Abstract
Fully protected 3,5-difluorotyrosine (F2Y), Fmoc-F2Y(tBu)-OH, is efficiently prepared by a chemo-enzymatic process and incorporated into individual peptides and combinatorial peptide libraries. The F2Y-containing peptides display similar kinetic properties toward protein tyrosine phosphatases (PTPs) to their corresponding tyrosine-containing counterparts, but are resistant to tyrosinase action. These properties make F2Y a useful tyrosine surrogate during peptide library screening for optimal PTP substrates.
Replacement of a hydrogen with fluorine results in a small increase in the molecular size but often dramatically different physical, chemical, and biological properties of the molecule.1 As such, fluorinated compounds provide useful mechanistic probes of enzyme-catalyzed reactions and other biological processes. For example, ring fluorinated analogs of tyrosine have been used to examine the catalytic mechanisms of tyrosinase,2 tyrosine phenollyase,3 protein tyrosine kinase and phosphatase,4,5 Δ5-3-ketosteroid isomerase,6 and ribonucleotide reductase.7
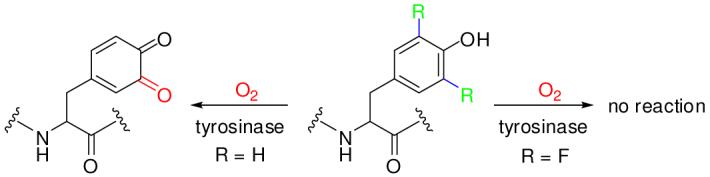
Our own interest in fluorotyrosines stems from our ongoing studies on protein tyrosine phosphatases (PTPs), a large family of enzymes that catalyze the hydrolysis of phosphotyrosine (pY) in proteins back to tyrosine and inorganic phosphate. A major challenge in the PTP field is to determine the physiological substrates and cellular function of these enzymes. We recently developed a combinatorial library method to profile the substrate specificity of PTPs and subsequently used the specificity information to predict the protein substrates of the PTPs.8 A key element of the technique involved selective derivatization of the reaction product (i.e., tyrosine) with a chemical tag (e.g., biotin). This was accomplished by first oxidizing the tyrosine side chain into an orthoquinone with O2 and tyrosinase, followed by conjugate addition with biotin-hydrazide. To prevent false positives from unreacted substrates, tyrosine was excluded from the library. However, PTPs may require tyrosine residues for optimal binding and catalysis. Thus, we envisaged the inclusion of a fluorinated tyrosine into the peptide library as a tyrosinase-resistant tyrosine surrogate. Among all of the known mono-, di-, and multiply fluorinated tyrosine analogs, we reasoned that 3,5-difluorotyrosine (F2Y, compound 1 in Scheme 1) should contain the minimal number of fluorine substitution to render it resistant to tyrosinase action. Because F and H atoms have similar van der Waals radii (1.35 Å for F and 1.10 Å for H), Tyr and F2Y are essentially isosteric, although the side chain of F2Y has a lower pKa value (7.2) than that of Tyr (9.9).9
Scheme 1.
Synthesis of Fully Protected F2Y
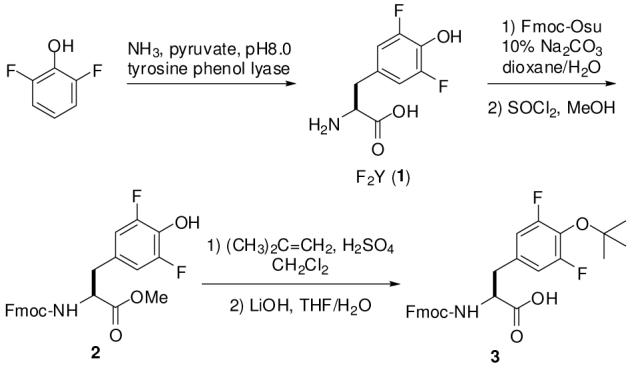
F2Y has previously been synthesized both chemically10 and enzymatically.3,4,7 Synthesis of F2Y-containing peptides employed side chain unprotected Fmoc-F2Y-OH and the resulting peptides were purified by HPLC.4,7 The reported peptides were either very short or contained F2Y near their N-termini (therefore no repeated coupling reactions after incorporation of F2Y) and each contained only a single F2Y residue. We felt that the unprotected F2Y side chain might be problematic with peptide library synthesis, during which more forcing coupling conditions are typically employed to drive reactions to completion, some library members will contain multiple F2Y residues, and HPLC purification is not an option. In this report, we describe the synthesis of fully protected F2Y, its incorporation into peptides and peptide libraries, and its activity against PTPs in comparison with the tyrosine counterparts.
