Abstract
Among several important considerations for implantation of a biomaterial, a main concern is the introduction of infection. We have designed a hydrogel scaffold from the self-assembling peptide, MAX1, for tissue regeneration applications whose surface exhibits inherent antibacterial activity. In experiments where MAX1 gels are challenged with bacterial solutions ranging in concentrations from 2 × 103 colony forming units (CFUs)/dm2 to 2 × 109 CFUs/dm2, gel surfaces exhibit broad-spectrum antibacterial activity. Results show that the hydrogel surface is active against Gram-positive (Staphylococcus epidermidis, Staphylococcus aureus, and Streptococcus pyogenes) and Gram-negative (Klebsiella pneumoniae and Escherichia coli) bacteria, all prevalent in hospital settings. Live–dead assays employing laser scanning confocal microscopy show that bacteria are killed when they engage the surface. In addition, the surface of MAX1 hydrogels was shown to cause inner and outer membrane disruption in experiments that monitor the release of β-galactosidase from the cytoplasm of lactose permease-deficient E. coli ML-35. These data suggest a mechanism of antibacterial action that involves membrane disruption that leads to cell death upon cellular contact with the gel surface. Although the hydrogel surface exhibits bactericidal activity, co-culture experiments indicate hydrogel surfaces show selective toxicity to bacterial versus mammalian cells. Additionally, gel surfaces are nonhemolytic toward human erythrocytes, which maintain healthy morphologies when in contact with the surface. These material attributes make MAX1 gels attractive candidates for use in tissue regeneration, even in nonsterile environments.
Introduction
Hydrogel materials are finding use in tissue regenerative applications as implantable extracellular matrix substitutes.1–10 These materials aid the healing of chronic and traumatic wounds by providing a hydrated environment suitable for host cell function. However, the microenvironment of hydrogel implants may be ideal not only for host cells but also for opportunistic bacteria.11 Even with the high levels of sterility common to the operating theater, a small number of bacteria introduced to the implant site could be problematic given the altered host immune response that exists at the wound site.12 Biomaterial-centered infections are common, accounting for about 45% of all nosocomial infections.12 Infection results in slowed tissue regeneration at the implant site, and in extreme cases, the implant must be removed. Implant preoperative sterilization helps to limit infection; however, there is valid concern that the harsh sterilization techniques commonly used such as irradiation or ethylene oxide treatment may alter the material properties and ultimate performance of the hydrogel implant.13
Hydrogel materials that display antibacterial activity have been prepared to address this potential problem.14,15 Typically, antimicrobial agents are either encapsulated within the gel for temporal release or covalently attached to the polymeric scaffold. Herein, we report a peptide-based hydrogel whose surface is inherently antibacterial; the gel shows broad-spectrum activity against both Gram-negative and -positive bacteria without incorporating exogenous antimicrobial agents. Hydrogel surfaces are nonhemolytic toward human red blood cells (hRBC’s), and co-culture experiments indicate that while the gel surfaces exert their action against bacteria, they allow mammalian cell proliferation.
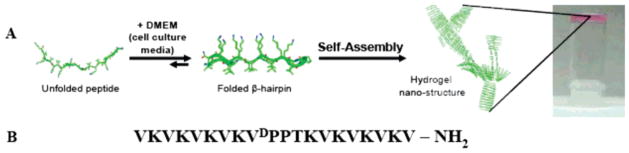
MAX1 is a 20-residue peptide capable of undergoing a solution–hydrogel phase transition in response to cell culture media enabling the triggered formation of hydrogel material for tissue regenerative applications.16 MAX1 consists of two eight-residue strands of alternating valine and lysine amino acids connected by a tetrapeptide sequence (–VDPPT–) (Figure 1). When dissolved in water, this peptide exists in an ensemble of random coil conformations rendering it fully soluble. Under these solution conditions, the lysine side chains are protonated and peptide folding is inhibited because of charge repulsion. However, the addition of an equal volume of DMEM cell culture media at pH 7.4, which contains approprimately160 mM mono-and divalent salts, effectively screens the side-chain charge and triggers peptide folding into an amphiphilic β-hairpin conformation.17 The folded state of MAX1 is characterized by a putative type II′ β-turn centered at –VDPPT–, which connects two amphiphilic β-strands, resulting in a hairpin with one lysine-rich face and one hydrophobic, valine-rich face.18 This conformation rapidly self-assembles, affording a mechanically rigid hydrogel. This triggered folding and consequent self-assembly mechanism allows hydrogel material formation with temporal resolution.16–21
Figure 1.
(A) Mechanism of folding and self-assembly for MAX1 hydrogel formation. Resulting gels are self-supporting as image at far right shows. (B) Sequence of MAX1 β-hairpin.
Extensive scattering, microscopy, and rheological data provide a working model of the gel’s nanostructure, which is characterized by a network of interconnected fibrils rich in β-sheet.18,22 Fibrils are composed of a bilayer of intermolecularly H-bonded hairpins (Figure 1) where bilayer formation is driven by the collapse of the valine-rich hydrophobic faces of individual hairpins. As a result, the interior of each fibril constituting the gel is hydrophobic, and, important to the work described herein, their exteriors display a large concentration of solvent-accessible lysine residues.
Polycationic polymers, such as ε-poly-L-lysine (N = 25–35), show broad-spectrum antibacterial activity when dissolved in aqueous solution.23–25 The mechanism of action is not fully understood but is thought to involve electrostatic adsorption of the polycation to the bacteria’s negatively charged cell surface, leading to the disruption of the bacteria’s outer membrane.25 Although ε-poly-L-lysine exerts its action when dissolved in solution, the expectation that the surface of a MAX1 hydrogel can exhibit antibacterial activity is not unreasonable given the gel’s polycationic, lysine-rich surface.
