Abstract
These studies examined how genetic differences that regulate architectural and bone material properties would be expressed during fracture healing and determine whether any of these features would affect rates of healing as defined by regain of strength. Controlled fractures were generated in three inbred strains of mice: A/J, C57Bl/6J (B6), and C3H/HeJ (C3H). Both the A/J and B6 strains showed faster healing than the C3H strain based on regains in strength and stiffness. Strain-specific architectural features such as moment of inertia, cross-sectional area, and cortical thickness were all recapitulated during the development of the callus tissues. None of these traits were directly relatable to rates of fracture healing. However, rates of healing were related to variations in the temporal patterns of chondrogenic and osteogenic lineage development. The B6 strain expressed the highest percentage of cartilage gene products and had the longest period of chondrocyte maturation and hypertrophy. The slowest healing strain (C3H) had the shortest period of chondrogenic development and earliest initiation of osteogenic development. Although the A/J strain showed an almost identical pattern of chondrogenic development as the C3H strain, A/J initiated osteogenic development several days later than C3H during fracture healing. Long bone growth plates at 28 days after birth showed similar strain-specific variation in cartilage tissue development as seen in fracture healing. Thus, the B6 strain had the largest growth plate heights, cell numbers per column, and the largest cell size, whereas the C3H columns were the shortest, had the smallest number of cells per column, and showed the smallest cell sizes. These results show that (1) different strains of mice express variations of skeletal stem cell lineage differentiation and (2) that these variations affect the rate of fracture healing.
Key words: genetics, fracture, stem cells, cartilage
INTRODUCTION
Fractures are one of the most common traumatic injuries in humans, and osteoporosis-related fractures are the fastest growing health care problem of aging.(1–3) Because bone healing after traumatic injury is a complex trait, it is necessary to examine bone healing in the context of how intact polygenic networks influence cellular activity during the healing process. Therefore, identifying aspects of genetic background and thus the biological controls that are both prognostic and or diagnostic of variation in the rate of fracture healing will have important clinical value in better defining when an individual can resume normal activities and weight bearing after fracture. Such information will also be important in defining individuals that will have an increased risk for complications after the traumatic fracture or during a period of healing after an orthopedic surgical procedure.
A number of inbred mouse strains have been studied that exhibit variable quantitative traits defining BMD, bone strength, cross-sectional morphology, matrix/mineral composition, and trabecular architecture. These traits have begun to be mapped to specific gene loci in various strains of mice and humans(4,5) and have been related to a hierarchy of subtraits that are both specific to sex and specific to bones such as the femur, tibia, and vertebrae.(4–12) Other studies have shown that these traits occur as the animal undergoes development with defined alterations in specific biological properties such as periosteal and endosteal rates of formation.(13–15) Finally, quantitative trait mapping between mice and humans has identified common loci that affect bone quality in both species.(16) Several studies have reported variability in soft tissue wound healing and bone healing after injury among many of these same inbred strains of mice.(17–20) These results suggest that genetic background also plays a contributory role in the processes associated with bone and soft tissue wound healing. However, the effect of genetic background on the development of a fracture callus has not been examined.
It is generally accepted that the molecular mechanism(s) that control the processes of bone healing recapitulates those that were operative during the embryological development of the skeleton.(21–25) We therefore hypothesized that these same developmental mechanisms that lead to genetic variability in bone quality would be activated during the temporal period of fracture healing and would contribute to variable rates of healing. Controlled fracture healing experiments were undertaken using three inbred strains of mice that have well-defined variation in specific traits associated with bone quality. Recent refinements in the understanding of skeletal tissue function has shifted away from simply considering the overall mineral content and density of a given bone toward a biomechanical-based definition that considers endpoint mechanical properties such as toughness, strength, and brittleness.(26) We tested whether genetic background affects how these traits vary during fracture healing and whether they are related to alterations in the rate of regain in mechanical function.
MATERIALS AND METHODS
Animals
Research was conducted in conformity with all Federal and USDA guidelines under an IACUC-approved protocol at the Boston University School of Medicine in Boston, MA, and the Mount Sinai School of Medicine in New York, NY. For fracture studies, 8- to 10-wk postbirth A/J, C57BL/6J (B6), and C3H/HeJ (C3H) male mice were obtained from the Jackson Laboratory and housed at the Boston University Medical Center animal housing facility for the duration of each study after 3 days of acclimation. Growth plate studies were carried out on 14- to 28-day postbirth male mice obtained from breeding stocks maintained at the Mount Sinai School of Medicine in New York, NY.
