SUMMARY
Tapasin is a glycoprotein critical for loading Major Histocompatibility Complex (MHC) class I molecules with high affinity peptides. It functions within the multimeric peptide-loading complex (PLC) as a disulfide-linked, stable heterodimer with the thiol oxidoreductase ERp57, and this covalent interaction is required to support optimal PLC activity. Here we present the 2.6 Å resolution structure of the tapasin/ERp57 core of the PLC. The structure reveals the basis for the stable dimerization of tapasin and ERp57 and provides the first example of a protein disulfide isomerase family member interacting with a substrate. Mutational analysis identified a conserved surface on tapasin that interacts with MHC class I molecules and is critical for the peptide loading and editing function of the tapasin-ERp57 heterodimer. By combining the tapasin/ERp57 structure with those of other defined PLC components we present a molecular model that illuminates the processes involved in MHC class I peptide loading.
INTRODUCTION
Major Histocompatibility Complex (MHC) class I molecules present peptide antigens recognized by CD8+ T cells and are critical for immune responses to viruses and tumors. The repertoire of peptides bound is extremely diverse, and the preferential loading of high affinity peptides contributes to the most effective stimulation of a T cell response. MHC class I molecules consist of heavy chain/β2microglobulin (HC/β2m) dimers that fold and assemble with 8–10 residue peptides in the endoplasmic reticulum (ER)(Cresswell et al., 2005). Final assembly and peptide binding take place within the peptide loading complex (PLC), where HC/β2m heterodimers undergo “peptide editing” to promote preferential loading and cell surface presentation of high affinity peptides (Elliott and Williams, 2005). In this final quality control step, tapasin, a specific chaperone critical for the stabilization and loading of empty MHC class I molecules, cooperates with other, more general, ER-resident chaperones. Tapasin is linked by a uniquely stable bond to an active site cysteine residue of the thiol oxidoreductase ERp57 (Dick et al., 2002; Peaper et al., 2005), and the tapasin/ERp57 heterodimer recruits MHC class I molecules as well as the lectin-like chaperone calreticulin to the PLC (Wearsch and Cresswell, 2007). Tapasin also binds to the Transporter associated with Antigen Presentation (TAP) (Ortmann et al., 1997), localizing empty MHC class I molecules near the source of antigenic peptides.
The discovery that tapasin forms an unusually stable heterodimer with ERp57 redirected our understanding of its structure and function (Dick et al., 2002; Peaper et al, 2005). ERp57 is a member of the protein disulfide isomerase (PDI) family of thiol oxidoreductases and normally facilitates disulfide bond formation during glycoprotein folding in conjunction with calreticulin and calnexin (Helenius and Aebi, 2004). It consists of four thioredoxin-like domains a, b, b′, and a′. The a and a′ domains have Cys-Gly-His-Cys active site motifs (C57-G58-H59-C60 and C406-G407-H408-C409) and are catalytically active (Ellgaard and Ruddock, 2005). A transient disulfide bond forms between the N-terminal cysteine in the catalytic motif and a substrate, which is subsequently resolved when the C-terminal cysteine attacks the N-terminal cysteine to release the substrate, a step called the ‘escape pathway’ (Walker and Gilbert, 1997). Within the PLC, Cys57, the N-terminal residue in the catalytic motif of the ERp57 a domain, forms a disulfide bond with Cys95 of tapasin (Dick et al., 2002). Unlike other ERp57 substrates, however, this disulfide bond is preserved by non-covalent interactions with tapasin, generating the stable heterodimer (Peaper et al., 2005). The formation of the tapasin/ERp57 ‘conjugate’ is required to maintain the structural stability of the PLC as well as facilitate optimal peptide loading in human cells (Dick et al., 2002; Peaper and Cresswell, 2008). Furthermore, the tapasin/ERp57 heterodimer, but not monomeric tapasin, supports PLC assembly, the stabilization of empty MHC class I molecules, and peptide editing in vitro (Wearsch and Cresswell, 2007).
Here, as a framework for understanding PLC function, we present the 2.6 Å structure of its tapasin/ERp57 core. The structure offers the first view of a protein disulfide isomerase family member interacting with its protein substrate while also explaining the molecular basis for the unusually stable association of ERp57 and tapasin. Further, we identify a conserved surface on tapasin that mediates the recruitment and peptide loading of MHC class I molecules, and we propose that interaction with this surface stabilizes the MHC class I peptide binding groove in an open, peptide-receptive conformation that facilitates peptide binding. The tapasin/ERp57 structure and MHC class I binding interface established here, combined with known structures of PLC components and previously established points of contact between them, have allowed us to develop a rational model of the specialized ER quality control complex that mediates peptide loading by MHC class I molecules.
RESULTS
Structure Overview
The heterodimer of the lumenal region of tapasin and ERp57 was expressed in Sf21 insect cells and purified as described previously (Wearsch and Cresswell, 2007). To obtain the high yields of conjugate required for crystallographic studies, we co-expressed tapasin with ERp57 bearing the C60A trapping mutation, which has identical in vitro activity to wild type ERp57 and improved stability during purification (Wearsch and Cresswell, 2007). We obtained two crystal forms of the purified conjugate. The structure was initially solved to 3.3 Å resolution by SAD using selenomethionine-substituted crystals containing one tapasin/ERp57 complex in each asymmetric unit. The resolution was extended to 2.6 Å using data from a second crystal form with two complexes in each asymmetric unit. In the final 2.6 Å model, one copy of ERp57 comprises residues 25–493, excluding the signal sequence (residues 1–24) and the C-terminus (residues 494–501); the second copy consists of residues 25–41 and 46–490. Residues 1–381 of tapasin are modeled, excluding poorly ordered regions (residues 12–19, 28–33, and 171–176 in one copy of tapasin and 12–17, 29–32, 173–176, and 316–320 in the second copy). The final model also contains 112 water molecules and the N-acetyl glucosamine groups of the glycan linked to Asn233 in tapasin. Data and refinement statistics are listed in Table 1.
Table 1.
Data Collection and Refinement Statistics
| Data Collection Spacegroup | P21 | P212121(semet substituted proteins) |
|---|---|---|
| wavelength | 0.9795 | 0.9792 |
| Cell parameters | a=74.1, b=72.3, c=200.2 Å, β=94.6° | a=72.6, b=74.8, c=202.4 Å |
| Resolution (Å) | 2.6 (2.69–2.60) | 3.3 (3.42–3.30) |
| Rsym (%) | 7.1 (20.7) | 9.6 (35.0) |
| I/σI | 19.4 (6.5) | 29.8 (6.3) |
| Completeness (%) | 98.1 (91.4) | 99.5 (100.0) |
| Redundancy | 3.7 (3.6) | 22.6 (22.0) |
| Refinement | ||
| Resolution (Å) | 20.0–2.6 | 20.0–3.3 |
| No. reflections | 63,580 | 31,455 |
| Rwork/Rfree (%) | 24.46/28.17* | 26.75/33.88 |
| No. atoms | 13,055 | 6,504 |
| Protein/carbohydrate | 12,915 | 6,504 |
| Water | 112 | - |
| Average B-factor (Å2) | 63.0** | 92.3 |
| R. m. s. Deviations | ||
| Bond lengths (Å) | 0.0073 | 0.0101 |
| Bond angles (°) | 1.44 | 1.60 |
| Ramachandran Plot | ||
| Most favored (%) | 88.2 | 68.6 |
| Allowed (%) | 11.2 | 29.5 |
| Generously Allowed (%) | 0.6 | 1.9 |
| Disallowed (%) | 0.1 | 0.0 |
Numbers in parenthesis refer to data in the highest resolution bin.