Results and Discussion
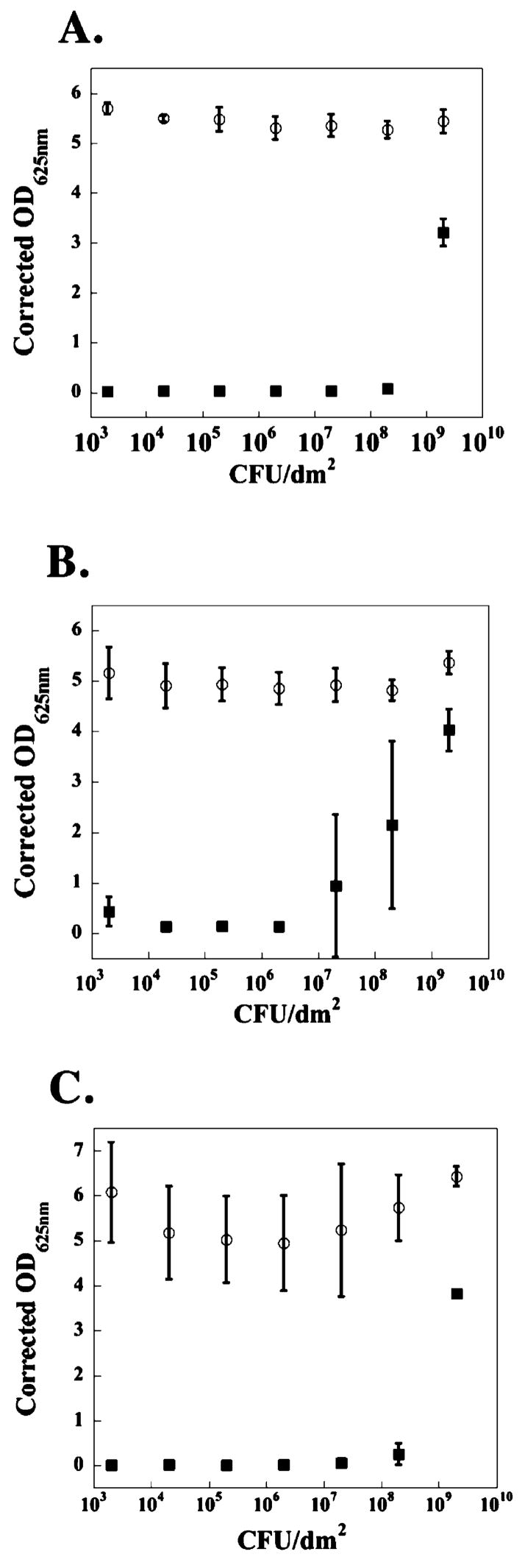
The surface of 2 wt % MAX1 hydrogel displays antibacterial activity against both Gram-negative and Gram-positive strains of bacteria common to hospital environments.26,27 For example, Figure 2 shows data resulting from an assay where the hydrogel surface is challenged with an increasing number of colony forming units (CFUs) of the Gram-negative bacteria Escherichia coli (panels A and B) and Klebsiella pneumoniae (panel C). In panel A, varying numbers of CFUs are introduced independently above the surfaces of distinct hydrogels as well as tissue culture treated polystyrene (TCTP) control surfaces and allowed to incubate for 24 h. In this time period, cells are able to proliferate in the solution above the gel’s surface28 and establish cell–cell contacts leading to cellular aggregation,29 and those cells that come into contact with the surface can establish cell–surface interactions leading to adhesion if the surface is permissive.30 Bacterial proliferation is quantified by measuring the optical density at 625 nm of the supernatant taken from above each surface after mixing to include any weakly adsorbed bacteria.
Figure 2.
TCTP control surface (○) and 2 wt % MAX1 hydrogel surface (■) challenged with an increasing number of CFUs of Gram-negative bacteria. (A) E. coli; 24 h. (B) E. coli; 48 h. (C) K. pneumoniae; 48 h. N = 3.
The data show that the MAX1 hydrogel surface (closed symbols) is capable of inhibiting E. coli proliferation when up to 2 × 108 CFUs/dm2 are initially introduced above the gel’s surface. When 2 × 109 CFUs/dm2 or greater are introduced, the surface is overwhelmed and loses its inhibitory capacity. In contrast, the control surface (open symbols) allows bacterial proliferation when challenged with even the lowest numbers of bacteria. Panel B shows the same assay but performed at 48 h to gain insight into the time-dependent behavior of the surface’s activity. Here, those bacteria that have yet to encounter the surface of the hydrogel have a greater amount of time to proliferate in the solution above the gel, resulting in an increased number of cells that the gel surface must address. This provides a more rigorous test of the gel’s surface activity compared to performing the assay at 24 h. As expected, the data show that the inhibitory activity of the surface apparently decreases and that the gel is active against up to 2 × 106 CFUs/dm2 initially introduced. The gel’s activity is not limited to E. coli. The data in Figure 2C show that the gel surface displays a slightly greater capacity to inhibit K. pneumoniae. Here, the gel is able to inhibit the proliferation of up to 2 × 108 CFUs/dm2 after incubating for 48 h.
