratagene Total RNA, Stratagene small RNA, Ambion Total RNA, Ambion Small RNA, Trizol LS per provided protocol, and Trizol LS with added −20C overnight precipitation step added. For Fig. 1C, each method is compared against each other on the left, using linear regression, in which higher correlation is depicted in red, and lower in gray. For each of the individual RNA isolation methods, the columns to the right of Fig 1C show the method which most closely correlates to that method; the median degree of correlation (R-squared; highest highlighted in yellow); and the average degree of correlation (highest highlighted in yellow). In sum, the overall R-squared are generally >0.90 but the correlations are highest between RNA samples that are designed to obtain total RNA with phenol/chloroform extraction (such as in Fig A). Small RNA isolating techniques show lower degree of correlation, as shown in Fig B.
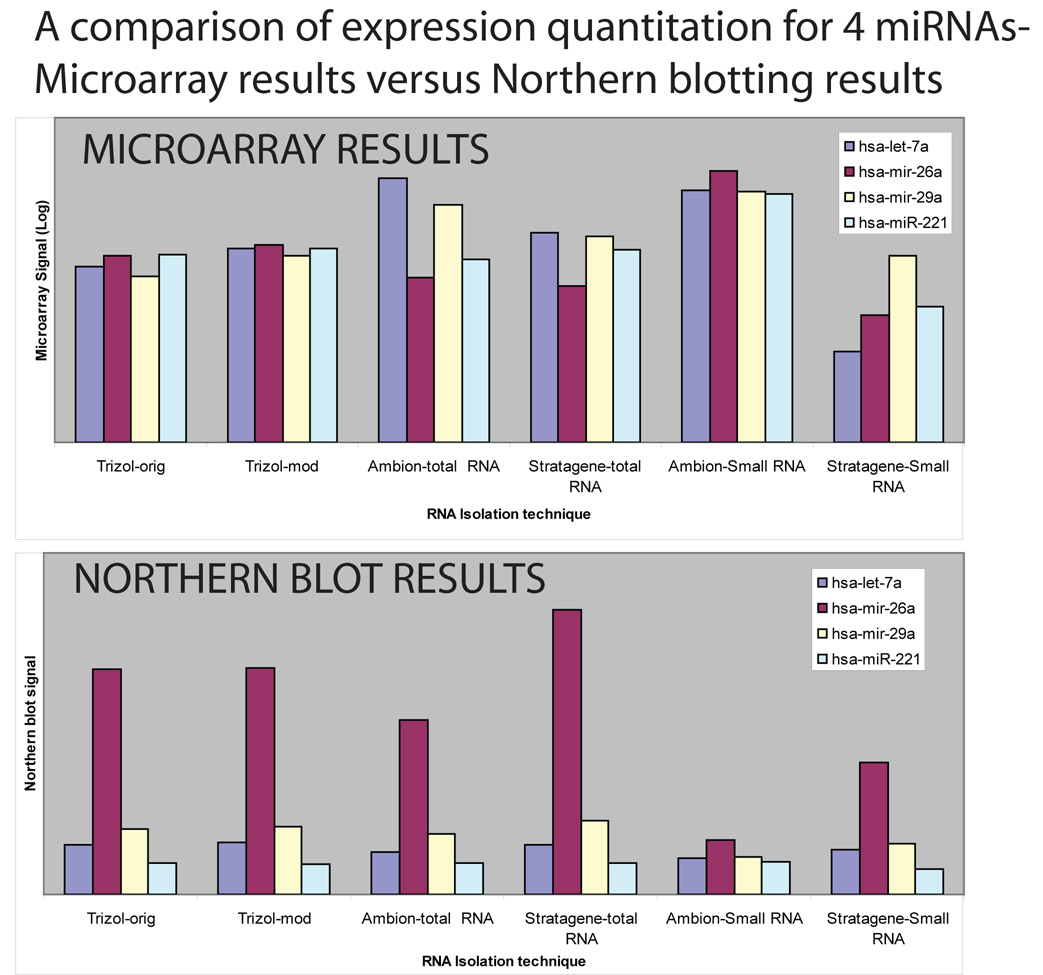
Figure 2.