Two-fold NCS restraints were used during positional refinement.
The average B-factors for ERp57 are 54.5 and 78.5 Å2, and for tapasin molecules they are 56.9 and 58.5 Å2.
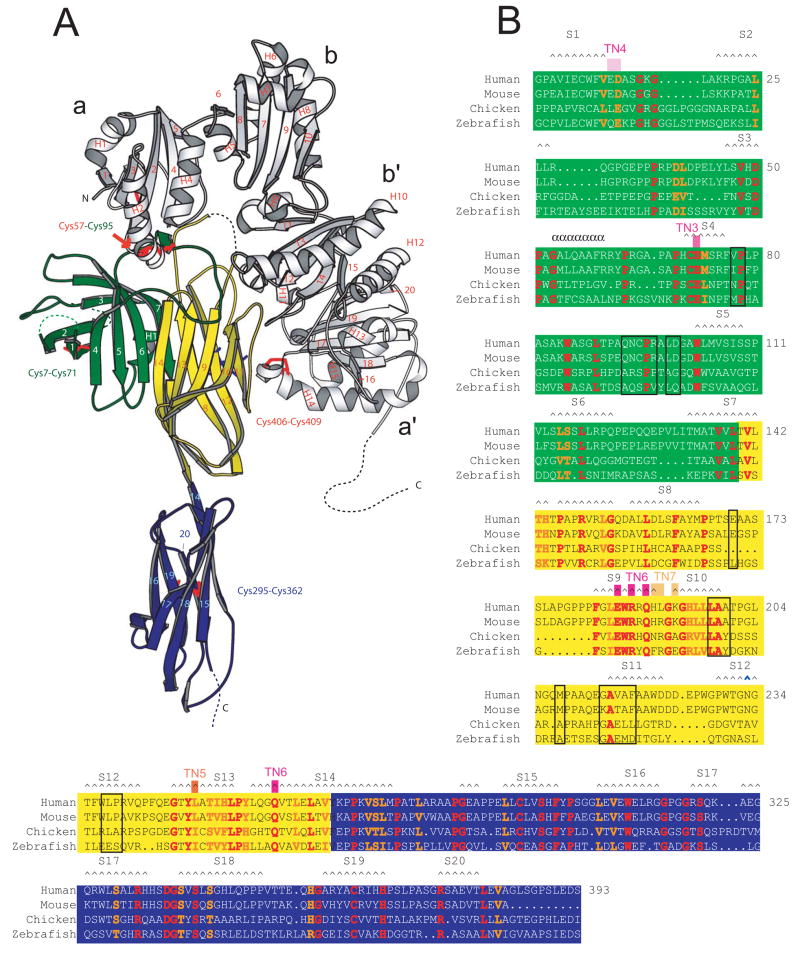
Tapasin forms an L-shaped structure, whereas the four domains of ERp57 are arranged in a twisted U. The a and a′ domains of ERp57 are at the ends of the U, with each contacting the N terminal domain of tapasin (Figure 1). The heterodimer measures ~140 Å in its longest direction and ~80 Å in the widest. Between the bb′ domains of ERp57 and the N-terminal domain of tapasin there is an elliptical cavity measuring ~10 by ~30 Å in diameter. Tapasin consists of two domains, comprising residues 1–269 and 270–381, respectively. The N-terminal domain is a fusion of a 7-stranded β-barrel (residues 1–147, β-strands S1–S7) and an Ig-like fold (residues 150–269, β-strands S8–S14) (Figure 1A, B). The connectivity within the β-barrel is novel with respect to other structures in the Protein Data Bank, as determined by DALI (Holm and Sander, 1996). The N-terminal domain can also be described as a 3-tiered β-sheet sandwich, where two smaller antiparallel β-sheets (S1-S2-S3 and S10-S9-S13-S14) are stacked on opposite sides of an extended central sheet (S4-S5-S6-S7-S8-S12-S11). The C-terminal domain of tapasin also forms an Ig-like fold. The N- and C-termini are both stabilized by disulfide bonds, between Cys7 and Cys71 and between Cys295 and Cys362, respectively, consistent with previous reports (Dick et al., 2002; Turnquist et al., 2004). The relative orientation of the N- and C-terminal domains is similar in two of the three independent copies of tapasin but differs by ~8° in the third (Figure S1A, B).
Figure 1. The structure of the tapasin/ERp57 heterodimer.
(A) Ribbon diagram with ERp57 in white. The N-terminal domain of tapasin is a fusion of a β-barrel (green) and an Ig-like domain (yellow); the C terminus is an Ig-like domain (blue). (B) Sequence alignment for tapasin. Residues in humans, mice, chickens, and zebrafish that are identical/similar are red/orange. Residues within 4.5 Å of an ERp57 atom are boxed in black. Secondary structure elements are labeled. Mutations that interfere with MHC class I interaction and function are indicated (TN3-TN6).
The arrangement of the four thioredoxin-like domains of ERp57 resembles that observed for yeast PDI (PDB ID 2B5E; Tian et al., 2006), which is similar in sequence (29% identity overall and 48% and 43% identity in a and a′, respectively). While the relative orientations of all four domains are similar in two of the three independent copies of the tapasin/ERp57 conjugate in the crystal structure, they differ in the third. Here, a and b, b and b′, and b′ and a′ are reoriented by ~7°, ~10°, and ~15°, respectively, suggesting flexibility between domains (Figure S1). A comparison with the crystal structure of the bb′ fragment alone (PDB ID 2H8L; Kozlov et al., 2006) shows the range of hinge motion is up to 14° between the b and b′ domains. The relative orientation and spacing of a and a′ are fixed by their interaction with tapasin; however, even in the absence of tapasin, ERp57 adopts a U-shape in solution (Kozlov et al., 2006).
Interactions between tapasin and ERp57
The interactions between tapasin and the a and a′ domains of ERp57 are similar in all three crystallographically independent copies of the complex, indicating that the interfaces are not crystallization artifacts. Interestingly, the a and a′ domains of ERp57 use equivalent surfaces to interact with tapasin, with the catalytic motifs in both domains buried in the interface (Figure 2A-C, Table S1). The a domain of ERp57 interacts with a long loop in the β-barrel portion of the tapasin N-terminal domain. The loop spans residues 77–102 (between S4 and S5), and, consistent with previous findings (Dick et al., 2002), there is a covalent bond between Cys95 in this loop and Cys57 in the catalytic motif. The a′ domain of ERp57 forms an interface with residues in S10 and S11 in the Ig-like portion of the tapasin N terminus. Cys406 and Cys409 in the a′ catalytic motif form an intramolecular disulfide bond, and neither is positioned so as to bond with a cysteine residue in tapasin.
Figure 2. Interactions between tapasin and ERp57.