We employed the enzymatic systhesis originally developed by Phillips and coworkers3 to prepare multigram quantities of F2Y from 2,6-difluorophenol, pyruvate, and NH3 (Scheme 1). Treatment of F2Y with N-(9-fluorenylmethylcarbonyloxy)succinimide in 10% sodium carbonate gave Nα-Fmoc-F2Y-OH, which was subsequently converted into its methyl ester 2 using thionyl chloride in refluxing methanol. The phenol group was next protected as a tert-butyl ether by treatment with isobutylene and H2SO4. Finally, hydrolysis of the methyl ester by LiOH in THF/H2O gave the desired Nα-Fmoc-F2Y(tBu)-OH (3) in 24% overall yield (from F2Y).
Three F2Y-containing peptides and their corresponding Tyr-containing counterparts were synthesized on the solid phase by using standard Fmoc/HBTU chemistry (Table 1, compounds 4-9). Peptide 5 is derived from a known pY motif of receptor protein tyrosine kinase erbB2,11 whereas peptides 7 and 9 are analogous to the prefered substrates of PTP1B (the prototypical PTP), previously identified from a peptide library.8 These peptides contain F2Y (or Tyr) at pY-3, pY-2, or pY-1 position (relative to pY, which is designated as position 0). Earlier studies have shown that residues at the N-terminal side of pY are most critical in defining the substrate specificity of PTPs.12 During peptide synthesis, Nα-Fmoc-F2Y(tBu)-OH was efficiently incorporated into peptides without incidence (as judged by ninhydrin tests), despite that it was used at only 1.5 equivalents (four equivalents were used for all other amino acids). Reversed-phase HPLC analysis of the crude peptides showed that in each case, the desired peptide was the major product (see Figure 1, for example). Peptides 4-9 were purified by preparative HPLC and their kinetic activities toward PTP1B (i.e., kcat, KM, and kcat/KM values) were determined at pH 7.4 (Table 1). The data show that substitution of F2Y for tyrosine resulted in minimal changes in kinetic constants (≤2-fold, which is within the margin of experimental error). Thus, F2Y is a good functional mimic of tyrosine, in terms of binding to the active site of PTPs.
Table 1.
Comparison of the Kinetic Constants of F2Y- and Tyr-Containing Peptides against PTP1B (pH 7.4)
| peptide | kcat (s-1) | KM (μM) | kcat/KM (μM-1 s-1) |
|---|---|---|---|
| DNLF2YpYWD-NH2 (4) | 36 ± 1 | 3.6 ± 0.4 | 10.0 |
| DNLYpYWD-NH2 (5) | 32 ± 1 | 5.8 ± 0.8 | 5.5 |
| DDTF2YDpYAA-NH2 (6) | 36 ± 2 | 13 ± 3 | 2.7 |
| DDTYDpYAA-NH2 (7) | 33 ± 1 | 14 ± 2 | 2.3 |
| REF2YEFpYAA-NH2 (8) | 44 ± 2 | 14 ± 2 | 3.2 |
| REYEFpYAA-NH2 (9) | 33 ± 1 | 6.0 ± 0.7 | 5.4 |
Figure 1.