The microarray and Northern results were not identical, and show that RNA isolation methods will impact differently on different profiling platforms. The top chart is microarray data for miR-26a, miR-29a, Let-7a, and miR-221. The bottom chart relates the data for miRNA expression from the same RNA samples via Northern blot. The microarray and Northern blot data are presented in Supplementary File 5. All the RNA was obtained as technical replicates. The microarray data represents the results of three different microarrays, the Northern blots a single experiment each. Two important differences can be seen relating the microarray results to Northern blots: firstly, the Northern blots appear to be relatively more sensitive for miR-26a, but less sensitive for miR-221 in all cases. Secondly, the overall pattern of expression is similar for between the RNA isolation techniques in the Northern blots. However, for the microarray results, the pattern varied for the different RNA isolation methods, which indicates that for this microarray platform the results are more sensitive (than Northern blots)to the RNA isolation method used.
The effect of different RNA isolation techniques on miRNA microarray results
Overall, RNA derived from a single tissue source was of differing quality depending upon the method used to isolate it. These results are demonstrated in Table 1. However, miRNAs are known to be relatively robust even in conditions that degrade other RNAs [42]. The important focus of concern was whether or not different RNA isolation techniques enabled consistent profiling results that could be validated by Northern blots. In our hands, there was little difference in the results (either by microarray or Northern blotting) when we introduced a longer, −20C isopropanol precipitation step in the Trizol LS processing, rather than using the manufacturer’s protocol exactly (Figure 2). Differences were seen in A260/230 ratios that we cannot readily explain. We included two RNA isolation methods that are nominally irrelevant to miRNA research: Ambion large RNA and Qiagen RNeasy. These methods both extract predominantly RNAs that are larger than miRNAs. These were included to test for unprocessed miRNA precursors that may be present in both large RNA and total RNA samples. However, we did not find these samples to provide contributory data. Hence, the results of the microarray data from RNA isolations for the larger RNA methods are included (see Table 1 and Supplemental File 5), but were not analyzed further.
In order to gain some insight into whether one or another of the RNA isolation methods was most desirable, we used two different strategies: 1. comparing microarray results from correlation between the different methods; and 2. comparing the microarray results from the different methods to a different profiling method—Northern blots.
In comparing the microarray results from different isolation methods, average signals results were used from three technical replicates of six different RNA isolation methods. The results are shown on Figure 1 and Figure 2. The techniques that showed the higher correlation coefficients tended to use the phenol/chloroform-based protocol (Trizol LS total and Stratagene total RNA kits).
In comparing the microarray results from different methods to Northern blots (Figure 2), we used only a selection of probes referent to miRNAs that were expressed at a high level by microarray. The digested results are presented as Figure 2. The raw data of the Northern blots and the comparison with microarray results are shown in Supplemental File 5. These results show differences between the microarray results and the Northern blots—the Northern blot appeared to be somewhat more sensitive for miR-26a and less sensitive for miR-221. Also, the pattern of relative signal strengths for the same miRNA in different extracts appeared to vary less in the Northern blot studies in comparison to the microarray studies. These results are discussed in greater detail below.
The importance of different cell types in the brain
Primary rat astrocytes versus neurons
Different CNS cell types express different miRNAs [43], and we sought to query the importance of this factor in microarray analyses. We first compared the results of miRNA expression profiling between primary rat cultured cells (E15–18) that were highly enriched either for neurons (hippocampal rat neurons) or astrocytes (cerebral cortical rat astrocytes). The results of these experiments are shown in Figures 4 and 5. There was a poor correlation between the miRNA repertoire from these two types of neuroepithelial-derived cells (Figure 3), indicating a cell type-specific expression of miRNAs.