(A) Tapasin residues within 4.5 Å of an ERp57 atom are labeled. (B) ERp57 has been pulled away from tapasin and rotated 180° about a vertical axis to expose the tapasin interaction surface. Residues at the tapasin interface are labeled. Contacts between tapasin and ERp57 are listed in Table S1. (C) Sequence alignment of ERp57 domains a and a′ and equivalent domains of other ER-resident PDIs. Residues in ERp57 within 4.5 Å of a tapasin atom are boxed green (domain a) or yellow (domain a′). Catalytic motifs are red. Secondary structure is indicated. (D) Domains a from yeast PDI (PDBID 2B5E) and ERp57 were superimposed. The catalytic motif in domain a of ERp57 (white) is not distorted when tapasin (green) is bound as compared with the equivalent motif in yeast PDI (red). Disulfide bonds are yellow.
Comparison of the domain a catalytic motifs in ERp57 and yeast PDI shows that tapasin binding does not significantly distort the geometry of the catalytic motif or sterically hinder the C-terminal Cys60 from attacking the Cys57ERp57-Cys95tapasin disulfide bond (Figure 2D), required for the escape pathway. Thus the stable association of ERp57 with tapasin does not appear to involve a direct effect on the mechanism of catalysis; rather the interaction is stabilized by cumulative protein-protein interactions. Because both catalytic domains interact with tapasin the interface is approximately doubled relative to that for a typical substrate, which would be expected to interact with only a single domain. The surface area occluded in the ERp57-tapasin interface, ~1580 Å2, falls in the range most frequently observed for protein-protein interactions, 1400–3000 Å2 (Reichmann et al., 2007). This larger interaction surface likely contributes to the lectin-independent recruitment of tapasin by ERp57 and the unusually stable association of the two proteins (Cresswell et al., 2005). Consistent with this, perturbing the interface between tapasin and the ERp57 a′ domain by mutation of ERp57 (C406A/C409A) significantly diminishes the efficiency of complex formation (Dick et al., 2002).
In vivo, tapasin forms a mixed disulfide bond with ERp57 but not the closely related ER-resident enzymes ERp72 and PDI (DP, unpublished data). Both ERp72 and PDI share high sequence identity with ERp57 in the interacting a and a′ domains (45–57%), and most of the tapasin contact residues are absolutely conserved in ERp57, PDI and ERp72 (Figure 2C). Thus, the sequences of the catalytic domains may not be the sole determinant for recognition. Another may be their relative orientation and spacing, dictated by their binding sites on tapasin. In the tapasin/ERp57 conjugate the distance between the two domains, measured by the distance between the Cα atoms in Cys57 and Cys406, measures ~34 Å. The distance between the equivalent atoms in yeast PDI, the only other PDI family member for which the complete structure is known (Tian et al., 2006), is shorter at 26.5 Å. While flexibility at the domain interfaces may allow each protein to sample a range of distances/orientations, the range may vary for different members of the PDI family. Domains a and a′ in ERp72 and PDI may not be able to accommodate the 34 Å distance while maintaining the orientation required for tapasin interaction.
A conserved surface in tapasin important for PLC function
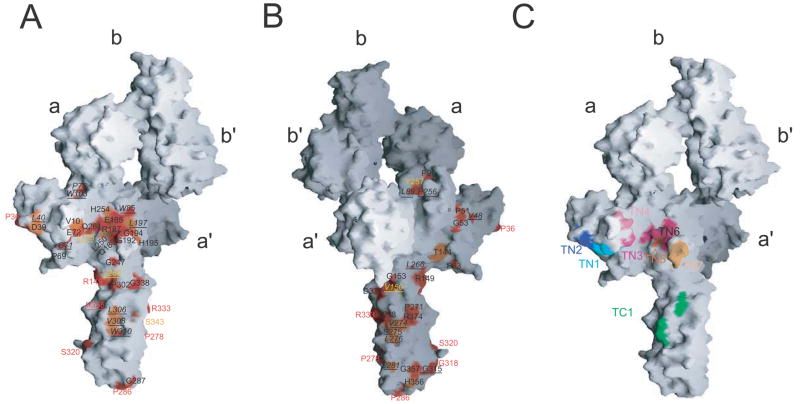
Within the PLC, the empty MHC class I heterodimer is thought to be stabilized and loaded with peptides via direct interactions with tapasin (Elliott, 1997; Wright et al., 2004; Chen and Bouvier, 2007). Mutants of the MHC class I heavy chain which affect PLC binding have been well established (Carreno et al., 1995; Peace-Brewer et al., 1996; Harris et al., 1998; Lewis & Elliot, 1998; Yu et al., 1999), but the functionally important interface on tapasin has yet to be defined. To map potential sites of interaction with the MHC class I molecule, we compared the sequences of tapasin from different species and identified a prominent conserved patch on the surface of the N-terminal domain (Figure 3A, B). Residues in this patch and other surfaces of tapasin were mutated (Table 2, Figure 3C) and tested for their effect on PLC function.
Figure 3. A conserved surface in tapasin is important for PLC assembly/function.
(A) Space filling representation of tapasin/ERp57 with surface residues colored red/orange as in Figure 1B. For underlined residues, the side chain forms part of the protein core. The complex is oriented as in Figure 1A. (B) The complex has been rotated 180° around a vertical axis. (C) Residues that were mutated are colored (Table 2). Mutations that affect PLC assembly/function are colored in shades of red. Others are blue/green.
Table 2.
In vitro activity of mutant tapasin/ERp57 conjugates
| Name | Mutation | Color in Fig. 3c | MHC Class I Association | Peptide Loading(%)* |
|---|---|---|---|---|
| WT | - | - | ++++ | 100 |
| TN1 | E6K/W8A | cyan | ++++ | 108±4 |
| TN2 | R37E/D39K | blue | ++++ | 114±8 |
| TN3 | E72K | medium pink | + | 58±3 |
| TN4 | E11K/D12R | light pink | +/− | 41±5 |
| TN5 | L250K | orange | +/− | 32±4 |
| TN6 | E185K/R187E/Q189S/Q261S | pink | - | 8±1 |
| TN7 | H190S/L191A/K193E | light orange | +/− | 53±2 |
| TC1 | S303W/E307K/E309K | green | ++++ | 99±13 |
% of WT activity ± SEM. Average of two experiments
Recombinant tapasin/ERp57 conjugate interacts with peptide-free MHC class I heavy chain-β2m heterodimers present in extracts of tapasin-negative cells and facilitates MHC class I binding of high affinity peptides in a cell-free system (Wearsch and Cresswell, 2007). Our initial screening of the mutants used this same approach to identify the region of tapasin that interacts with MHC class I molecules. We co-expressed eight different tapasin mutants (Table 2, Figure 3C) with C60A ERp57 in Sf21 insect cells and purified the recombinant proteins. All tapasin mutants were stably expressed and efficiently formed the conjugate with ERp57 (Figure S2). To analyze for the MHC class I interaction, we incubated extracts from tapasin-negative .220. B*0801 cells, which are enriched for empty MHC class I heterodimers, with purified conjugates immobilized on beads using the tapasin-specific mAb PaSta1. Tapasin-associated MHC class I molecules were identified by pulldown and immunoblotting. As shown in Figure 4A, we observed a normal interaction between MHC class I heavy chain and the recombinant conjugates containing wild type tapasin and two of the mutants in the amino terminal region (TN1, TN2) that are most distant to the conserved patch and the single mutant in the carboxy terminal region (TC1). The five other tapasin mutants had variable effects (Figure 4A). The TN6 mutation, where residues central to the conserved patch were altered, completely abrogated MHC class I binding. Only trace amounts of MHC class I heavy chains were found to interact with mutants TN3, TN4, TN5, and TN7, which are located in or around the conserved patch.