Fracture model
Unilateral fractures were produced in the right femur of 8- to 10-wk-old male mice using a modification(25) of the method developed for the rat.(27) The location and quality of fractures were assessed by X-ray analysis while animals were still anesthetized after surgery. Fracture configurations that were comminuted or were not localized to the mid-diaphyseal region were excluded from the study.
Two time points during fracture healing were chosen for the biomechanical and μCT analyses (see below). Day 21 after fracture was examined because it is the earliest time at which mechanical properties can be measured and is representative of the key transition point between the endochondral and primary bone formation periods. Day 35 after fracture was examined because it is representative of when the bones have regained their prefracture strength and are undergoing secondary bone formation and remodeling. Contralateral, nonfractured femora were used as controls. Two times points were examined in the histological studies. Day 14 postfracture calluses were examined, because our previous studies in the B6 strain had shown that this strain would be undergoing resorption and transition to primary bone at this time.(23) Day 21 was examined because any changes in the cartilage to bone transition would be seen at day 14 could be related to the initial stages of bone formation at day 21. The histological assessments at day 21 after fracture also provided a tissue comparison with the μCT data and mechanical assessment data obtained at this time point. For molecular biological studies of fracture healing, mRNA was extracted from callus tissues at day 0 (no fracture) and days 3, 5, 10, 14, and 21 after fracture.
Demineralized histomorphometry analysis
For histological assessment of fractures, femora with surrounding muscle and soft tissues were fixed, decalcified, sectioned, and stained as previously described.(26,28) For growth plate assessments, femora from either 14- or 28-day-old mice (n = 6–8) were fixed in 4% paraformaldehyde containing ruthenium hexamine trichloride.(29,30) The cartilaginous components of the growth plates were separated into the reserve zone, proliferative zone, and hypertrophic zone based on established cell morphology criteria.(30,31) Total area, matrix area, cell area, mean cellular area, and cellularity were all measured using ImageJ (Version 1.36b) for the growth plate as a whole and for each zone of the growth plate separately. The secondary growth centers from 14-day postbirth distal femora were also examined.
μCT
3D images of fractured and nonfractured contralateral femora were obtained using an eXplore Locus SP PreClinical Specimen μCT system (GE Healthcare, London, Ontario, Canada). TMDn was calculated by converting the grayscale output of bone voxels in Hounsfield units (HU) to mineral values (in terms of mg/ml of HA) through the use of a calibration phantom containing air, water, and hydroxyapatite (SB3; Gamex RMI, Middleton, WI, USA). TMDn was defined as the average bone voxel HU value divided by the average HA phantom HU value multiplied by 1130 mg/ml (HA physical density). The same calibration phantom was used in each scan to normalize mineral density measurements and to account for possible variability among scan sessions. Scans were performed at an 8.7-μm voxel resolution. Samples were filtered to remove extraneous voxels using a gaussian smoothing algorithm and individually thresholded using a standard thresholding algorithm(32) to segment bone and nonbone voxels. Thresholded μCT images were used to measure callus diameter, area of mineralized callus (spatial distribution of bone), polar moment of inertia of mineralized callus, subperiosteal diameter, area of original bone, and polar moment of inertia of original bone. Measurements were obtained for four cross-sections just proximal and distal to the fracture site, and the values were averaged. Measures of cross-sectional morphology (area, moment of inertia) were also determined for the mid-diaphyses of the nonfractured, contralateral femora.
Mechanical testing
After μCT analysis, fractured and nonfractured femora were subjected to torsional testing to assess biomechanical properties. Proximal and distal metaphyses were placed in square brass pots and held rigid with acrylic cement. The femora were aligned relative to the loading axis using a custom-made jig that centers the proximal and distal ends of the femoral shaft relative to the center of the square pot. Femora were loaded to failure in torsion at 90°/s using a modified Burstein-Frankel mechanical testing device(33) that was adapted to accommodate mouse bones. Torsional stiffness and failure torque were measured from torque-rotation curves as described previously.(34)
Isolation of mRNA and molecular biology procedures
Tissues were collected, and RNA was processed as previous described.(35) mRNA levels were assessed by either ribonuclease protection analysis (RPA)(35,36) or real-time PCR as previously described.(37) Quantitative RT-PCR was used to assess the relative temporal expression of a series of mRNAs for the primary transcription factors that are known to be determinants of the chondrogenic and osteogenic lineages,(38) as well as a series of mRNAs that are indicative of the differentiated state of these cell lineages. Although qRT-PCR provides a reproducible means of assessing the relative temporal expression of individual mRNAs, it does not provide a direct analysis of the absolute levels of expression between individually expressed mRNAs. This can only be achieved by establishing individual standard curves with known concentrations of the target sequence for each gene that is assessed. Absolute levels of mRNA expression were addressed using ribonuclease protection analysis, a solution hybridization approach. Because probes for the multiple mRNAs are labeled in common and the hybridization of the multiple mRNAs is in the same solution, the only potential variable that effects these measurements might be labeling bias for the individual probe sequences. Thus, the intensity of each of the protected products is representative of the absolute expression levels of the individual mRNAs.