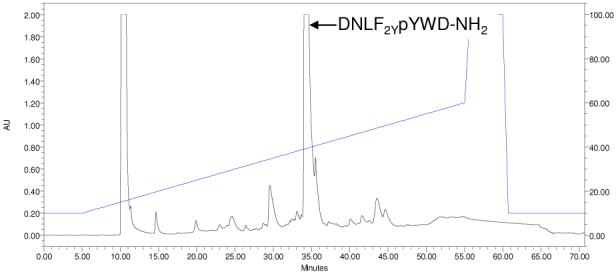
HPLC analysis of peptide 4 on a C-18 column, eluted with a linear gradient of 10-60% CH3CN in H2O containing 0.1% trifluoroacetic acid over 50 min.
Next, a pair of Tyr- and F2Y-containing peptides (peptides 8 and 9 in Table 1) were tested for activity against tyrosinase. The peptides were individually treated with tyrosinase in the presence of atmospheric O2 and excess biotin hydrazide. Reversed-phase HPLC/MS analysis of the reaction mixtures revealed that the Tyr-containing peptide REYEFpYAA was quantitatively converted into a new species, which had an increased retention time (from 32.0 to 34.5 min) and molecular mass (from m/z 1168.4. to 1440.7) (Figure S1 in Supporting Information). The increase of 272.3 amu in molecular mass is consistent with the addition of a single biotin hydrazide molecule to the tyrosine side chain. As expected, the F2Y-containing peptide (8) was completely resistant to tyrosinase action.
Finally, to test whether F2Y is compatible with peptide library synthesis, screening, and post-screening sequence identification, we designed a pY peptide library containing five random positions immediately N-terminal to pY [SAXXXXXpYAABBRM-resin, where B is β-alanine and X is F2Y, norleucine (Nle), or any of the 17 proteinogenic amino acids excluding Met, Cys and Tyr]. The library was synthesized on polyethylene glycol polyacrylamide (PEGA) resin13 by the split-and-pool method.14 A portion of the resulting one-bead-one-compound (OBOC) library (∼40,000 beads) was treated with 1.0 nM PTP1B for 20 min (at pH 7.4), followed by incubation with 1.2 μM mushroom tyrosinase in the presence of atmospheric O2 and 6 mM 3-methyl-2-benzothiazolinonehydrazone (MBTH). Under these conditions, only beads that carried the most preferred substrates of PTP1B were dephosphorylated (usually only partially). The exposed tyrosine side chain was subsequently oxidized by the excess tyrosinase activity into an orthoquinone, which was trapped by reaction with MBTH to form a dark pink pigment (Figure 2a).15 Thus, as a result of this reaction cascade, a bead carrying a preferred PTP1B substrate would turn pink/red, whereas beads carrying poor substrates would not. This was indeed the case (Figure 2b). The pink/red colored beads were removed from the library and individually sequenced by partial Edman degradation/mass spectrometry16 to reveal the most preferred PTP1B substrates (Table 2). Figure 2c shows an example of the MS spectrum derived from one of the pink beads (bead No. 2 in Table 2), carrying the sequence of SAVITDF2YpYAABBRM. Our results are in agreement with earlier reports that PTP1B prefers acidic residues at the N-terminal side of pY, especially at the pY-2 position.8,12,17 A control experiment without the PTP1B treatment resulted in no pink/red colored beads at all (data not shown). Since ∼25% of the library members contain at least one F2Y residue in the random region, the lack of any pink/red beads in the control provides further evidence that F2Y is completely resistant to tyrosinase action. Most importantly, F2Y is compatible with every aspect of library synthesis, screening, and sequence identification.
Figure 2.