Figure 4. Characterization of tapasin mutants.
(A) In vitro assay for the association of MHC class I molecules with wild type (WT) and mutant tapasin/ERp57 conjugates. Digitonin extracts from .220. B*0801 cells were incubated with 250 nM of the indicated recombinant conjugates immobilized on beads. Samples were washed, analyzed bv SDS-PAGE and subjected to immunoblotting (IB) for the MHC class I heavy chain. Data are representative of at least 3 independent experiments. (B) In vitro assay comparing the ability of the recombinant tapasin/ERp57 conjugates to facilitate MHC class I peptide loading. Extracts were prepared from .220. B*0801 cells and incubated with 250 nM recombinant conjugate and [125I]-NP(380-387L). Immunoprecipitation of MHC class I molecules were then performed with the W6/32 Ab and peptide loading was measured by gamma counting. Activities were normalized to that of the wild type tapasin/ERp57 conjugate. Data are an average of 2 independent experiments +/− the S. E. M. (C) The ability of the indicated tapasin constructs to restore HLA-B*4402 expression in tapasin-deficient cells was analyzed by FACS. .220. B*4402 cells transduced with the indicated constructs were stained with anti-HLA-A, B, C and cell surface expression was quantitated for EGFP+ cells. Data are representative of 2 independent experiments. (D) PLC composition in transduced .220. B*4402 cells. Immunoprecipitations were performed from digitonin extract, analyzed by non-reducing SDS-PAGE, and transferred to membranes for immunoblotting with Abs to TAP and MHC class I heavy chain. (E) The PaSta2 mAb does not bind to MHC class I-associated tapasin. Extracts of .220. B*0801 cells were supplemented with the indicated concentrations of purified tapasin/ERp57 conjugate and subjected to immunoprecipitation with either PaSta1 or PaSta2 and immunoblotting for the MHC class I heavy chain. (F) The TN7 mutation of tapasin eliminates recognition by PaSta2. Extracts were prepared from either uninfected Sf21 cells (−) or those infected with baculovirus for the indicated tapasin/ERp57 conjugates were immunoprecipitated with either PaSta1 or PaSta2 coupled beads and analyzed by non-reducing SDS-PAGE. Proteins were visualized by staining with Coomassie Blue.
The relative abilities of the mutants to bind to MHC class I molecules closely matched their relative capacities for mediating peptide loading. Using the assay previously described (Wearsch and Cresswell, 2007) we measured the ability of the recombinant conjugates to promote the binding of the HLA-B8-specific peptide NP(380-387L) to empty MHC class I molecules present in .220. B*0801 extracts (Figure 4B). Again, the most dramatic effect was observed for TN6, which retained less than 10% of the activity observed for the wild type tapasin/ERp57 conjugate (Table 2). For the TN3, TN4, TN5, and TN7 mutants that displayed impaired MHC class I binding, the ability to catalyze peptide loading was reduced to between 42% and 68% of wild type levels while TN1, TN2 and TC1 were normal.
As a further test of functionality, we used retroviral transduction to introduce the TN6 mutant into the tapasin-negative .220. B*4402 cell line, along with TN1, as a control, and C95A tapasin (Dick et al., 2002; Peaper and Cresswell, 2008), which does not form the conjugate or support optimal PLC function. Cell surface expression of HLA-B*4402 is highly tapasin-dependent, consistent with the role of tapasin in facilitating peptide loading (Peh et al., 1998). FACS analysis of the transduced cells showed that the TN6 mutant failed to restore surface expression of HLA-B*4402 (Figure 4C) even though its level of intracellular expression was similar to that of wild type tapasin (data not shown). The TN1 mutant and wild type tapasin restored surface expression, while the C95A tapasin mutant, as shown previously (Dick et al., 2002), only partially restored class I expression. This level of restoration, ~60% of wild type, may be a result of a low, but significant, interaction between free tapasin and MHC class I not reflected in our in vitro measurements of activity, combined with the known stabilization of TAP levels by tapasin association (Bangia et al. 1998; Garbi et al., 2003) which increases the supply of peptides in the ER. Nevertheless, consistent with the data obtained with recombinant tapasin/ERp57 conjugates (Figure 4A), no MHC class I heavy chains were detectably associated by co-immunoprecipitation with the PLC in TN6-expressing cells, although this mutant associated normally with TAP (Figure 4D). Thus, the conserved surface in the N-terminal domain of tapasin likely represents the dominant site of interaction with MHC class I molecules.
Further evidence that the region of tapasin defined by mutational analysis is responsible for the MHC class I HC interaction was obtained using a second tapasin-specific mAb, PaSta2. This antibody recognizes free tapasin/ERp57 conjugates just as well as PaSta1 (Figure 4F and data not shown), but does not immunoprecipitate MHC class I-associated tapasin. This behavior was demonstrated when purified wild type tapasin/ERp57 conjugate were incubated with extracts from .220. B*0801 cells and then subjected to immunoprecipitation with either PaSta1 or PaSta2 (Figure 4E). Dose-dependent reconstitution of the MHC class I/tapasin interaction was confirmed by immunoprecipitation with PaSta1, but these complexes could not be isolated using PaSta2. Thus, the tapasin/MHC class I interaction appears to prevent antibody access to the PaSta2 epitope. In an attempt to localize the epitope, we screened the recombinant conjugates containing the tapasin mutants (Table 2) for binding to PaSta2. Immunoprecipitation for all of the mutants with PaSta1 and PaSta2 was equal except for the TN7 mutant (data not shown and Figure 4F), which is adjacent to the highly conserved tapasin surface (Figure 3C) and is partially defective for MHC class I association and peptide loading (Figure 4A, B). The triple mutation generated in TN7 is H190S/L191A/K193E. Notably, mouse tapasin, which has an arginine in place of leucine at residue 191 (Figure 1B), is not recognized by PaSta2 (data not shown). Thus, the PaSta2 epitope, which is blocked by the MHC class I interaction, is proximal to the highly conserved surface of tapasin that interacts with MHC class I molecules.
DISCUSSION
Speculation regarding the mechanism by which tapasin regulates peptide binding to MHC class I molecules has often centered on comparisons with that of HLA-DM, which performs a similar function for MHC class II (Busch et al., 2005; Sadegh-Nasseri et al., 2008). While superficially the processes are similar, tapasin function is clearly more complex in that it operates in cooperation with a number of other components, including its heterodimeric partner, ERp57. The structure of the tapasin/ERp57 heterodimer revealed here increases our appreciation for the differences between the two; DM is a structural homologue of class II molecules themselves (Fremont et al., 1998; Mosyak et al., 1998), while tapasin lacks any resemblance to DM or MHC molecules except that all are members of the immunoglobulin superfamily. Determination of the tapasin/ERp57 structure allows us to address several outstanding questions regarding the architecture and function of the PLC. In particular, we evaluate two central issues. First, what is the mechanism and function of covalent sequestration of ERp57 by tapasin? Second, how do tapasin and the other PLC components interact with empty MHC class I molecules to facilitate their stabilization and loading with high affinity peptides?