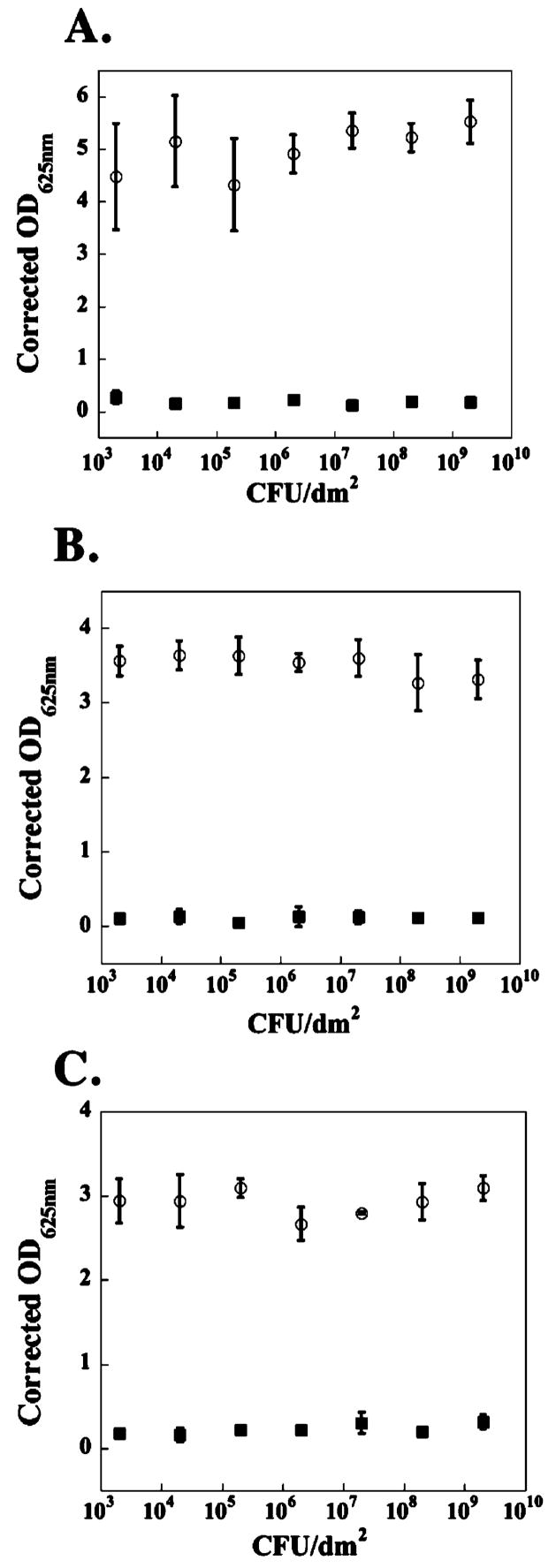
Similar assays were performed using the Gram-positive strains, Staphylococcus aureus, Staphylococcus epidermidis, and Streptococcus pyogenes. The data in Figure 3 show that the gel’s surface is very active against these strains. Even when the surface is challenged with up to 2 × 109 CFUs/dm2 for 48 h, bacterial proliferation is inhibited. Again, the control surface allows proliferation when challenged with even small numbers of bacteria. The reason for the apparent increased sensitivity of Gram-positive strains is not known but may be due to several factors such as differences in cell surface composition, proliferation rates, and the number of cells present in their respective stationary phases.31 For example, S. aureus has a much slower doubling time than E. coli (Supporting Information) and thus proliferates to a lesser degree in the solution above the gel before these cells sediment to the gel surface. In contrast, E. coli initially added to the solution above the gel proliferates more quickly, resulting in an increased number of bacteria the gel surface must address.
Figure 3.
TCTP control surface (○) and 2 wt % MAX1 hydrogel surface (■) challenged with an increasing number of CFUs of Gram-positive bacteria for 48 h. (A) S. aureus. (B) S. epidermidis. (C) S. pyogenes. N = 3.
One question that arises from this data is what level of antibacterial activity must a material possess to be useful in the clinic. Savino et al. have published the suggested acceptable levels of total air microbial counts that may exist for an active operating theater.32 Optimal sterility conditions exist if 0–60 CFUs/dm2 are detected on a 9-cm blood agar Petri dish left open to theater air for 1 h. Acceptable conditions exist if 61–90 CFUs/dm2 are detected. If greater than 91 CFUs/dm2 are detected, theater conditions are deemed unacceptable for use. The data in Figures 2 and 3 indicate that the MAX1 gel surface easily provides effective activity against the levels of bacterial infection expected in an active operating theater. In fact, even after 48 h of exposure to E. coli, the surface is active against 20 000 times the number of bacteria (2 × 106 CFUs/dm2) that it would be expected to encounter in a surgical theater operating under unacceptable conditions containing 100 CFUs/dm.2 This simple calculation also exemplifies the fact that the number of bacteria used in the assays described in this report is exceedingly high. These data suggest that MAX1 gels would be effective at inhibiting advantageous infections that may occur during material implantation.
Insight into the Mechanism of Antibacterial Activity
Bacterial proliferation is monitored in Figures 2 and 3 using an assay that measures the OD of the solution above the hydrogel surface. Even after washing the surface to displace adsorbed bacteria, it is possible that living bacteria bind tightly to the surface and evade detection using this assay. To test this possibility and gain insight into the mechanism by which hydrogel surfaces exert their activity, cell viability assays were performed at the hydrogel’s surface employing a live–dead assay. In this assay, cells are introduced above the hydrogel surface and a control borosilicate surface and allowed to incubate for 24 h under growth conditions. After 24 h, Syto 9 and propidium iodide dyes are added.33 Syto 9 binds to the nucleic acid of both living and dead cells and fluoresces green when excited at 488 nm. Propidium iodide enters the cytoplasm of only dead cells whose membranes have been compromised, binds nucleic acid, and displays red fluorescence. When used in appropriate concentrations, live cells fluoresce green and dead cells fluoresce red. Laser scanning confocal microscopy (LSCM) enables one to visualize cells strictly at the surface of the gel.