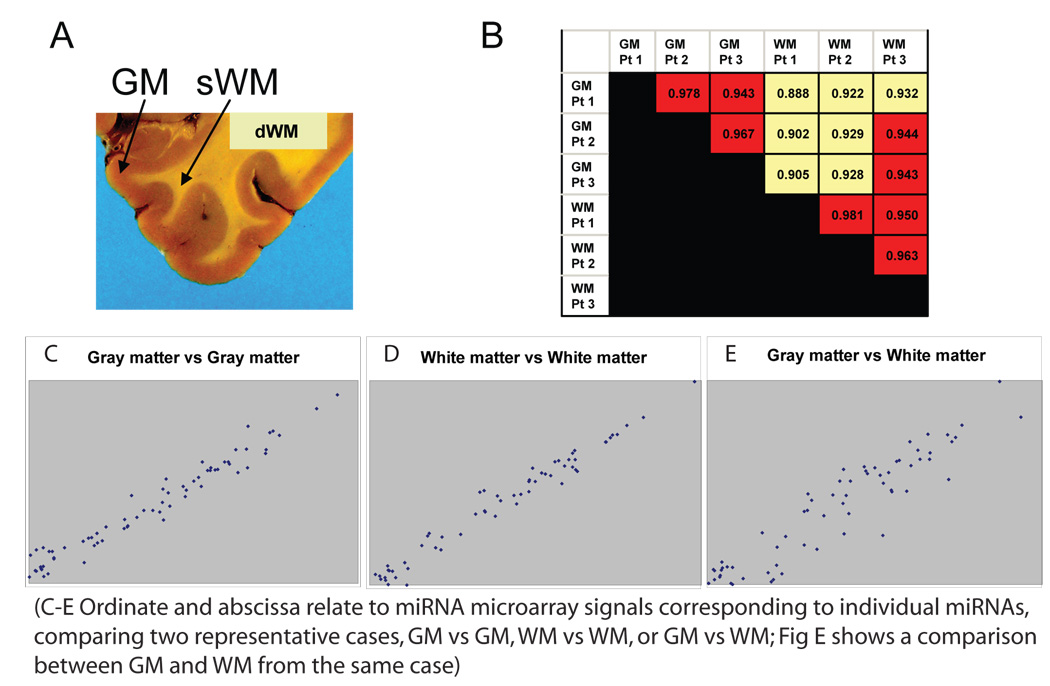
Figure 4.
Human gray matter and white matter express different miRNA repertoire, which poses a challenge to tissue sampling of human cerebral cortex. These figures comparing miRNA profiles derived from RNA isolated from gray matter (GM) and superficial white matter (sWM; as opposed to deep white matter dWM) of human brain (gross coronal section for illustration’s purposes). MiRNAs from GM and sWM from the superior and middle temporal cortex from three different human patients were compared. These biological replicates tended to have a high degree of expression correlation across different miRNAs when gray matter was compared with gray matter, or white matter to white matter, from different patients. However, when miRNA profiles were compared between white matter and gray matter (whether within or between patients) there was less correlation. Figs 4 C–E are charts that show the correlation in miRNA expression for representative cases of gray matter and white matter. Note that the correlation in Figs 4 C and D are tighter in comparison to Fig 4E, despite the fact that only figure 4E is comparing the miRNA profiling results within the same patient.
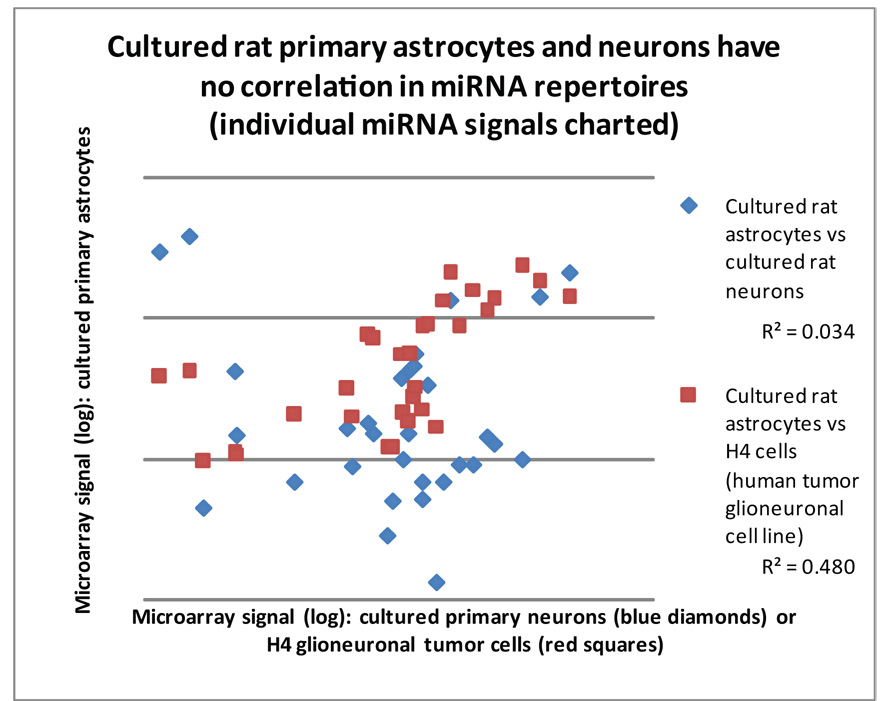
Figure 3.