All reagents for the qRT-PCR analysis were from Applied Biosystems, and plate assays were read on an ABI 7700 Sequence Detector (Applied Biosystems, Foster City, CA, USA). One microgram of total RNA was used for each preparation of cDNA. All cDNA preparations were generated by random hexamer priming. All external and internal primers were from commercial sets that are available from Applied Biosystems. Pertinent sequence information, and amplicon sizes are available for each target gene from Applied Biosystems. Each plate contained two negative controls and a positive control probe. All mRNA levels were normalized to β-actin, and each analysis was run in triplicate. The fractional cycle number at which the fluorescence passes the fixed threshold (CT values) was used for quantification by using a comparative CT method. This method is described within the Applied Biosystems instruction manual for the instrumentation. Sample values were normalized to the threshold value for β-actin (Actb) for each time-point: ΔCT = X CT (exp) – X CT (Actb). The CT value for day 0 was used as a reference. ΔΔCT = X CT (exp) – X CT (exp day 0). The fold change in mRNA expression for each time point was plotted in a graph using day 0 as a reference: 2−ΔΔCT (day 0) = 1.
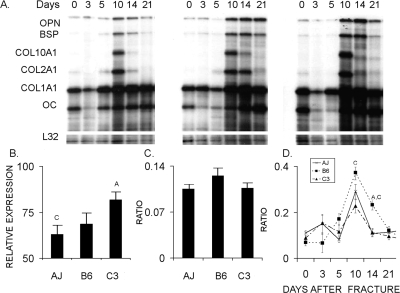
RPA assays were carried out to quantify the overall anabolic activities of the three strains of mice and for the purposes of assessing the absolute ratios of bone to cartilage tissue development. For these studies, the expression of osteopontin (OPN), bone sialoprotein (BSP), collagen α1 type II (Col 2A1), collagen α1 type X (Col10A1), and osteocalcin (OC) were used. Band intensity was assessed by direct γ-counting on a flat bed γ-counter. Replicate counts from four separate gels were used. Individual cpms of each of the mRNA bands were normalized to the cpms of the protected bands for L32 ribosomal protein mRNA. Total measures of anabolic activity were based on the additive mean values of normalized cpms of all of the expressed mRNAs across the entire time course. To assess the total anabolic activity devoted to either cartilage tissue development or bone development, normalized cpm values for Col2A1 plus Col10A1 and Col1A1 plus OC were used, respectively, for the two tissues.
Statistical analysis
Trait means and SDs for the mechanical and μCT measurements were calculated for each genotype and time point. Differences in trait values between genotype and time points were determined by a two-way ANOVA. Posthoc t-tests were conducted to test for specific differences among groups. Posthoc t-tests were conducted in the analysis of differences between individual groups for the RNA expression data and growth plate histomorphometry data.
RESULTS
Genetic variability in the regain of mechanical functionality during fracture healing
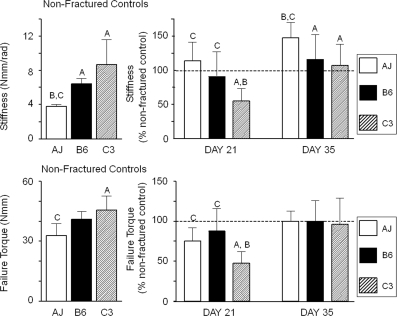
Torsional tests to failure were carried out on fractured and nonfractured femora from three inbred mouse strains to quantitatively assess genetic variation in the regain of mechanical function (Fig. 1). Table 1 presents a summary of all the primary biomechanical and structural values from which the comparative data in Fig. 1 was derived. Significant differences in torsional stiffness and failure torque of the nonfractured contralateral femora were observed among the inbred strains, with A/J femora showing the smallest torsional stiffness and failure torque among the three strains (left panels). Variation in the regain of mechanical function was assessed by comparing the mechanical properties of the fractured femora to that of the nonfractured, contralateral femora (right panels). A two-way ANOVA showed significant differences relative to genotype (p < 0.02) and time (p < 0.0001) for both stiffness and strength. At 21 days after fracture, the fractured A/J and B6 femora had regained nearly 100% of the prefracture stiffness and 75–88% of the prefracture failure torque. In contrast, C3H had only regained 47–55% of the prefracture mechanical properties. A comparison among individual strains showed that C3H was significantly different from A/J and B6, indicating that the fractured C3H femora showed a delay in the regain of mechanical function. By 35 days of age, all mechanical properties had returned to prefracture values for all three strains, and in the case of A/J, this strain exceeded its prefracture stiffness. Thus, the C3H strain was the slowest healer, whereas the A/J and B6 strains healed at approximately the same rates, based on regain of prefracture mechanical properties.