We previously postulated that the disulfide bond formed between tapasin and ERp57 is preserved as a consequence of non-covalent interactions (Peaper et al., 2005). The structure provides no evidence that the escape pathway is inhibited by perturbation of the a domain active site. Instead it appears that the heterodimer is stabilized by the association of tapasin with both of the active sites of ERp57. This finding has several implications. It suggests that tapasin has evolved and acquired these non-covalent interactions as the primary means for stabilizing what would otherwise be a transient mixed disulfide between ERp57 and substrate. It is also consistent with the evidence that the stable, covalent association with ERp57 has a direct effect on the activity of tapasin (Wearsch and Cresswell, 2007) and speculation that it might affect tapasin conformation. Lastly, the association of tapasin with both active sites of ERp57 has strong implications regarding the role of ERp57 in the PLC.
ERp57 functions in the early oxidative folding of the MHC class I heavy chain (Zhang et al., 2006), and some studies have also suggested a role for its enzymatic activity within the PLC, i.e. in reduction/oxidation of the disulfide bond at the base of the MHC class I peptide binding groove (Dick et al., 2002; Lindquist et al., 2001; Santos et al., 2007). In the crystal structure, however, both ERp57 active sites are engaged in contacts with tapasin and inaccessible to the MHC class I molecule (Figure 2). While we cannot exclude that ERp57 may assume an alternative conformation within the PLC, the structures described here suggest that tapasin-associated ERp57 does not exhibit redox activity. In further support of this conclusion, we recently found by mutational analysis that the cysteines in the ERp57 a′ catalytic motif are not required for efficient MHC class I peptide loading (Peaper and Cresswell, 2008). Although the structure is not inconsistent with the conclusion by Dick and co-workers (Kienast et al., 2007) that tapasin inhibits the redox activity of ERp57 to prevent reduction of class I heavy chains, we favor an alternate explanation for the role of tapasin/ERp57 conjugation and a positive role for ERp57 in peptide loading. Other studies (Garbi et al., 2006; Peaper and Cresswell, 2008; Wearsch and Cresswell, 2007) better support a model in which tapasin has adapted the quality control machinery by covalently sequestering ERp57 in order to enhance the recruitment and stability within the PLC of empty MHC class I molecules associated with calreticulin, which interacts independently with the class I glycan and the bb′ domains of ERp57 (Helenius and Aebi, 2004).
Two long helices, α1 and α2, form the walls of the MHC class I peptide binding groove. Molecular dynamics simulations as well as the crystal structure of a peptide-free MHC class I-like molecule (Olson et al., 2005; Zacharias and Springer, 2004) support the proposal that in the absence of peptide, the binding groove adopts an “open” conformation characterized by mobility in the N-terminal segment of helix α2, α2-1 (Elliot, 1997). Tapasin is thought to stabilize a peptide-receptive, open conformation of MHC class I, and mutations on the same side as α2-1 affect tapasin binding (Figure 5A, B)(Carreno et al., 1995; Peace-Brewer et al., 1996; Lewis & Elliot, 1998; Yu et al., 1999). We suggest that the conserved, functionally important surface of tapasin that we have identified serves to stabilize helix α2-1 while the binding groove maintains an open conformation. Both tapasin and MHC class I are tethered to the ER membrane by C-terminal transmembrane segments, and, given this restraint, the two molecules can be juxtaposed so that the conserved surface in tapasin is accessible to α2-1 (Figure 5A, B). The shallow trough within which the conserved residues are located (Figure 3A, C) could easily accommodate and stabilize a short helix such as α2-1. Within the PLC, the MHC class I molecule would then remain primarily in an open conformation until peptide binds with high affinity, locking it into the “closed” conformation observed in peptide-bound structures. This weakens the interaction with tapasin, releasing the class I molecule from the PLC and allowing it to move to the plasma membrane for recognition by CD8+ T cells.
Figure 5. In silico model of the lumenal subcomplex of the PLC.
Structures of tapasin (grey) and MHC class I (green) are juxtaposed in (A, B), so that residues important for their interaction are at or near the protein interface. (A) Mutations in tapasin (space filling model) that affect PLC assembly are colored as in Figure 3 and Table 2. The heavy chain is dark green and β2m is lighter. Helices α1, α2-1, α2-2 that define the MHC class I peptide binding groove are indicated, and a red line marks the groove. (B) The MHC class I molecule (space filling model) is juxtaposed with tapasin. Residues in the heavy chain that are important for interactions with tapasin (Carreno et al., 1995; Peace-Brewer et al., 1996; Lewis & Elliot, 1998; Yu et al., 1999) are highlighted yellow. (C) Side view of the assembled sub-complex, with tapasin, ERp57, MHC class I, and calreticulin. Residues in the ERp57 b′ domain that interact with the calreticulin P-loop are blue (Kozlov et al., 2006; Russell et al., 2004). The 64-3-7 epitope of MHC class I heavy chains (H2-Ld) that is accessible in the PLC is white. (D) Top view of the assembled sub-complex. The MHC class I molecule is shown with a semi-transparent surface. The site of glycosylation at Asn86 in the heavy chain is indicated. This glycan is bound in the calreticulin glucose-binding pocket. Calreticulin residues important for glucose binding are labeled blue (Thomson and Williams, 2005).
Atomic resolution structures of the peptide-bound form of MHC class I (we used PDB ID 1DUY (Khan et al., 2000) for HLA-A*0201) and of the calreticulin homolog calnexin are available (PDB ID 2JHN (Schrag et al., 2001)), as is a vast body of biochemistry regarding the interactions of the MHC class I molecule with tapasin and calreticulin and of calreticulin/calnexin with ERp57 (Wright et al., 2004). Combining these data with the tapasin/ERp57 structure and the MHC class I interaction site identified here allowed us to generate a rational model of the sub-complex of the PLC containing the lumenal components (Figure 5C, D). Class I heavy chain residues known to interact with tapasin (Carreno et al., 1995; Peace-Brewer et al., 1996; Lewis & Elliot, 1998; Yu et al., 1999) are modeled at the tapasin/class I interface, and helix α2-1 is near the conserved, functionally important surface of tapasin we have identified (Figure 5A, B). For calreticulin, we have substituted the calnexin structure but with an appropriately shortened proline-rich P-domain. The lectin-binding site of calreticulin (Thomson and Williams, 2005) is placed proximal to the N-linked glycan at Asn86 of the MHC class I heavy chain which, in the monoglucosylated form, is known to interact with calreticulin (Harris et al., 1998; Wearsch et al., 2004). The P-domain is flexible (Ellgard et al., 2002), and it was incorporated into the model so that its tip is adjacent to the basic surface of ERp57 b′ domain known to interact with this region of calnexin and calreticulin (Kozlov et al., 2006; Russell et al., 2004). The conformation of the P-domain indicated in the model is one of a number that are possible, but in all likelihood it forms an arm that encloses the empty MHC class I peptide binding groove. This could protect the incompletely assembled MHC class I molecule from reduction by free, non-PLC-associated, ERp57, or other thiol oxidoreductases in the ER such as PDI. It may also protect the still improperly folded empty peptide binding groove from recognition by other ER chaperones that might target the peptide-free MHC class I molecule for degradation.