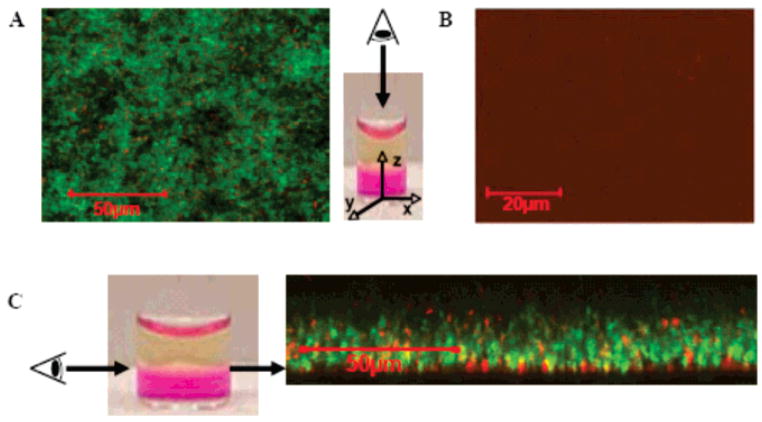
Figure 4 shows the results of this experiment performed with E. coli, a representative Gram-negative strain. In panel A, 2.5 × 103 CFUs/dm2 were introduced above the TCTP control surface and allowed to incubate for 24 h. Considering the light scattering data in Figure 2A, one would expect unencumbered proliferation and a predominance of green fluorescent cells at the control surface. This is, in fact, what is observed in Figure 4A, which shows an image of the control surface parallel to its z-axis. A small number of dead (red) cells are also observed because of natural attrition. In panel B, the same number of cells (2.5 × 103 CFUs/dm2) was introduced above the surface of a 2 wt % MAX1 hydrogel and allowed to incubate for 24 h. This image of the gel’s surface, again taken parallel to its z-axis, clearly shows that within the limits of detection nearly all of the cells are fluorescing red. Since propidium iodide is known to penetrate the compromised membranes of dead cells,33 these confocal experiments strongly suggest that the gel surface is bactericidal and that when bacteria come into contact with the surface, they undergo cell death via a mechanism that involves cell membrane disruption. The data in Figure 4B are internally consistent with the light scattering data presented in Figure 2A that also show that the hydrogel surface is active against this level of infection.
Figure 4.
LSCM xy projections taken of 2.5 × 103 CFUs/dm2 E. coli incubated on a borosilicate control surface (A) and 2 wt % MAX1 hydrogels (B) after 24 h. Gel is viewed parallel to the z-axis. Green fluorescence denotes live cells, and red fluorescence denotes dead cells with compromised membranes. (C) LSCM xy projections taken of 2.5 × 109 CFUs/dm2 E. coli incubated on a 2 wt % MAX1 hydrogel surface viewed perpendicular to the z-axis. Arrows denote the gel–bacterial interface.
The limiting case is shown in Figure 4C. Here, 2.5 × 109 CFUs/dm2 of E. coli have been introduced above the gel’s surface. The light scattering data in Figure 2A show that, at this level of infection, the gel’s surface is overwhelmed and the bacteria are able to proliferate. The confocal image in panel C provides insight into how the bacteria are capable of eluding the effects of the gel surface at high cell numbers. Here, the gel surface is viewed perpendicular to its z-axis, which allows one to visualize three distinct regions simultaneously: the bulk gel, the interface between the gel and the tryptic soy broth media (e.g., the gel surface), and the media above the gel. The bottom of the image shows the bulk gel in black. The top of the image shows the media (also in black). On careful observation of the gel surface (indicated by the arrows in panel C), one observes a thin layer of dead (red) cells. On top of this layer exists a thick layer of mostly live cells. This data suggest that, when high numbers of bacteria are introduced to the gel surface, those that first contact the surface are killed. The cellular debris that results serves to shield the remaining bacteria from the gel’s surface and is used as a matrix for cell attachment. Similar live–dead assays were performed using S. pyogenes, a representative Gram-positive strain. The results show that when this strain is introduced to the gel’s surface, nearly all of the cells are killed upon contact regardless of the number of cells introduced, consistent with the light scattering data shown in Figure 3C (see Supporting Information for LSCM data). The LSCM images shown in Figure 4 and in the Supporting Information are reproducible and are representative of multiple experiments.
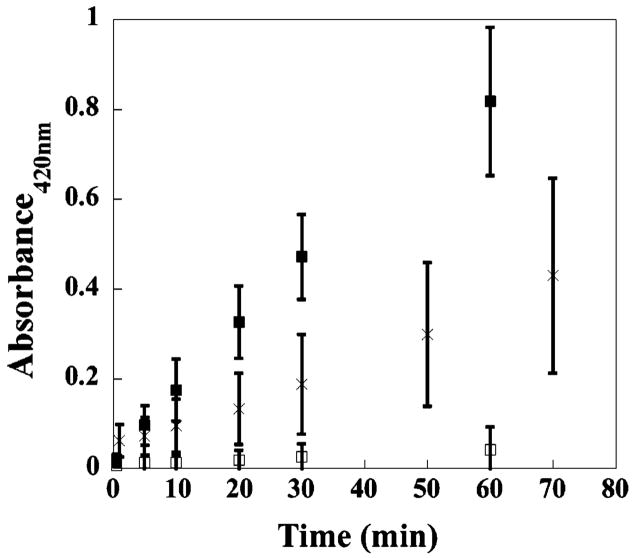
The fact that propidium iodide is able to bind to the nucleic acid of E. coli, which has come into contact with the gel’s surface, strongly suggests that both the inner and outer membranes of this bacteria have been compromised. Although remote, the possibility that propidium iodide itself influences membrane integrity cannot be ruled out. Therefore, instead of measuring the ability of a dye to enter the cell, membrane disruption is commonly assessed by measuring the cellular components that leak from the cell.34,35 Figure 5 shows data from an experiment employing lactose permease-deficient E. coli ML-35.36 This strain is incapable of transporting lactose into the cytoplasm. Therefore, if lactose is added to the media, any β-galactocidase activity measured must be due to enzyme that has leaked from the cytoplasm. In these experiments, o-nitrophenol-β-D-galactopyranoside (ONPG) is used as a lactose mimic whose hydrolysis product is UV active. For the negative control, 4.0 × 109 CFUs/dm2 of E. coli ML-35 are introduced onto TCTP control surfaces and allowed to incubate for 2 h (□), after which time, ONPG is introduced to the media and absorbance monitored over time. As expected, the cells remain healthy with intact membranes and no ONPG hydrolysis is detected above background over 60 min. For the positive control, the same number of cells is sonicated to ensure complete membrane disruption (■). Here, ONPG hydrolysis is clearly evident, and this rate of hydrolysis is assumed to represent maximal membrane disruption and full β-galactocidase activity.35 When the same number of cells is introduced onto the gel’s surface and allowed to incubate for 2 h, β-galactocidase activity is clearly evident with the rate of ONPG hydrolysis intermediate with respect to the positive and negative controls. The level of enzymatic activity observed in Figure 5 for cells introduced to the gel surface is consistent with the data in Figures 2A and 4C that show that when high numbers of bacteria are introduced above the material’s surface, only those cells that come into contact with the surface are affected. The data in Figure 5 suggest that, after 2 h, about half the cells have come into contact with the gel’s surface.