Neurons and astrocytes express distinct subsets of miRNAs and this can be used to help ‘genotype’ a human cell line. A comparison of rat E15–18 derived primary cortical cultures—hippocampal neurons versus cerebral cortical astrocytes—shows that each has a relatively different miRNA repertoire. Each data point represents a particular miRNA, correlating the microarray single intensity from 3 combined and averaged microarray experiments each. Note that there is a very poor correlation (R2 correlation coefficient = 0.034; blue diamonds) between the miRNA expression profile of these primary cultured cells. Human H4 glioneuronal cancer line cells show a far better correlation with cultured primary rat astrocytes (R2= 0.48; red squares) than primary cultured rat neurons (R2=.016; data not shown).
Since different neural cell populations show distinct miRNA profiles, we sought to test a cell line to see if those differences could be biologically informative. We tested the H4 “glioneuronal” cell line [40]. The miRNA repertoire for H4 cells was much closer to the miRNA repertoire of primary cultured rat astrocytes than to that of primary cultured rat neurons. This is despite the fact that both the neuronal and astrocytes primary cells were from rat whereas the H4 cells are a tumor cell line from humans.
Human cerebral cortical gray matter versus white matter
Gray matter and white matter are terms that are used to distinguish areas in the CNS, as shown in Figure 4. MiRNA expression profiles were compared from human superior and medial temporal isocortex tissue from age-matched and clinically-matched patients from the University of Kentucky ADC Brain Bank. Gray matter and immediately subjacent white matter were dissected from tissue portions that had been snap-frozen in liquid nitrogen, and RNA was isolated and profiled using a microarray as above. The results indicate that the miRNA repertoire of superficial white matter tends to correlate across different patients to a higher degree than the miRNA repertoire correlates with that of the immediately overlying gray matter in the same patient. Hence, closely-juxtaposed gray matter and white matter express different miRNA profiles, and these differences are relatively consistent between different patients.
Discussion
The details of RNA isolation can have profound -- and even surprising -- impact on experimental results, particularly in the context of miRNA microarray data. Important focus points include details of tissue processing, RNA isolation techniques, and the significance of different miRNA repertoires in different neuroepithelial cell types.
A limitation of this study is that detailed analyses were performed using only a single miRNA profiling platform. It provided a large amount of data—these results comprise dozens of different arrays. However, there now many different miRNA profiling platforms in the scientific community and it is impossible to predict how a particular tissue processing step will differentially affect the results across miRNA microarray platforms, much less versus PCR-based or sequencing-based platforms. Pre-analytical parameters must be evaluated for each and every platform separately for this reason. Another limitation of this study was that the neuronal and astrocyte populations that were being compared were not perfectly analogous. The astrocytes were from cerebral isocortex and the neurons from hippocampus which is also cortical, but somewhat distinct embryologically and cytoarchitectonically. Perhaps more importantly, even in culture, mature astrocytes divide and neurons do not. It is likely that some of the miRNA expression changes are more specific to dividing cells versus non-dividing cells, however, this difference may actually reflect the normal miRNA repertoire in situ, where, again, astrocytes unlike neurons divide into adulthood. Another potential limitation of this study is that some of the comparisons were made between miRNAs with paralogs. For example, miRs-29a, -26a, and let-7a each have relatively closely-related but distinct miRNA genes in humans. We have found large differences in reported expression for miR-29a/b, miR-26a/b, and let-7a/b, for example, but, further work may incorporate RTqPCR to better delineate differences in expression for these and other paralogous miRNAs.
This study focused on variables of RNA isolation, including the specific RNA isolation technique, as well as miRNA expression differences in some of the different cell types in the CNS, and between gray and subjacent white matter of adult humans. These data suggest that the method used to isolate RNA can impact the results, and this effect appears to be more marked in the context of a microarray platform than in Northern blots. These data do not in themselves recommend a particular RNA isolation method as being absolutely ‘superior’ to others. However, we found the application of Trizol LS somewhat easier to use with more consistent results than other methods in obtaining total RNA. In our hands, there was only a small difference in the results (either on microarray or Northern blotting) when we introduced a longer, −20C alcohol precipitation step in the Trizol LS processing. As expected, in per-microgram of RNA, there was greater signal for the miRNA microarray when we used ‘small RNA’ derived from the Ambion MiRVANA kit.