Additional evidence in support of the model structure indicated in Figure 5 is provided by an MHC class I mAb epitope that is the only one known to be expressed and readily accessible on MHC class I molecules present in the PLC. The 64-3-7 epitope has been used to define the ‘open’ conformation of class I putatively present in the PLC. It is present in H2-Ld molecules, can be transferred to other MHC alleles, and corresponds to residues 46–52 in the MHC class I heavy chain (Hansen et al., 2005). This sequence, which lies towards the N-terminal end of the α1 helix, would be easily available to an antibody in the structure proposed (Figure 5C). Tapasin residue 191, which is part of the PaSta2 epitope obscured by MHC class I binding, is buried in the MHC class I/tapasin interface.
Data from Shastri and co-workers (Kanaseki et al., 2006), have argued that binding of N-terminally extended peptides to MHC class I molecules followed by trimming by the enzyme ERAAP/ERAP-1 is likely to occur during peptide loading. In the MHC class I orientation shown in the model in Figure 5, N-terminally extended peptides transiently associated with MHC class I molecules within the PLC would be accessible to ERAAP/ERAP-1. This suggests that such peptide trimming need not be restricted to MHC class I molecules that are freely diffusing in the ER membrane.
More than a decade has passed since the discovery of tapasin and the PLC, and yet much work remains before we fully understand the mechanisms by which they facilitate peptide loading and editing. A major obstacle to designing experiments to probe this mechanism has been the lack of structural information regarding the PLC and, in particular, the MHC class I-specific chaperone tapasin. Thus, the structure of the tapasin/ERp57 heterodimer and the model for the lumenal sub-complex of the PLC that we present here is not only a major step toward better understanding the architecture/function of the PLC, but, equally importantly, will serve as a powerful springboard for further mechanistic probing.
EXPERIMENTAL PROCEDURES
Protein expression and purification
The lumenal domain of human tapasin with a 6x His tag was cloned into the pFastBac Dual vector (Invitrogen) along with full length, human ERp57 bearing the C60A mutation (Wearsch and Cresswell, 2007). The native proteins were co-expressed in Sf21 insect cells and resulting ‘C60A conjugate’ was purified as described (Wearsch and Cresswell, 2007). Following the final chromatography step, buffer was exchanged and protein concentrated to ~ 10 mg/ml in 150 mM NaCl, 10 mM Tris, pH 7.4, 5% glycerol.
Selenomethionine (SeMet) substituted proteins were expressed in Hi-5 insect cells and purified by modified protocols. Insect cells were initially grown in EX-CELL 405 serum free medium (SAFC Biosciences), than transferred to ESF 921 methionine-free medium (Expression Systems). Baculovirus stock was added when cells reached a density of 1.6–1.8×106 cells/ml. 100 mg of DL-SeMet (Sigma) were added per liter of cell culture 10 hrs and again 36 hrs after infection. 68–72 hrs after infection, the cells were removed from the medium, which contained secreted tapasin/ERp57, by centrifugation. The medium was supplemented with 5 mM CaCl2, 1mM NiSO4, and 50 mM Tris-Cl, pH 8.0 and stirred at room temperature for 20 min. The solution was centrifuged at 10,000xg, filtered, and loaded onto Ni-NTA beads (Qiagen) that were pre-equilibrated with 300 mM NaCl, 20 mM imidazole, and 20 mM Tris-Cl, pH 8.0. Protein was eluted with buffer supplemented with 200 mM imidazole. Proteins were further purified by ion exchange chromatography and by gel filtration, then concentrated, as described for the native protein.
Crystallization and data collection
The tapasin/ERp57 heterodimers used for crystallization contain ERp57 missing only the secretion sequence (residues 25–501) and the soluble, lumenal domain of tapasin (residues 1–392). The complex was crystallized at 22°C by the hanging drop method. The reservoir solution contained 100 mM HEPES (pH 7.5), 20% PEG 3K, and 200 mM CaCl2. Initial small, needle-like crystals were streak-seeded into freshly set up drops that also contained 100 mM guanidine hydrochloride. Small rod-like crystals obtained by microseeding were then used for macroseeding, eventually yielding single crystals that measure 0.1x0.3x0.8 mm. Crystals were transferred to solutions containing increasing concentrations of glycerol (finally 20% (v/v) glycerol), loop mounted and flash-frozen in liquid nitrogen.
Most crystals of the native protein belonged to spacegroup P212121 (a=72.57, b=74.80, c=202.45 Å) and typically diffracted to 3.3–3.5 Å resolution. One of several hundred crystals screened belonged to spacegroup P21 (a=74.10, b=72.30, c=200.19 Å, β=94.61°) and diffracted to 2.6 Å. Data were collected at 24-ID-C at the Advanced Photon Source (APS), Argonne National Laboratory. The SeMet substituted protein was crystallized using the same conditions as for native protein and all crystals belonged to the same P212121 spacegroup. Harvest and cryoprotection protocols were as for the native crystals, except that 0.1 % (v/v) of H2O2 was added to the cryoprotection solution one minute before the crystals were flash frozen. The addition of H2O2 converts all seleniums to their oxidized state (Sharff et al., 2000), which results in a larger anomalous signal. Crystals treated this way diffracted to 3.3 Å resolution. A complete anomalous data set was collected at the selenium edge, at 24-ID-E at the APS. All data were integrated and scaled using HKL2000 (Otwinowski and Minor, 1997 (Academic Press)).
Structure determination
The structure was initially solved at 3.3 Å resolution using data collected from SeMet substituted crystals. These crystals have one tapasin/ERp57 complex per asymmetric unit. All thirteen selenium sites were located using SHELXD (Schneider and Sheldrick, 2002). The positions were refined and phases calculated with SHARP (de La Fortelle and Bricogne, 1997 (Academic Press)). The phasing power and figure of merit were 0.92 and 0.59, respectively. We first built ERp57, using available structures of domains a and a′ of PDI (PDB ID 2B5E) and domains bb′ of ERp57 (PDB ID 2H8L) as guides. The partial model was used for phase combination in SHARP, and the improved electron density maps were used to model tapasin (Figure S3A, B). COOT (Emsley and Cowtan, 2004) was used for model building. Cycles of manual rebuilding, torsion angle dynamics, and individual B-factor refinement in CNS (Brunger et al., 1998) resulted in Rwork/Rfree values of 26.8% and 33.9%. The final model contains residues 25–493 of ERp57, excluding the C-terminus (residues 494–501), and residues 1–11, 18–27, 34–172, 177–381 of tapasin.