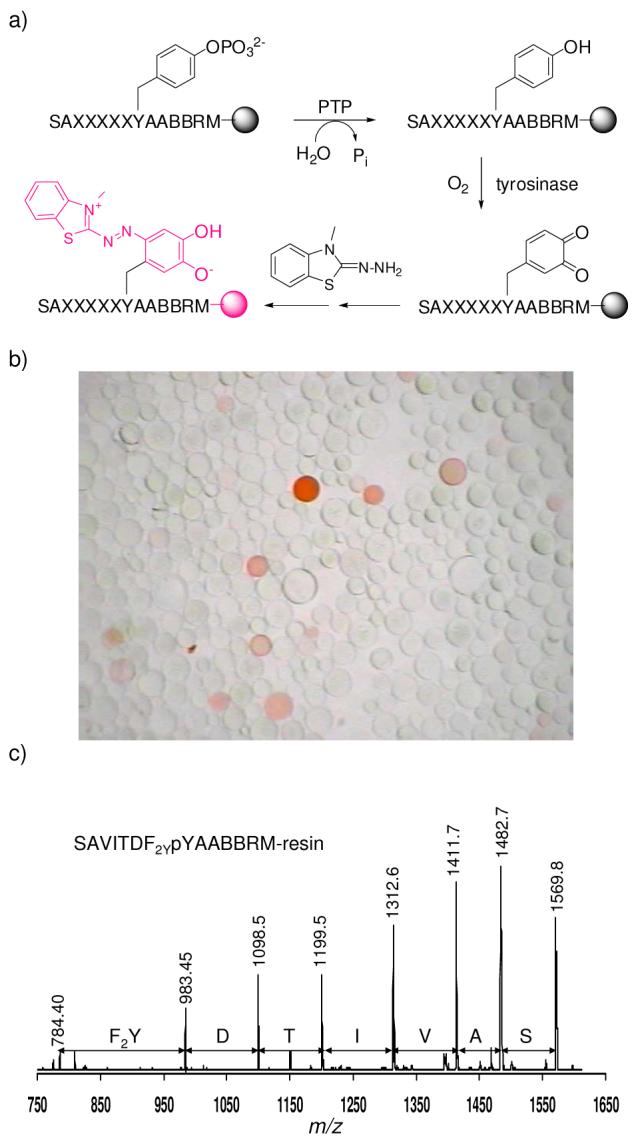
a) Reactions involved in library screening against PTP and bead coloration; b) A portion of the OBOC pY library after treatment with PTP1B (1.0 nM) and tyrosinase (1.2 μM) (viewed under a disecting microscope); c) MALDI-TOF spectrum of peptide SAVITDF2YpYAABBRM* and its truncation products after partial Edman degradation (derived from a single PEGA resin bead). M*, homoserine lactone.
Table 2.
A Partial List of the Selected PTP1B Substrates
| Bead No. | Peptide sequence |
|---|---|
| 1 | QDVDApYAA |
| 2 | VITDF2YpYAA |
| 3 | LQF2YDNpYAA |
| 4 | ITMDQpYAA |
| 5 | WGTDSpYAA |
| 6 | SSFDVpYAA |
| 7 | SRHEWpYAA |
| 8 | ETDFApYAA |
| 9 | NDLF2YEpYAA |
| 10 | FTSGLpYAA |
M, norleucine.
In summary, we have found that F2Y is a functional, tyrosinase-resistant mimetic of tyrosine for applications such as defining the substrate specificity of PTPs through combinatorial library screening. An efficient method has been developed for the chemoenzymatic synthesis of fully protected F2Y and its incorporation into peptides and peptide libraries. In addition, an improved PTP screening assay has been developed by employing MBTH as the coupling reagent. Because the pink/red pigment is only formed on the beads that have undergone PTP reaction, the current method does not generate any false positives and is operationally straightforward. Application of the methodologies to systematically profile the substrate specificity of PTPs is currently ongoing in our laboratory and will be reported elsewhere in due course.
Supplementary Material
Acknowledgment
This work was supported by National Institutes of Health (GM062820).