It is noteworthy that the total RNA isolation techniques did not appear biased toward any RNA that was also present in the large RNA samples. This is an important bias to rule out, but may be differentially present or absent in different profiling platforms.
This study demonstrates that identifying an optimal RNA isolation technique for miRNA microarray studies is non-trivial. Whether or not there is a conventionally-assumed “gold standard”, we feel that there is no absolute standard for miRNA expression profiling. We have received (as yet unpublished) accounts from a number of colleagues describing discrepant results from different profiling modalities (microarrays, Northern blots, PCR, sequencing), and, even theoretically, this is not surprising, because each would be predicted to have unique biases. In our hands, different profiling platforms can have differential sensitivities to specific miRNAs. For example, relative to the microarray experiments, the Northern blots “report” a higher relative amount of miR-26a, and a lower relative amount of miR-221. Furthermore, the Northern blot also showed a greater degree of constancy in the results, irrespective of the RNA isolation technique. By contrast, microarray results appeared to vary more according to which RNA isolation technique was used. This was not apparently due to the presence or absence of larger miRNA precursors in the total RNA isolation samples, because the techniques for isolating only larger RNA fragments (RNEasy and Ambion large RNA isolation) did not detect miRNA signals that were present in the total RNA isolation techniques but absent using the small RNA isolation techniques. In fact, miRNA profiles using the total RNA isolation techniques (Trizol LS and Stratagene total RNA isolation) appeared to have a relatively robust correlation with Northern blotting.
Our interpretation of these results provide the basis for some recommendations for isolating RNA for miRNA expression profiling:
It is important to be methodologically consistent across different tissues and samples; never compare RNA against each other that were isolated using different techniques.
When ‘positive’ findings are indicated by miRNA microarray, they must be carefully validated using different modalities.
The various biases of each technique should be investigated (including for Northern blots!).
When reporting the results of a microarray experiment, the methods involved in RNA isolation should be carefully documented.
In addition to studying the effects of the RNA isolation technique per se, this study provided some insights into the significance of the cell and tissue variables in assessing neural tissues. One aspect of these studies involved a comparison between primary cultures of rat E15–18 neurons and astrocytes. The miRNA repertoires of these cell types showed differences. Some of these differences have been reported previously in an excellent study by Smirnova et al [43]. Specifically, these researchers reported that miR-23a and miR-26a were highly enriched in astrocytes, whereas mir-124a and miR-128 were specific to neurons. We and others have also previously shown that miR-124a is selectively expressed in neurons [44, 45]. These results give an added measure of confidence in the degree to which these results can be generalized.
We also found differences in the miRNA profiles in the gray matter and the white matter that is immediately underlying gray matter. Gray matter and white matter are terms that are used to distinguish areas in the CNS. Gray matter includes neurons and white matter is the domain with a preponderance of myelinated axons and oligodendrocytes. Many astrocytes are present in both gray matter and white matter. As a practical consideration, the junction between gray matter and white matter is not absolutely distinct, and particularly in some portions of the cerebral cortex (e.g., motor cortex) the transition can be impossible to delineate exactly. However, in the present context, the question is whether it is important to dissect away – and/or analyze separately – the white matter even in a relatively small sample of human brain (deeper white matter may be expected to have a more clear-cut difference in expression profiles). Our results, showing that gray matter and superficial white matter have differences in miRNA expression, are at least partly due to the different cell types that are present in gray versus white matter, the latter having more oligodendrocytes and less neurons. This outcome would probably differ across distinct areas of the cerebral cortex, for practical reasons such as the fact that in temporal cortex there are greater numbers of neurons within white matter than in many other areas of the cerebral cortex.
Taken together, the finding of different miRNA profiles in primary cultures of neurons versus astrocytes, and also the difference between gray matter and nearby white matter, provides specific considerations for dissection of CNS tissue for RNA isolation in the context of miRNA microarrays:
When comparing different tissue types, it is important to consider the biases inherent to tissues with different ratios of neurons to glial cells (e.g. gliomas versus normal brain, or infarctions versus non-infarcted tissue).