Figure 5.
Membrane permeabilization assay monitoring the activity of cytoplasmic β-galactosidase released from E. coli ML-35 incubated on a TCTP control surface, □; TCTP control surface after cell sonication, ■; 2 wt % MAX1 hydrogel surface, ×. N = 3.
The possibility exists that the observed antibacterial activity may not be due to the material’s surface but may be the result of soluble peptide that has diffused from the hydrogel’s surface. MAX1 adopts a polycationic amphiphilic β-hairpin when folded. There is a wealth of literature that describes the antibacterial action of soluble, polycationic, amphiphilic peptide hairpins.32,37–42 Although their mechanisms of action vary, membrane permeation is a common theme.
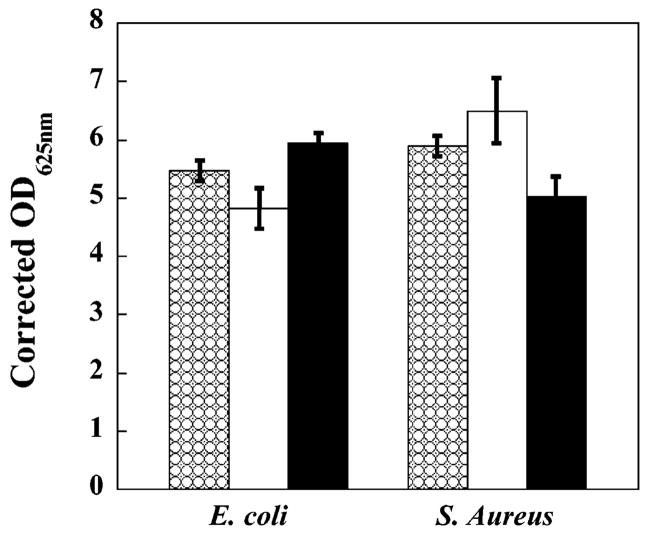
When MAX1 gels are initially formed, a small amount of residual folded peptide (<30 μM; measured by HPLC) does not incorporate into the gel and diffuses into solution. After the gel is washed, no additional leached peptide can be detected. However, under the conditions of the antibacterial assays shown in Figures 2 and 3, this soluble peptide would be present since no additional washes were performed on the gel before introducing the bacteria. At low micromolar concentrations, MAX1 folds and self-assembles to form soluble aggregates and not hydrogel; hydrogelation is only realized at greater than 0.5 wt % (1.5 mM) peptide concentrations. The soluble aggregates may, in fact, be the active species. However, this possibility was ruled out by measuring the proliferation of E. coli and S. aureus, representative Gram-negative and Gram-positive strains, in the presence of 100 μM MAX1. This concentration is well above that which would be expected to be present in solution. Figure 6 shows proliferation data resulting from the addition of 2.0 × 106 CFUs/dm2 of these strains to TCTP surfaces using media with and without added MAX1. Bacterial proliferation in the presence of soluble MAX1 is comparable to that observed in the absence of the peptide, indicating that soluble peptide is most likely not the active species. Last, since hydrogels are prepared from the trifluoroacetate (TFA) salt of MAX1, it is possible that the TFA counterions of MAX1 could be leaching from the gel and are the causative agent. To examine this possibility, the proliferation of E. coli and S. aureus was measured on TCTP surfaces using tryptic soy broth supplemented with trifluoroacetic acid. A 1.7-fold excess of TFA with respect to the theoretical maximal amount of TFA counterion that could be present during the antibacterial assays shown in Figures 2 and 3 was used. The data shown in Figure 6 (black bars) show that the proliferation of these strains is not substantially influenced by TFA at a concentration commensurate with 2 wt % peptide gels.
Figure 6.
Proliferation of 2 × 106 CFUs/dm2 E. coli and S. aureus on a TCTP control surface in the absence (checkered bars), in the presence of 100 μM soluble MAX1 (white bars), and in the presence of 38.7 mM TFA (black bars). N = 3.
Although the exact mechanism is not yet known, the data presented in Figures 2–6 suggest that the observed antibacterial activity is established by the gel’s surface. In addition, direct cellular interaction with the surface may be a necessary requirement that leads to membrane disruption and ultimate cell death.