FIG. 1.
Biomechanical characteristics of fractured callus in response to torsion testing. The data depicted in each of the panels is denoted in the figure. Group designations in all figures are A/J (AJ), C57Bl/6J (B6), and C3H/HeJ (C3). Left panels depict biomechanical properties of the unfractured contralateral bones. Right panels depict the regain of each of the biomechanical properties as a percentage of the contralateral values. The 100% value is denoted in the figure with a dashed line. The bars are as indicated in the legend. Error bars depict SD. Two-way ANOVA for genotype and age: stiffness (p = 0.0001, p = 0.0002) and torque to failure (p = 0.0194, p = 0.0002). Letters above each figure indicate the individual group comparisons showing statistical differences (A = A/J; B = C57Bl/6J; C = C3H/HeJ).
Table 1.
Variation in Structural and Biomechanical Properties of Intact and Fractured Femora Among Three Inbred Mouse Strains
| Stiffness (Nmm/rad) | Torque (Nmm) | Total area (mm2) | Moment of intertia (mm4) | |
| AJ intact | 3.8 ± 0.3B,C | 32.9 ± 6.1C | 1.23 ± 0.11B,C | 0.25 ± 0.05B,C |
| AJ day 21 | 4.3 ± 1.0 | 24.7 ± 5.7 | 5.37 ± 1.49 | 5.13 ± 2.82 |
| AJ day 35 | 5.5 ± 0.9* | 33.0 ± 4.4 | 2.47 ± 0.51 | 2.47 ± 0.51 |
| B6 intact | 6.4 ± 0.6A | 41.0 ± 4.0 | 2.05 ± 0.25A,C | 0.74 ± 0.18A,C |
| B6 day 21 | 5.8 ± 2.3 | 36.2 ± 11.5 | 9.66 ± 1.65 | 16.11 ± 5.06 |
| B6 day 35 | 7.4 ± 2.3 | 41.0 ± 10.6 | 6.81 ± 2.09 | 6.81 ± 2.09 |
| C3H intact | 8.7 ± 2.9A | 45.5 ± 7.2A | 1.75 ± 0.16A,B | 0.54 ± 0.10A,B |
| C3H day 21 | 4.8 ± 1.6* | 21.4 ± 7.0* | 7.45 ± 1.64 | 9.98 ± 4.16 |
| C3H day 35 | 9.3 ± 2.7 | 43.8 ± 15.1 | 5.57 ± 1.34 | 5.57 ± 1.34 |
Superscript letters indicate the strains showing significantly different traits for intact femora (p < 0.05, posthoc t-tests). Letters denotations are A = A/J, B = C57Bl/6J, and C = C3H/HeJ.
* Difference relative to intact (nonfractured) femora.