An important difference between mature neuronal and glial cells are that only the latter tend to divide, and so some of the miRNA expression differences between them may subserve either promoting (glia) or suppressing (neurons) cell division.
Dissect away white matter from gray matter, as it will increase the specificity of the results; for this reason it is logical to have the dissection performed by a person with expertise.
As we have reported previously [45], tissue-level miRNA microarray analyses should, where possible, be complemented with cellular and subcellular level analyses (in situ hybridization) of particular miRNAs.
Prior studies have focused upon the impact of RNA isolation variables, in the context of mRNA, but, miRNAs may have different stability issues. For example, miRNAs appear to be more stable in formalin-fixed, paraffin-embedded tissue than are mRNAs [32, 42]. Many unresolved issues remain. For example, the impact of premortem variables (for example, patients being treated with morphine, or chemotherapy, or other highly biologically active medication?; or, pre-mortem pain or stress?). Also, post-mortem effects (post-mortem interval, tissue pH, tissue homogenization variables, and freezing and thawing) may be important. The relative sensitivity and specificity differences among the various miRNA profiling techniques may lead to discrepant results. It is imperative to further investigate the significance of these variables to lay a solid foundation for further studies.
It must be acknowledged that the study of miRNAs in the brain is in its infancy, and the technical aspects of miRNA isolation for expression profiling is likewise in an early stage. These issues merit further study. This does not of course preclude obtaining very interesting – and possibly biologically and/or medically important -- results with currently available miRNA expression profiling technology.
Supplementary Material
Supplemental File 1: Microarray methods
Supplemental Files 2–4: Bioanalyzer results
Supplemental File 5: Northern blot and microarray raw data
Table 2. Microarray signals from cells in culture for comparison’s sake.
The miRNAs expressed in rat primary E15–E18 cultures show a differing expression profile for cortical neurons and astrocytes. The highly-expressed miRNAs are shown. The human H4 glioneuronal cell line shows an expression profile that is not exactly like either rat primary cell type, however, it more closely resembles the rat astrocytes. These data represent three technical replicates each.
| Primary rat astrocytes | Primary rat neurons | H4 cells | |
|---|---|---|---|
| miR-124b | 12.3 | 2924.5 | 380.6 |
| miR-124a | 18.2 | 3805.5 | 414.5 |
| miR-20 | 22.1 | 44.7 | 94.8 |
| miR-128 | 33.6 | 420.6 | 108.9 |
| miR-219 | 35.3 | 147.4 | 108.5 |
| miR-151* | 76.7 | 67.9 | 206.1 |
| miR-213 | 155.2 | 163.9 | 314.7 |
| miR-16 | 168.7 | 87.8 | 201.0 |
| miR-100 | 208.6 | 177.2 | 750.2 |
| miR-320 | 222.0 | 152.4 | 712.0 |
| miR-143 | 266.0 | 28.2 | 119.3 |
| miR-143a | 288.4 | 49.7 | 119.3 |
| miR-99a | 328.0 | 374.3 | 557.5 |
| miR-145 | 334.2 | 97.7 | 211.2 |
| miR-26a | 358.9 | 149.0 | 181.5 |
| miR-99 (ctrl) | 370.1 | 433.9 | 555.7 |
| miR-181a | 382.8 | 458.7 | 270.0 |
| miR-107 | 397.7 | 546.5 | 326.3 |
| miR-27 | 429.2 | 51.3 | 873.7 |
| miR-22 | 430.8 | 67.9 | 225.5 |
| miR-125a | 460.1 | 329.5 | 899.8 |
| miR-193 | 526.4 | 13.2 | 169.3 |
| miR-23b | 576.7 | 67.9 | 1347.5 |
| let-7a | 629.2 | 1338.0 | 2098.4 |
| miR-24-1,2 | 706.9 | 89.4 | 864.6 |
| miR-23a | 861.6 | 89.4 | 1592.0 |
| miR-29 | 1035.2 | 144.1 | 1120.0 |
| miR-221 | 1135.4 | 127.5 | 1364.3 |
| miR-21 | 1662.4 | 97.7 | 2359.3 |
| let-7bL | 2101.5 | 1424.2 | 1824.9 |
| miR-125b | 3146.6 | 2121.3 | 1406.8 |
Table 3. miRNAs that are more expressed in either gray matter or white matter in human temporal lobe isocortex (n=3 for WM, n=6 for GM).