Models of ERp57 and tapasin from the 3.3 Å resolution structure were used in molecular replacement with the 2.6 Å data set collected from a native crystal. This crystal has two tapasin/ERp57 complexes in the asymmetric unit. We used Phaser (McCoy et al., 2007) to locate the first copy of both ERp57 and tapasin. We built the second copy of each protein by locating domains individually: the N- and C-terminal domains of tapasin and domains a, b, b′, and a′ of ERp57. Rigid body refinement was used to optimize the position of each of the domains in both copies of the tapasin/ERp57 complex. After a round of torsion angle dynamics at 5000K (Brunger et al., 1998), a Fo-Fc difference map was calculated and missing residues were built into the difference density. Composite omit maps (Figure S3C, D) were calculated and used to further improve the model. Cycles of manual rebuilding, torsion angle dynamics, and individual B-factor refinement and the final addition of water molecules resulted in Rwork/Rfree values of 24.5%/28.1%. NCS restraints between equivalent domains in the two copies of tapasin/ERp57 were used in all stages of refinement, except for B factors. All refinement statistics are included in Table 1. The programs Molscript ((Carson, 1997 (Academic Press)), Pymol (DeLano), and Grasp (Nicholls) were used to make the figures.
Characterization of tapasin mutants
These assays are described in detail in the Supplemental Experimental Procedures. Briefly, for in vitro assays mutants of soluble tapasin were co-expressed with C60A ERp57 in Sf21 insect cells and recombinant proteins were isolated using beads coupled to the tapasin-specific mAb PaSta1 (Dick et al., 2002). In vitro assays for MHC class I binding and peptide loading were performed with .220. B*0801 cell extracts essentially as described (Wearsch and Cresswell, 2007), except that the tapasin/ERp57 complexes were immobilized by the PaSta1 mAb, which had minimal effects on tapasin function. The experiments shown in Figure 4E, F used the PaSta2 mAb, which was produced in the same fusion that generated PaSta1 from mice immunized against recombinant, soluble human tapasin expressed in insect cells (Dick et al., 2002). For analysis in mammalian cells, wild type and mutant tapasin were cloned into the pBMN-IRES-EGFP retroviral expression vector and transduced as described (Peaper and Cresswell, 2008) into 721.220 cells expressing HLA-B*4402 (Peh et al., 1998). Transduced cells were stained with mouse anti-HLA-A, B, C Ab (Becton Dickinson) to evaluate MHC class I surface expression by FACS. For the analysis of PLC composition, immunoprecipitation of tapasin and immunoblotting for associated TAP and MHC class I HC was performed as described (Peaper et al., 2008).
Supplementary Material
Acknowledgments
The authors thank Hongwen Zhou for sharing her expertise regarding the production of selenomethionine substituted proteins in insect cells and Hong Zheng and Susan Mitchell for their technical help. We thank the staff of ID24 at the Advanced Photon Source for help with data collection, and K. M. R thanks D. W. Rodgers for discussions. This work was supported by NIH grant GM070521 to K. M. R.; P. C. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Accession Numbers
Coordinates and data have been deposited in the Protein Data Bank with ID code XXXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bangia N, Lehner PJ, Hughes EA, Surman M, Cresswell P. The N-terminal region of tapasin is required to stabilize the MHC class I loading complex. Eur J Immunol. 1998;6:1858–1870. doi: 10.1002/(SICI)1521-4141(199906)29:06<1858::AID-IMMU1858>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Busch R, Rinderknecht CH, Roh S, Lee AW, Harding JJ, Burster T, Hornell TM, Mellins ED. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol Rev. 2005;207:242–260. doi: 10.1111/j.0105-2896.2005.00306.x. [DOI] [PubMed] [Google Scholar]
- Carreno BM, Solheim JC, Harris M, Stroynowski I, Connolly JM, Hansen TH. TAP associates with a unique class I conformation, whereas calnexin associates with multiple class I forms in mouse and man. J Immunol. 1995;155:4726–4733. [PubMed] [Google Scholar]
- Carson M. (Academic Press) in Methods in Enzymology. 1997;277:493–505. [PubMed] [Google Scholar]
- Chen M, Bouvier M. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 2007;26:1681–1690. doi: 10.1038/sj.emboj.7601624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P, Ackerman AL, Giodini A, Peaper DR, Wearsch PA. Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunol Rev. 2005;207:145–157. doi: 10.1111/j.0105-2896.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E, Bricogne G. (Academic Press) in Methods in Enzymology. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMolMolecular Graphics System DeLano Scientific; San Carlos, USA: http://www.pymol.org. [Google Scholar]
- Dick TP, Bangia N, Peaper DR, Cresswell P. Disulfide bond isomerization and the assembly of MHC class I-peptide complexes. Immunity. 2002;16:87–98. doi: 10.1016/s1074-7613(02)00263-7. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgard L, Bettendorff P, Braun D, Herrmann T, Fiorito F, Jelesarov I, Guntert P, Helenius A, Wuthrich K. NMR structures of 36 and 73-residue fragments of the calreticulin P-domain. J Mol Biol. 2002;322:773–784. doi: 10.1016/s0022-2836(02)00812-4. [DOI] [PubMed] [Google Scholar]
- Elliot T. How does TAP associate with MHC class I molecules? Immunol. Today. 1997;18:375–379. doi: 10.1016/s0167-5699(97)01097-9. [DOI] [PubMed] [Google Scholar]
- Elliott T, Williams A. The optimization of peptide cargo bound to MHC class I molecules by the peptide-loading complex. Immunol Rev. 2005;207:89–99. doi: 10.1111/j.0105-2896.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fremont DH, Crawford F, Marrack P, Hendrickson WA, Kappler J. Crystal Structure of mouse H2-M. Immunity. 1998;9:385–393. doi: 10.1016/s1074-7613(00)80621-4. [DOI] [PubMed] [Google Scholar]
- Garbi N, Tiwari N, Momburg F, Hammerling GJ. A major role for tapasin as a stabilizer of the TAP peptide transporter and consequences for MHC class I expression. Eur J Immunol. 2003;33:264–273. doi: 10.1002/immu.200390029. [DOI] [PubMed] [Google Scholar]
- Garbi N, Tanaka S, Momburg F, Hammerling GJ. Impaired assembly of the Major Histocompatibility Complex class I peptide-loading complex in mice deficient in the oxidoreductase ERp57. Nat Immunol. 2006;7:93–102. doi: 10.1038/ni1288. [DOI] [PubMed] [Google Scholar]
- Hansen TH, Lybarger L, Yu L, Mitaksov V, Fremont DH. Recognition of open conformers of classical MHC by chaperones and monoclonal antibodies. Immunol Rev. 2005;207:100–111. doi: 10.1111/j.0105-2896.2005.00315.x. [DOI] [PubMed] [Google Scholar]