Genetic variation in structural traits contribute to the regain of strength
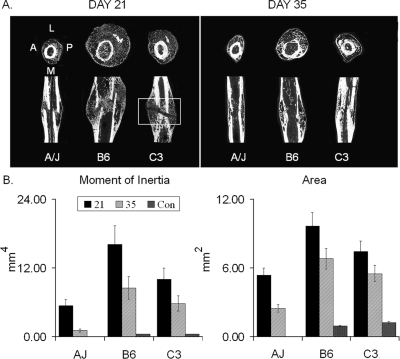
μCT was carried out to assess the primary architectural variations in the same set of fracture calluses before mechanical testing described above. These studies attempted to explain the differences observed in the rate of regain of mechanical functionality based on variation in the structure of the fracture calluses. Representative μCT images of the three strains at 21 and 35 days after fracture are presented (Fig. 2A). Significant differences in both the size and structure of the calluses were observed among the three strains (p < 0.0001, two-way ANOVA) at both time points (p < 0.0001, two-way ANOVA). The average total area (amount of tissue) and polar moment of inertia (distribution of tissue) of the 21- and 35-day calluses are shown in comparison with the cross-sectional morphology of the intact femora (Fig. 2B). The area and polar moment of inertia of the fracture calluses for the A/J, B6, and C3H femora were respectively, 321% and 1022%, 525% and 2603%, and 484% and 2286% greater compared with the intact bone at day 21. For each strain, the callus size (area, moment of inertia) was significantly greater at 21 days compared with 35 days after fracture, indicating that the resorption/consolidation phase of callus development had begun between these two time points. The μCT images also showed a number of easily discernible differences in the quality of the newly formed bone. The most obvious was an area of X-ray translucency in the central region of the C3H callus at day 21, which is indicative of a lack of bony bridging. Because this was not observed for the A/J and B6 fracture calluses, this X-ray translucency may partly explain the delayed regain of stiffness and strength of the C3H femora. This translucence would be indicative of cartilage, unmineralized bone, or fibrous tissue (see histological characterization; Fig. 3). Interestingly, except for the central regions of lesser mineral density, the trabecular bone volume fraction (BV/TV) and thickness (TbTh) were not statistically different between the strains at 21 days after fracture (data not shown). However, tissue mineral density (TMDn) was 12–16% greater in the A/J and C3H fracture calluses compared with B6 (p < 0.05, t-tests). By 35 days after fracture, all three strains of mice showed bony bridging of all fracture calluses. These data indicate that whole callus properties defining the percent bone fraction and mineral density can not explain the delay in regain of mechanical function seen for the C3H strain relative to A/J and B6 strains.
FIG. 2.
Structural comparisons of fracture calluses from three inbred strains of mice. (A) Representative μCT images in both transverse and longitudinal orientations taken at 21 and 35 days after fracture. The orientation of the μCT images is denoted for the left transverse section of the A/J strain. Boxed area of C3H day 21 images shows area that remained unbridged by bone. (B) Calculated moments of inertia and mean cross sectional areas based on μCT measurements. The control (Con) is from an averaged set of values taken from identical mid-diaphyseal segments of the unfractured contralateral bones. The key in the figure denotes time after fracture and is presented in comparison with the contralateral bone that is used as the control. Error bars depict SD. Individual letter denotations are not included because differences were significant between all groups and times. Two-way ANOVA for genotype and age indicated: moment of inertia (p = 0.0001, p = 0.0001) and area (p = 0.0047, p = 0.0002) between all groups and time.
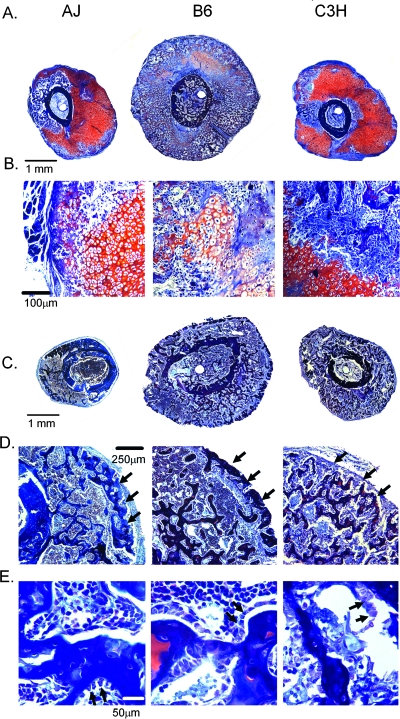
FIG. 3.
Histological assessment of callus tissues at 14 and 21 days after fracture. All sections were stained with aniline blue safranin O. Cartilage stains red. Both strain and times are indicated in the figure. Microscope magnifications and absolute scales of the images are denoted in the figure. (A) Montage of a composite set of micrographs from the central regions of the fracture callus from the three strains of mice at 14 days. Eight to 15 micrographs were overlapped to generate each montage. Photomicrographs that were used to generate the montage were taken at a total microscopic magnification of ×40. (B) Selected micrographs from day 14 postfracture callus tissues showing representative cartilage and bone tissue and cellular morphologies. Total microscopic magnifications were at ×200. (C) Montage of a composite set of micrographs from the central regions of the fracture callus from the three strains of mice at 21 days. Eight to 15 micrographs were overlapped to generate each montage. Photomicrographs that were used to generate the montage were taken at a total microscopic magnification of ×40. (D) Selected micrographs of the peripheral surfaces of 21 day postfracture calluses. Arrows denote the rim of newly formed cortical bone bridging the outer surface of each callus. Total microscopic magnifications were at ×200. (E) Selected micrographs from day 21 postfracture callus tissues showing representative trabecular tissue, hematopoietic, stromal/mesenchymal, and cellular morphologies. Arrows in this image indicate individual cells lining callus trabecular surfaces. Total microscopic magnifications were at ×400X. Scale bars are included in each panel.