A comparison between human temporal lobe cerebral cortical tissue from gray matter (GM) and from white matter (WM) shows some miRNAs that are relatively concentrated in GM or WM. The particular miRNAs that are shown are from among the 50 miRNAs that are show highest expression in human brain. Those with a GM/WM ratio <0.75 or >1.5 are shown. These results indicate the importance of technical expertise in sampling tissue from human cerebral cortex for miRNA expression profiling.
| GM/WM | |
|---|---|
| White matter concentrated | Ratio |
| miR-100 | 0.49 |
| miR-151* | 0.50 |
| miR-213 | 0.51 |
| let-7bL | 0.53 |
| miR-23b | 0.62 |
| miR-191 | 0.66 |
| miR-181a | 0.68 |
| miR-320 | 0.72 |
| miR-342 | 0.73 |
| Gray matter concentrated | |
| miR-143 | 1.65 |
| miR-145 | 1.74 |
| miR-124 | 1.83 |
| miR-29a | 1.89 |
| miR-172 | 1.98 |
| miR-185 | 2.09 |
| miR-128a | 2.16 |
| miR-143 | 2.23 |
| miR-128ash | 2.27 |
| miR-128b | 2.31 |
| miR-126 | 2.39 |
| miR-145sh | 2.48 |
| miR-29b | 2.87 |
Acknowledgements
We thank very kindly Dr. James Geddes and Dr. Christopher Norris for their provision of rat primary cultures (hippocampal neurons and cerebral cortical astrocytes), as well as Ms. Irina Artiuthin for technical assistance in the rat cultures. We thank Dr. Guiliang Tang for technical and theoretical perspectives in miRNA microarray work. We also thank Ms. Willa Huang for technical help in RNA isolation. Funding was provided through NIH K08 NS050110
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends in biochemical sciences. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome biology. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA (New York, N.Y. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA (New York, N.Y. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer research. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 6.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 7.Kosik KS, Krichevsky AM. The Elegance of the MicroRNAs: A Neuronal Perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochemical and biophysical research communications. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome biology. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Molecular cell. 2006;24:157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 11.Bilen J, Liu N, Bonini NM. A new role for microRNA pathways: modulation of degeneration induced by pathogenic human disease proteins. Cell cycle (Georgetown, Tex. 2006;5:2835–2838. doi: 10.4161/cc.5.24.3579. [DOI] [PubMed] [Google Scholar]
- 12.Davidson TS, Johnson CD, Andruss BS. Analyzing micro-RNA expression using microarrays. Methods in enzymology. 2006;411:14–34. doi: 10.1016/S0076-6879(06)11002-2. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic acids research. 2004;32:e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA (New York, N.Y. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonetta L. Gene expression: an expression of interest. Nature. 2006;440:1233–1237. doi: 10.1038/4401233a. [DOI] [PubMed] [Google Scholar]
- 16.Yu Z, Jian Z, Shen SH, Purisima E, Wang E. Global analysis of microRNA target gene expression reveals that miRNA targets are lower expressed in mature mouse and Drosophila tissues than in the embryos. Nucleic acids research. 2007;35:152–164. doi: 10.1093/nar/gkl1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic acids research. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA (New York, N.Y. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, Lobenhofer EK, Sharon E, Shiboleth YM, Shtutman M, Bentwich Z, Einat P. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome research. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 21.Tang F, Hajkova P, Barton SC, Lao K, Surani MA. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic acids research. 2006;34:e9. doi: 10.1093/nar/gnj009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 23.Fu HJ, Zhu J, Yang M, Zhang ZY, Tie Y, Jiang H, Sun ZX, Zheng XF. A novel method to monitor the expression of microRNAs. Molecular biotechnology. 2006;32:197–204. doi: 10.1385/MB:32:3:197. [DOI] [PubMed] [Google Scholar]