Cytotoxic Selectivity: Mammalian versus Bacterial Cells
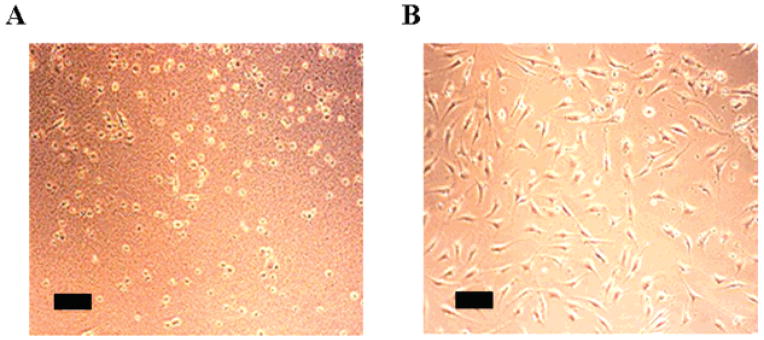
Although previous work employing monocultures of mammalian cells demonstrated that the surface of MAX1 hydrogel is cytocompatible,16 a co-culture experiment was performed to assess the selectivity of the material’s surface when both mammalian and bacterial cells are introduced simultaneously. This experiment more closely resembles the clinical situation where bacteria attempt to infect a hydrogel that has been implanted into a wound site containing mammalian cells. In this co-culture experiment, murine NIH 3T3 fibroblasts and two species of Gram-negative bacteria (Alcaligenes xylosoxidans (xylosoxidans), commonly referred to as Achromobacter xylosoxidans, + Stenotrophomonas maltophilia) were mixed and introduced onto both a control TCTP surface (panel A) and the surface of a 2 wt % MAX1 hydrogel (panel B). These airborne bacterial strains are commonly found in hospitals, having been detected in operating theaters,43 on dialysis machines,44 nebulizers,44 and even in solutions of disinfectant.45 Cell adhesion and proliferation was visually assessed at 25, 32, and 47 h. Figure 7 shows data for the 32-h time point. Panel A shows that the mammalian cells have died on the control surface (note the rounded cell morphologies), and although not clearly visible in the image, the bacteria have proliferated as assessed by visual inspection. However, panel B shows that when a mixture of fibroblasts and bacteria cells is introduced to the hydrogel surface, the bacteria do not proliferate, yet the mammalian cells attach to the gel surface, adopt healthy spread morphologies, and proliferate to near confluency. At 72 h, the fibroblasts have reached confluency. These data show that the gel’s surface shows selective cytotoxic activity toward common strains of airborne bacteria, yet are compatible toward mammalian cells.
Figure 7.
Co-culture of NIH 3T3 murine fibroblasts, A. xylosoxidans (xylosoxidans) and S. maltophilia, on a TCTP control surface (A) and on a 2 wt % MAX1 hydrogel surface (B) after 32 h. (Scale bar = 100 μm)
Commenting on the molecular basis for the observed selectivity would be speculative at this point. One could resort to the classic explanation often given for antimicrobial peptides, which is that differences in membrane composition between bacterial and mammalian cells are the determining factor.32,46,47 The membranes of bacteria, such as E. coli, are mainly composed of negatively charged phospholipids such as phosphatidylglycerol, cardiolipin, and phosphatidylserine, whereas mammalian cells, such as human erythrocytes, are largely composed of net neutral phospholipids such as phosphatidylcholine, phosphatidyethanolamine, and sphingomyelin. Polycationic antimicrobial peptides preferentially engage bacterial membranes via electrostatics and subsequently insert into the membrane because of their amphiphilicity. After this step, mechanisms vary, but all lead to cell death. This explanation may not be ideal for polycationic surfaces when one considers the extracellular membrane-bound biomolecules that would actually first engage the material’s surface for each cell type. Interestingly, the surface of Gram-negative bacteria is densely covered by lipopolysac-charides whose most solvent-exposed portion, O-antigen, is actually neutral.48,49 Conversely, nearly all mammalian cell surfaces are decorated with heparan sulfate glycosaminoglycans, which carry negative charge.50 These apparent contradictions seem to indicate that MAX1 surface-mediated activity may involve a mechanism that is quite different than those of classic antimicrobial peptides.15,51,52 Further investigations into the mechanism exerted by this surface should shed light on the basis of the observed selectivity.
Hemolytic Activity
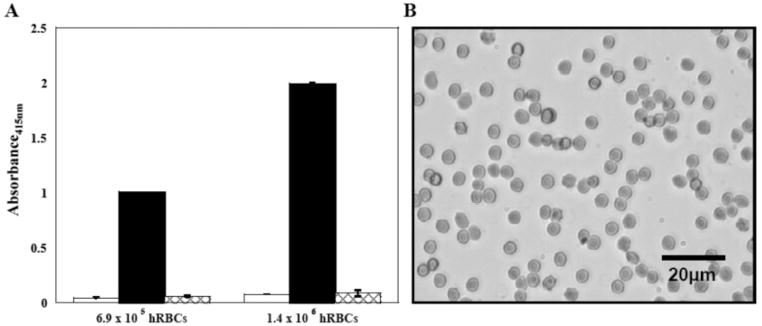
Soluble compounds and material surfaces that exert antibacterial activity by disrupting membranes have the potential to act not only on bacterial membranes but also on the membranes of red blood cells. Hemolytic activity of the hydrogel’s surface was investigated under shear flow conditions by introducing hRBC’s above the material’s surface and rocking the gels in an orbital shaker. Cell lysis is monitored by measuring the absorbance of hemoglobin released from hRBC’s that have been lysed (Figure 8A). Solutions containing 6.9 × 105 hRBC’s introduced on a control TCTP surface for 1 h show minimal absorbance at 415 nm, indicating that the cells are intact (data at left, white bar). A positive control was performed where 1% Triton-X-100 was added to hRBC’s introduced to the TCTP surface. This surfactant causes rapid cell lysis, resulting in maximal hemoglobin absorbance (black bar). Finally, hRBC’s were introduced onto a 2 wt % MAX1 surface and incubated under shear flow conditions for 1 h. The data (checkered bar) indicate that the cells are intact and that the gel surface is nonhemolytic. These experiments were also performed at twice the cell loading density (1.4 × 106, data at right) to help detect any small percentage of lysed hRBC’s. However, the data clearly show that the hydrogel surface is no more hemolytic than the control surface. As expected, the positive control responds in an hRBC concentration-dependent manner. The micrograph in Figure 8B shows hRBC’s resting on a MAX1 gel surface after incubation for 5 h. Cells display disc-shaped morphologies common to healthy hRBC’s, providing further evidence that the gel’s surface is nonhemolytic.