Variation in tissue development during fracture healing
A qualitative histological assessment was carried out to determine whether there was observable tissue or cellular differences in the calluses among the three strains (Fig. 3). A comparison of the overall callus tissue morphology is seen in the montage of overlapping micrographs that provides a panoramic view of the entire cross sectional area of the callus tissues (Fig. 3A). Differences in the overall cross-sectional areas were observed between the three strains consistent with the μCT analysis. The most striking differences in tissue morphologies were seen at 14 days after fracture. The callus tissues for the B6 strain showed the fastest progression in endochondral bone formation with large areas undergoing resorption compared with the A/J and the C3H strains. In contrast, the A/J and C3H strains showed large areas of unresorbed or nonhypertrophic cartilage based on the areas of red safranin O–stained tissues. At this time, the B6 calluses had a very extensive, well-developed network of mixed cartilage and boney trabeculae on which new osteoid formed. The A/J calluses had an intermediate level of these structures with fewer trabeculae. Few of these tissue structures were seen in the C3H calluses. Figure 3B depicts the definable cellular differences seen in chondrocytes and tissue structures at day 14. A comparison of chondrocyte morphology showed that cells in the B6 calluses appeared to be the largest, whereas cells in the A/J callus had an intermediate size compared with those seen in the C3H calluses, which appeared the smallest. The B6 cartilage tissues also showed less intense Safranin O red staining than either the C3H or A/J tissues, indicating that the underlying proteoglycans within the B6 tissue were undergoing degradation. At day 21 after fracture, no large areas of cartilage tissue remained in any of the calluses, and an extensive number of trabecular structures had developed in the central areas of the calluses for all three strains (Fig. 3C). Whereas a well-developed cortical shell that almost completely surrounded the calluses was seen in sections from both the B6 and A/J strains, this outer rim of new bone was much less developed in the C3H calluses. This difference is clearly emphasized in the higher magnification of the outer callus shell (Fig. 3D) and is denoted by the arrows. Higher magnifications examining the cellular details of the tissue morphologies for day 21 calluses are seen in Fig. 3E. At this postfracture time, no cartilage cells remained in the callus tissue from the three strains of mice, although remnants of mineralized cartilage cores were seen in the trabeculae in the B6 calluses. Comparisons across the three different strains showed that both the A/J and B6 tissues had well-organized arrangements of hematopoietic cells between the trabecular elements with cuboidal shaped cells lining the trabecular surfaces indicated with arrows. In contrast, the hematopoietic cells were much less organized between the trabeculae in the C3H calluses, and there was no apparent arrangement of cells lining the trabecular surfaces in the C3H calluses.