- 24.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Molecular cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ro S, Park C, Jin J, Sanders KM, Yan W. A PCR-based method for detection and quantification of small RNAs. Biochemical and biophysical research communications. 2006;351:756–763. doi: 10.1016/j.bbrc.2006.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legendre M, Lambert A, Gautheret D. Profile-based detection of microRNA precursors in animal genomes. Bioinformatics (Oxford, England) 2005;21:841–845. doi: 10.1093/bioinformatics/bti073. [DOI] [PubMed] [Google Scholar]
- 27.Ge X, Wu Q, Wang SM. SAGE detects microRNA precursors. BMC genomics. 2006;7:285. doi: 10.1186/1471-2164-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA (New York, N.Y. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krichevsky AM. MicroRNA profiling: from dark matter to white matter, or identifying new players in neurobiology. ScientificWorldJournal. 2007;7:155–166. doi: 10.1100/tsw.2007.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonard S, Logel J, Luthman D, Casanova M, Kirch D, Freedman R. Biological stability of mRNA isolated from human postmortem brain collections. Biological psychiatry. 1993;33:456–466. doi: 10.1016/0006-3223(93)90174-c. [DOI] [PubMed] [Google Scholar]
- 31.Johnson SA, Morgan DG, Finch CE. Extensive postmortem stability of RNA from rat and human brain. Journal of neuroscience research. 1986;16:267–280. doi: 10.1002/jnr.490160123. [DOI] [PubMed] [Google Scholar]
- 32.Van Deerlin VM, Gill LH, Nelson PT. Optimizing gene expression analysis in archival brain tissue. Neurochemical research. 2002;27:993–1003. doi: 10.1023/a:1020996519419. [DOI] [PubMed] [Google Scholar]
- 33.Barton AJ, Pearson RC, Najlerahim A, Harrison PJ. Pre- and postmortem influences on brain RNA. Journal of neurochemistry. 1993;61:1–11. doi: 10.1111/j.1471-4159.1993.tb03532.x. [DOI] [PubMed] [Google Scholar]
- 34.Cummings TJ, Strum JC, Yoon LW, Szymanski MH, Hulette CM. Recovery and expression of messenger RNA from postmortem human brain tissue. Mod Pathol. 2001;14:1157–1161. doi: 10.1038/modpathol.3880451. [DOI] [PubMed] [Google Scholar]
- 35.Harrison PJ, Heath PR, Eastwood SL, Burnet PW, McDonald B, Pearson RC. The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: selective mRNA vulnerability and comparison with their encoded proteins. Neuroscience letters. 1995;200:151–154. doi: 10.1016/0304-3940(95)12102-a. [DOI] [PubMed] [Google Scholar]
- 36.Mattson MP, Cheng B, Baldwin SA, Smith-Swintosky VL, Keller J, Geddes JW, Scheff SW, Christakos S. Brain injury and tumor necrosis factors induce calbindin D-28k in astrocytes: evidence for a cytoprotective response. Journal of neuroscience research. 1995;42:357–370. doi: 10.1002/jnr.490420310. [DOI] [PubMed] [Google Scholar]
- 37.Norris CM, Kadish I, Blalock EM, Chen KC, Thibault V, Porter NM, Landfield PW, Kraner SD. Calcineurin triggers reactive/inflammatory processes in astrocytes and is upregulated in aging and Alzheimer's models. J Neurosci. 2005;25:4649–4658. doi: 10.1523/JNEUROSCI.0365-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bu J, Bruckner SR, Sengoku T, Geddes JW, Estus S. Glutamate regulates caveolin expression in rat hippocampal neurons. Journal of neuroscience research. 2003;72:185–190. doi: 10.1002/jnr.10556. [DOI] [PubMed] [Google Scholar]
- 39.Krex D, Mohr B, Hauses M, Ehninger G, Schackert HK, Schackert G. Identification of uncommon chromosomal aberrations in the neuroglioma cell line H4 by spectral karyotyping. Journal of neuro-oncology. 2001;52:119–128. doi: 10.1023/a:1010680920087. [DOI] [PubMed] [Google Scholar]
- 40.Arnstein P, Taylor DO, Nelson-Rees WA, Huebner RJ, Lennette EH. Propagation of human tumors in antithymocyte serum-treated mice. Journal of the National Cancer Institute. 1974;52:71–84. doi: 10.1093/jnci/52.1.71. [DOI] [PubMed] [Google Scholar]
- 41.Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA (New York, N.Y. 2004;10:387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nature methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 43.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. The European journal of neuroscience. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 44.Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- 45.Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA (New York, N.Y. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File 1: Microarray methods
Supplemental Files 2–4: Bioanalyzer results
Supplemental File 5: Northern blot and microarray raw data