- Harris MR, Yu YY, Kindle CS, Hansen TH, Solheim JC. Calreticulin and calnexin interact with different protein and glycan determinants during the assembly of MHC class I. J Immunol. 1998;160:5404–5409. [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Kanaseki T, Blanchard N, Hammer GE, Gonzalez F, Shastri N. ERAAP synergizes with MHC class I molecules to make the final cut in the antigenic peptide precursors in the endoplasmic reticulum. Immunity. 2006;25:795–806. doi: 10.1016/j.immuni.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AR, Baker BM, Ghosh P, Biddison WE, Wiley DC. The structure and stability of an HLA-A*201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J Immunol. 2000;164:6398–6405. doi: 10.4049/jimmunol.164.12.6398. [DOI] [PubMed] [Google Scholar]
- Kienast A, Preuss M, Winkler M, Dick TP. Redox regulation of peptide receptivity of Major Histocompatibility Complex class I molecules by ERp57 and tapasin. Nat Immunol. 2007;8:864–872. doi: 10.1038/ni1483. [DOI] [PubMed] [Google Scholar]
- Kozlov G, Maattanen P, Schrag JD, Pollock S, Cygler M, Nagar B, Thomas DY, Gehring K. Crystal structure of the bb’ domains of the protein disulfide isomerase ERp57. Structure. 2006;14:1331–1339. doi: 10.1016/j.str.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Lewis JW, Elliot T. Evidence for successive peptide binding and quality control stages during MHC class I assembly. Curr Biol. 1998;8:717–720. doi: 10.1016/s0960-9822(98)70280-5. [DOI] [PubMed] [Google Scholar]
- Lindquist JA, Hammerling GJ, Trowsdale J. ER60/ERp57 forms disulfide-bonded intermediates with MHC class I heavy chain. FASEB J. 2001;15:1448–1450. doi: 10.1096/fj.00-0720fje. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosyak L, Zaller DM, Wiley DC. The structure of HLA-DM, the peptide exchange catalyst that loads antigen onto class II MHC molecules during antigen presentation. Immunity. 1998;9:377–383. doi: 10.1016/s1074-7613(00)80620-2. [DOI] [PubMed] [Google Scholar]
- Nicholls A. GRASP: graphical representation and analysis of surface properties (computer program) Columbia University; New York: [Google Scholar]
- Olson R, Huey-Tubman K, Dulac C, Bjorkman P. Structure of a pheromone receptor-associated MHC molecule with an open and empty groove. PLoS Biology. 2005;3:1436–1448. doi: 10.1371/journal.pbio.0030257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann B, Copeman J, Lehner PJ, Sadasivan B, Herberg JA, Grandea AG, Riddell SR, Tampe R, Spies T, Trowsdale J, Cresswell P. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 1997;277:1306–1309. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. (Academic Press) in Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Peace-Brewer AL, Tussey LG, Matsui M, Li G, Quinn DG, Frelinger JA. A point mutation in HLA-A*201 results in failure to bind the TAP complex and to present virus-derived peptides to CTL. Immunity. 1996;4:505–514. doi: 10.1016/s1074-7613(00)80416-1. [DOI] [PubMed] [Google Scholar]
- Peaper DR, Cresswell P. The redox activity of ERp57 is not essential for its functions in MHC class I peptide loading. Proc Natl Acad Sci U S A. 2008;105:10477–10482. doi: 10.1073/pnas.0805044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaper DR, Wearsch PA, Cresswell P. Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. EMBO J. 2005;24:3613–3623. doi: 10.1038/sj.emboj.7600814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peh CA, Burrows SR, Barnden M, Khanna R, Cresswell P, Moss DJ, McCluskey J. HLA-B27-restricted antigen presentation in the absence of tapasin reveals polymorphism in mechanisms of HLA class I peptide loading. Immunity. 1998;8:531–542. doi: 10.1016/s1074-7613(00)80558-0. [DOI] [PubMed] [Google Scholar]
- Reichmann D, Rahat O, Cohen M, Neuvirth H, Schreiber G. The molecular architecture of protein-protein binding sites. Curr Opin Struct Biol. 2007;17:67–76. doi: 10.1016/j.sbi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Ruddock LW, Salo KE, Oliver JD, Roebuck QP, Llewellyn DH, Roderick HL, Koivunen P, Myllyharju J, High S. The primary substrate binding site in the b′ domain of ERp57 is adapted for endoplasmic reticulum lectin association. J Biol Chem. 2004;279:18861–18869. doi: 10.1074/jbc.M400575200. [DOI] [PubMed] [Google Scholar]
- Sadegh-Nasseri S, Chen M, Narayan K, Bouvier M. The convergent roles of tapasin and HLA-DM in antigen presentation. Trends Immunol. 2008;29:141–147. doi: 10.1016/j.it.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SG, Campbell EC, Lynch S, Wong V, Antoniou AN, Powis SJ. Major Histocompatibility Complex class I-ERp57-tapasin interactions within the peptide-loading complex. J Biol Chem. 2007;282:17587–17593. doi: 10.1074/jbc.M702212200. [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Schrag JD, Bergeron JJ, Li Y, Borisova S, Hahn M, Thomas DY, Cygler M. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol Cell. 2001;8:633–644. doi: 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- Sharff AJ, Koronakis E, Luisi B, Koronakis V. Oxidation of selenomethionine: some MADness in the method! Acta Crystallogr D Biol Crystallogr. 2000;56:785–788. doi: 10.1107/s090744490000370x. [DOI] [PubMed] [Google Scholar]
- Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Thomson SP, Williams DB. Delineation of the lectin site of the molecular chaperone calreticulin. Cell Stress Chaperones. 2005;10:242–251. doi: 10.1379/CSC-126.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnquist HR, Petersen JL, Vargas SE, McIlhaney MM, Bedows E, Mayer WE, Grandea AG, III, Van Kaer L, Solheim JC. The Ig-like domain of tapasin influences intermolecular interactions. J Immunol. 2004;172:2976–2984. doi: 10.4049/jimmunol.172.5.2976. [DOI] [PubMed] [Google Scholar]
- Walker KW, Gilbert HF. Scanning and escape during protein-disulfide iosmerase-assisted folding. J Biol Chem. 1997;272:8845–8848. doi: 10.1074/jbc.272.14.8845. [DOI] [PubMed] [Google Scholar]
- Wearsch PA, Cresswell P. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat Immunol. 2007;8:873–881. doi: 10.1038/ni1485. [DOI] [PubMed] [Google Scholar]
- Wearsch PA, Jakob CA, Vallin A, Dwek RA, Rudd PM, Cresswell P. Major histocompatibility complex class I molecules expressed with monoglucosylated N-linked glycans bind calreticulin independently of assembly status. J Biol Chem. 2004;279:25112–25121. doi: 10.1074/jbc.M401721200. [DOI] [PubMed] [Google Scholar]
- Wright CA, Kozik P, Zacharias M, Springer S. Tapasin and other chaperones: models of the MHC class I loading complex. Biol Chem. 2004;385:763–768. doi: 10.1515/BC.2004.100. [DOI] [PubMed] [Google Scholar]
- Zacharias M, Springer S. Conformational flexibility of the MHC class I α1-α2 domain in peptide bound and free states: a molecular dynamics simulation study. Biophys J. 2004;87:2203–2214. doi: 10.1529/biophysj.104.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YY, Turnquist HR, Myers NB, Balendiran GK, Hansen TH, Solhein JC. An extensive region of an MHC class I a2 domain loop influences interaction with the assembly complex. J Immunol. 1999;163:4427–4433. [PubMed] [Google Scholar]
- Zhang Y, Baig E, Williams DB. Functions of ERp57 in the folding and assembly of Major Histocompatibility Complex class I molecules. J Biol Chem. 2006;281:14622–14631. doi: 10.1074/jbc.M512073200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.