Genetic variability in skeletal stem cell differentiation during fracture healing
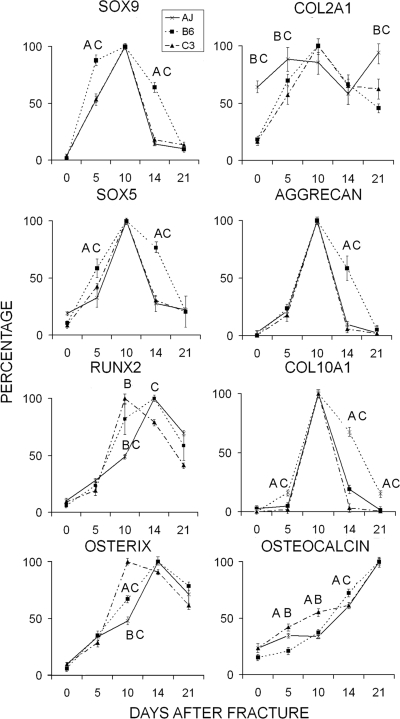
The molecular mechanisms regulating cellular development within the callus tissues were examined using two approaches based on the expression of mRNAs that represent the temporal progression of skeletal progenitor cells and their subsequent differentiation into chondrogenic and osteogenic cells. Quantitative RT-PCR was used to assess the relative temporal progression of skeletal cell differentiation (Fig. 4). Four transcription factors were first compared: SOX 9, which commits stem cells to the skeletal lineage and is common to both chondrogenic and osteogenic cells; SOX 5, which subsequently contributes to the commitment and maturation of cells of the chondrogenic lineage; osterix, which directs the lineage commitment to osteogenesis; and RUNX2, which is involved in both terminal hypertrophic chondrocyte differentiation and is necessary with osterix to promote osteogenesis (left panels). Large temporal differences in the patterns of SOX 9 and SOX5 mRNA expression were observed for the B6 strain compared with the A/J and C3H strains. The B6 strain showed the most prolonged period of expression for both SOX 9 and SOX5, suggesting that the skeletal progenitors in the B6 strain underwent an earlier and longer period of chondrogenesis compared with A/J and C3H. In contrast, osterix and RUNX2 mRNA expression appeared earlier in the C3H strain compared with the A/J and B6 strains, suggesting there was an earlier commitment toward osteogenic differentiation in C3H. Equally informative to the variations in the patterns of skeletal cell differentiation within the callus tissues was the expression of RUNX2. In the B6 strain, RUNX2 expression was induced at an intermediate time to C3H and A/J in which it was seen to peak either earlier or later, respectively, in these two strains. Because RUNX2 expression is equally important to both the hypertrophic development and mineralization of cartilage tissues as to osteogenic differentiation, these results are consistent with the B6 strain undergoing an earlier and longer period of chondrocyte hypertrophy compared with the other strains.
FIG. 4.
Temporal profiling of the relative mRNA expression for the major transcription factors and extracellular matrix proteins unique to different stages of chondrogenesis and osteogenesis. Error bars = SD of each set of replicate measurements. Legend denotes the strains. Single letters over the individual points denote the strains in a graph that are statistically different from the one which is denoted. Letters denotations are A = A/J, B = C57Bl/6J, and C = C3H/HeJ. mRNA levels were determined from three to four replicate RT-PCR trials. Each mRNA is denoted in the figure. All values were normalized to the levels of β-actin. Relative values were determined from 2 − ΔΔCT (day 0) = 1 Max2 − ΔΔCT/Min2 − ΔΔCT × 100, with the highest expressed level set as Max.
The temporal expression of specific mRNAs that encode extracellular matrix proteins that are both targets of the transcriptional regulators described above and are themselves markers of various stages of lineage progression of chondrocytes and osteoblasts are shown in the right set of panels of Fig. 4. These data further corroborate the conclusions made from both the histology and the expression profiles of the various transcription factors. The B6 mice showed the longest period of expression for both aggrecan and type X (Col01A1) collagen. Although the A/J and C3H strains showed the same pattern for the aggrecan expression, Col10A1expression continued to show low levels of expression in the A/J strain until day 14. This is in contrast to the C3H strain, which showed a very sharp decline after day 10. Osteocalcin, which was used as a definitive marker for osteogenesis, showed the earliest induction in the C3H strain, whereas its induction was the slowest in the B6 strain.
In the second analysis, the overall anabolic activities of the specific types of cells in the callus tissues were assessed by RPA measurements of cartilage and bone specific extracellular matrix protein mRNAs (Fig. 5). The temporal pattern of expression observed by RPA confirmed those observed by qRT-PCR. Total anabolic activity of the callus tissues was calculated across the whole 21-day time course based on an analysis of the additive values of the cpm(s) for all mRNA bands normalized by the cpm(s) of the L32 bands. This set of quantitative measurements showed that the C3H strain had the greatest anabolic activity of the three strains and was statistically larger than the A/J strain. These data showed that the B6 strain contributed the highest percentage of its anabolic activity to cartilage tissue formation, whereas the C3H strain and A/J strains showed lesser and comparable levels of cartilage mRNA expression, although these results were not statistically significant (Fig. 5C). At 10 and 14 days after fracture, the B6 strain had statistically higher levels of cartilage to bone expression than either the C3H or A/J strains (Fig. 5D). In summary, the results obtained from the two mRNA analyses showed primary differences between the three strains related to both the timing of the differentiation of the chondrogenic and osteogenic lineages and to the percentage of skeletal progenitor cells that develop into cartilage and bone tissues.
FIG. 5.