References
- (1).Hagmann WK. J. Med. Chem. 2008;51:4359–4369. doi: 10.1021/jm800219f. [DOI] [PubMed] [Google Scholar]
- (2).Battaini G, Monzani E, Casella L, Lonardi E, Tepper AWJW, Canters GW, Bubacco L. J. Biol. Chem. 2002;277:44606–44612. doi: 10.1074/jbc.M207829200. [DOI] [PubMed] [Google Scholar]
- (3).Phillips RS, Von Tersch RL, Secundo F. Eur. J. Biochem. 1997;244:658–663. doi: 10.1111/j.1432-1033.1997.00658.x. [DOI] [PubMed] [Google Scholar]
- (4).Kim K, Cole PA. J. Am. Chem. Soc. 1998;120:6851–6858. [Google Scholar]
- (5).Martin B, Pallen CJ, Wang JH, Graves DJ. J. Biolo. Chem. 1985;260:14932–14937. [PubMed] [Google Scholar]
- (6).Brooks B, Philips RS, Benisek WF. Biochemistry. 1998;37:9738–9742. doi: 10.1021/bi980454x. [DOI] [PubMed] [Google Scholar]
- (7).Yee CS, Chang MCY, Ge J, Nocera DG, Stubbe J. J. Am. Chem. Soc. 2003;125:10506–10507. doi: 10.1021/ja036242r. [DOI] [PubMed] [Google Scholar]
- (8).Garaud M, Pei D. J. Am. Chem. Soc. 2007;129:5366–5367. doi: 10.1021/ja071275i. [DOI] [PubMed] [Google Scholar]
- (9).Seyedsayamdost MR, Reece SY, Nocera DG, Stubbe J. J. Am. Chem. Soc. 2006;128:1569–1579. doi: 10.1021/ja055926r. [DOI] [PubMed] [Google Scholar]
- (10).Kirk KL. J. Org. Chem. 1980;45:2015–2016. [Google Scholar]
- (11).Yamamoto T, Ikawa S, Akiyama T, Nomura N, Miyajima N, Saito T, Toyoshima K. Nature. 1986;319:230–234. doi: 10.1038/319230a0. [DOI] [PubMed] [Google Scholar]
- (12).(a) Zhang Z-Y, Thieme-Sefler AM, Maclean D, McNamara DJ, Dobrusin EM, Sawyer TM, Dixon JE. Proc. Natl. Acad. Sci. U. S. A. 1993;90:4446–4450. doi: 10.1073/pnas.90.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang Z-Y, Maclean D, McNamara DJ, Sawyer TK, Dixon JE. Biochemistry. 1994;33:2285–2290. doi: 10.1021/bi00174a040. [DOI] [PubMed] [Google Scholar]
- (13).Meldal M. Tetrahedron Lett. 1992;33:3077–3080. [Google Scholar]
- (14).(a) Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]; (b) Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Nature. 1991;354:84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]; (c) Furka A, Sebestyen F, Asgedom M, Dibo G. Int. J. Peptide Protein Res. 1991;37:487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- (15).Winder AJ, Harris H. Eur. J. Biochem. 1991;198:317–326. doi: 10.1111/j.1432-1033.1991.tb16018.x. [DOI] [PubMed] [Google Scholar]
- (16).Thakkar A, Wavreille A-S, Pei D. Anal. Chem. 2006;78:5935–5939. doi: 10.1021/ac0607414. [DOI] [PubMed] [Google Scholar]
- (17).(a) Pellegriini MC, Liang H, Mandiyan S, Wang K, Yuryev A, Vlattas I, Sytwu T, Li Y-C, Wennogle LP. Biochemistry. 1998;37:15598–15606. doi: 10.1021/bi981427+. [DOI] [PubMed] [Google Scholar]; (b) Vetter SW, Keng Y-F, Lawrence DS, Zhang Z-Y. J. Biol. Chem. 2000;275:2265–2268. doi: 10.1074/jbc.275.4.2265. [DOI] [PubMed] [Google Scholar]; (c) Walchli S, Espanel X, Harrenga A, Rossi M, Cesareni G, van Huijsduiinen RH. J. Biol. Chem. 2004;279:311–318. doi: 10.1074/jbc.M307617200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