Figure 8.
(A) Hemolytic activity of a TCTP control surface and a 2 wt % MAX1 hydrogel surface toward 6.9 × 105 and 1.4 × 106 hRBC’s under shear flow conditions. TCTP control surface (white bars); TCTP control surface + 1% Triton-X-100 (black bars); and 2 wt % MAX1 hydrogel surface (checkered bars). (B) Image of hRBC’s resting on a 2 wt % MAX1 hydrogel surface after 5 h of incubation at 37 °C. N = 3.
Conclusion
MAX1 hydrogel surfaces were shown to exhibit broad-spectrum antibacterial activity against E. coli, K. pneumoniae, S. aureus, S. epidermidis, and S. pyogenes. Although the exact mechanism is not yet known, live–dead assays employing LSCM as well as β-galactosidase leakage experiments indicate that when E. coli comes into contact with the gel’s surface their cell membranes become compromised, ultimately resulting in cell death. Co-culture experiments showed that, when NIH 3T3 fibroblasts and a mixture of A. xylosoxidans (xylosoxidans) and S. maltophilia were introduced onto the hydrogel, its surface inhibited bacterial proliferation yet allowed mammalian cell adhesion and proliferation, indicating that the surfaces are selective. Last, hemolysis experiments showed that the gel surfaces are nonhemolytic toward hRBC’s.
This work brings to light an interesting question. Can the copious lessons learned from soluble antimicrobial peptides, such as structure–activity relationships, be used to design self-assembled materials that use these peptides as building blocks?53 If so, perhaps the attributes that are currently enjoyed by antimicrobial peptides, such as decreased susceptibility to resistance,47 can be conferred to materials. In any event, the material attributes of the MAX1 gel described in this report make it an attractive candidate for use in tissue regenerative therapies, even in nonsterile environments.
Acknowledgments
This work was supported by the NIH-National Institute of Dental and Craniofacial Research Grant R01 DE01638601. We also thank Professor Robert Hancock for the kind gift of E. coli ML-35.
Footnotes
Supporting Information Available: The Experimental Section, analytical RP-HPLC chromatogram of purified MAX1 and its corresponding mass spectral data, calculation for the reported corrected OD625nm, live–dead LSCM data for S. pyogenes, and growth curves for S. aureus and E. coli. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.For recent reviews, see refs 1–10: Drury JL, Mooney DJ. Biomaterials. 2003;24:4337–4351. doi: 10.1016/s0142-9612(03)00340-5.
- 2.Fairman R, Akerfeldt KS. Curr Opin Struct Biol. 2005;15:453–463. doi: 10.1016/j.sbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman AS. Adv Drug Delivery Rev. 2002;54:3–12. doi: 10.1016/s0169-409x(01)00239-3. [DOI] [PubMed] [Google Scholar]
- 4.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 5.Mart RJ, Osborne RD, Stevens MM, Ulijn RV. Soft Matter. 2006;2:822–835. doi: 10.1039/b607706d. [DOI] [PubMed] [Google Scholar]
- 6.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Adv Mater. 2006;18:1345–1360. [Google Scholar]
- 7.Xu CY, Kopecek J. Polym Bull. 2007;58:53–63. [Google Scholar]
- 8.Zhang SG. Biotechnol Adv. 2002;20:321–339. doi: 10.1016/s0734-9750(02)00026-5. [DOI] [PubMed] [Google Scholar]
- 9.Woolfson DN, Ryadnov MG. Curr Opin Chem Biol. 2006;10:559–567. doi: 10.1016/j.cbpa.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 10.Rajagopal K, Schneider JP. Curr Opin Struct Biol. 2004;14:480–486. doi: 10.1016/j.sbi.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Gristina AG. Science. 1987;237:1588–1595. doi: 10.1126/science.3629258. [DOI] [PubMed] [Google Scholar]
- 12.Schierholz JM, Beuth J. J Hosp Infect. 2001;49:87–93. doi: 10.1053/jhin.2001.1052. [DOI] [PubMed] [Google Scholar]
- 13.An YH, Alvi FI, Kang Q, Laberge M, Drews MJ, Zhang J, Matthews MA, Arciola CR. Int J Artif Organs. 2005;28:1126–1137. doi: 10.1177/039139880502801110. [DOI] [PubMed] [Google Scholar]
- 14.For comprehensive reviews, see refs 14 and 15: Kenawy ER, Worley SD, Broughton R. Biomacromolecules. 2007;8:1359–1384. doi: 10.1021/bm061150q.
- 15.Worley SD, Sun G. Trends Polym Sci. 1996;4:364–370. [Google Scholar]
- 16.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Biomaterials. 2005;26:5177–5186. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 17.Ozbas B, Kretsinger J, Rajagopal K, Schneider JP, Pochan DJ. Macromolecules. 2004;37:7331–7337. [Google Scholar]
- 18.Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. J Am Chem Soc. 2002;124:15030–15037. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]
- 19.Haines LA, Rajagopal K, Ozbas B, Salick DA, Pochan DJ, Schneider JP. J Am Chem Soc. 2005;127:17025–17029. doi: 10.1021/ja054719o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haines-Butterick L, Rajagopal K, Branco M, Salick D, Rughani R, Pilarz M, Lamm MS, Pochan DJ, Schneider JP. Proc Natl Acad Sci USA. 2007;104:7791–7796. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pochan DJ, Schneider JP, Kretsinger J, Ozbas B, Rajagopal K, Haines L. J Am Chem Soc. 2003;125:11802–11803. doi: 10.1021/ja0353154. [DOI] [PubMed] [Google Scholar]
- 22.Ozbas B, Rajagopal K, Schneider JP, Pochan DJ. Phys Rev Lett. 2004;93:268106/1–4. doi: 10.1103/PhysRevLett.93.268106. [DOI] [PubMed] [Google Scholar]
- 23.Shih IL, Shen MH, Van YT. Bioresour Technol. 2006;97:1148–1159. doi: 10.1016/j.biortech.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida T, Nagasawa T. Appl Microbiol Biotechnol. 2003;62:21–26. doi: 10.1007/s00253-003-1312-9. [DOI] [PubMed] [Google Scholar]
- 25.Shima S, Matsuoka H, Iwamoto T, Sakai H. J Antibiot. 1984;37:1449–1455. doi: 10.7164/antibiotics.37.1449. [DOI] [PubMed] [Google Scholar]
- 26.Weber DJ, Raasch R, Rutala WA. Chest. 1999;115:34S–41S. doi: 10.1378/chest.115.suppl_1.34s. [DOI] [PubMed] [Google Scholar]
- 27.Weinstein RA. Emerging Infect Dis. 1998;4:416–420. doi: 10.3201/eid0403.980320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiloach J, Fass R. Biotechnol Adv. 2005;23:345–357. doi: 10.1016/j.biotechadv.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Eboigbodin KE, Ojeda JJ, Biggs CA. Langmuir. 2007;23:6691–6697. doi: 10.1021/la063404z. [DOI] [PubMed] [Google Scholar]
- 30.Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. J Bacteriol. 2002;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeaman MR, Yount NY. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 32.Pasquarella C, Pitzurra O, Savino A. J Hosp Infect. 2000;46:241–256. doi: 10.1053/jhin.2000.0820. [DOI] [PubMed] [Google Scholar]
- 33.Boulos L, Prevost M, Barbeau B, Coallier J, Desjardins R. J Microbiol Methods. 1999;37:77–86. doi: 10.1016/s0167-7012(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 34.Falla TJ, Karunaratne DN, Hancock REW. J Biol Chem. 1996;271:19298–19303. doi: 10.1074/jbc.271.32.19298. [DOI] [PubMed] [Google Scholar]
- 35.Scocchi M, Zelezetsky I, Benincasa M, Gennaro R, Mazzoli A, Tossi A. FEBS J. 2005;272:4398–4406. doi: 10.1111/j.1742-4658.2005.04852.x. [DOI] [PubMed] [Google Scholar]
- 36.Martinez RJ, Carroll SF. Infect Immun. 1980;28:735–745. doi: 10.1128/iai.28.3.735-745.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulet P, Stocklin R, Menin L. Immunol Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- 38.Epand RM, Vogel HJ. Biochim Biophys Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 39.Frecer V, Ho B, Ding JL. Antimicrob Agents Chemother. 2004;48:3349–3357. doi: 10.1128/AAC.48.9.3349-3357.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai JR, Huck BR, Weisblum B, Gellman SH. Biochemistry. 2002;41:12835–12842. doi: 10.1021/bi026127d. [DOI] [PubMed] [Google Scholar]
- 41.Powers JPS, Rozek A, Hancock REW. Biochim Biophys Acta. 2004;1698:239–250. doi: 10.1016/j.bbapap.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 42.Tang M, Waring AJ, Lehrer RI, Hong M. Biophys J. 2006;90:3616–3624. doi: 10.1529/biophysj.105.062075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Igrasiegman Y, Chmel H, Cobbs C. J Clin Microbiol. 1980;11:141–145. doi: 10.1128/jcm.11.2.141-145.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denton M, Kerr KG. Clin Microbiol Rev. 1998;11:57–80. doi: 10.1128/cmr.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukhopadhyay C, Bhargava A, Ayyagari A. J Clin Microbiol. 2003;41:3989–3990. doi: 10.1128/JCM.41.8.3989-3990.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glukhov E, Stark M, Burrows LL, Deber CM. J Biol Chem. 2005;280:33960–33967. doi: 10.1074/jbc.M507042200. [DOI] [PubMed] [Google Scholar]
- 47.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 48.Kotra LP, Golemi D, Amro NA, Liu GY, Mobashery S. J Am Chem Soc. 1999;121:8707–8711. [Google Scholar]
- 49.Seltmann G, Holst O. The Bacterial Cell Wall. Springer; Berlin: 2002. [Google Scholar]
- 50.Bishop JR, Schuksz M, Esko JD. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 51.Lewis K, Klibanov AM. Trends Biotechnol. 2005;23:343–348. doi: 10.1016/j.tibtech.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Tiller JC, Liao CJ, Lewis K, Klibanov AM. Proc Natl Acad Sci USA. 2001;98:5981–5985. doi: 10.1073/pnas.111143098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Lopez S, Kim HS, Choi EC, Delgado M, Granja JR, Khasanov A, Kraehenbuehl K, Long G, Weinberger DA, Wilcoxen KM, Ghadiri MR. Nature. 2001;412:452–455. doi: 10.1038/35086601. [DOI] [PubMed] [Google Scholar]