Quantitative analysis of chondrogenic and osteogenic activities in the callus tissues based on extracellular matrix mRNA expression. Error bars = SD of each set of replicate measurements. Legend denotes the strains. Single letters over individual points or bars in a graph represent those strains that are statistically different from the one which is denoted. Letters denotations are A = A/J, B = C57Bl/6J, and C = C3H/HeJ. (A) Representative image of an RPA gel. Individual bands for each of the mRNA species are denoted in the figure. (B) Graphical representation of the total anabolic activities of the callus tissues over the entire time course of 21 days based on analysis of the cpm(s) of all mRNA bands/cpm(s) of the L32 bands. (C) Graphical representation of ratio of the anabolic activities of the chondrogenic vs. osteogenic cells in the callus tissues over the total time course of 21 days based on the ratio Col2A1 plus Col10a1 to Col1A1 plus osteocalcin mRNAs. (D) Graphical representation of the anabolic activities of the chondrogenic vs. osteogenic tissue formation at individual time points over the 21-day time course based on analysis of Col2A1 plus Col10a1 mRNAs.
Genetic variation in normal endochondral bone development
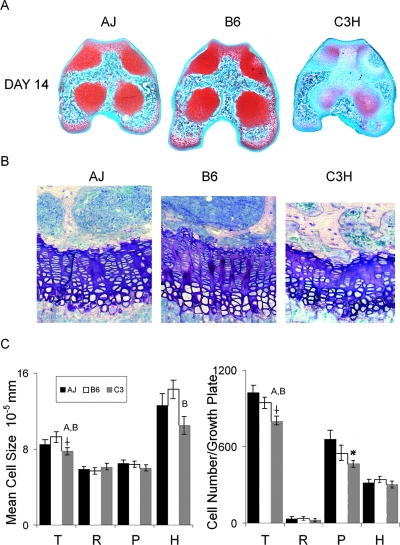
The data accumulated from the fracture healing studies led to the question of whether the variability in biological mechanisms of cartilage tissue development was an intrinsic genetic feature of these various strains. This question was addressed by examining the progression of endochondral bone formation during the development of the secondary centers of the distal femur at 14 days of age and the distal femoral growth plates at 28 days of age. Transverse sections from identical anatomical positions in the middle of the secondary growth centers of 14-day postbirth femora were examined (Fig. 6A). These analyses showed that bone formation was well progressed in both the A/J and B6 strains. In contrast, the central regions within the distal femur of the C3H strain showed little marrow infiltration and lacked the Safranin O staining for mature proteoglycan that was observed within cartilage tissues of the A/J and B6 strains (Fig. 6A). The femoral distal growth plates from 28 days of age were examined to further assess whether differences in endochondral development persisted into later stages of development (Figs. 6B and 6C). At this age, the epiphyseal growth plates were well developed and were representative of the linear arrangement of cells that are most commonly examined in growing long bones. As can be seen, the mean cell size was greatest for the B6 strain and smallest for the C3H strain (p = 0.024, t-test). When the differences in cell sizes were examined individually in the different zones of the growth plates, significant differences were observed between B6 and C3Hfor the cells localized to the hypertrophic zone (p = 0.01). Furthermore, the overall number of chondrocytes in the growth plates varied significantly among the strains (p = 0.05, ANOVA). A comparison between strains showed that C3H showed the smallest number of cells compared with A/J and B6 (p = 0.05, t-tests). When these differences were broken down by zones, the proliferative zone showed a strong trend toward being different (p = 0.064).
FIG. 6.
Variation in postnatal cartilage maturation in three inbred strains of mice. Legend denotes the strains. (A) Identical midregion sections of the secondary growth centers localized between the joint surface and the top of the distal epiphyseal growth plate of 14 day after birth femora were used for comparison. Representative sections from a group of five that were examined are depicted. Mature proteoglycan was detected by staining with safranin O fast green. Mature cartilage that stained bright red is seen as darker gray areas in the images. (B) Representative histological variations seem in 28-day postbirth distal femur growth plate. Centrally localized regions are depicted. (C) Morphometric assessments (N = 6–8) for the growth plates depicted in B were carried out to determine average cell size and total cell number per growth plate. Letters over individual bars in a graph represent those groups that are statistically different from the one that is denoted. Letters denotations are A = A/J, B = C57Bl/6J, and C = C3H/HeJ. rSignificance at p ≤ 0.05 ANOVA between the three strains. *Trend toward being different (p = 0.064). Each parameter is denoted in the graphics. T, total growth plate; R, resting zone; P, proliferative zone; H, hypertrophic zone.
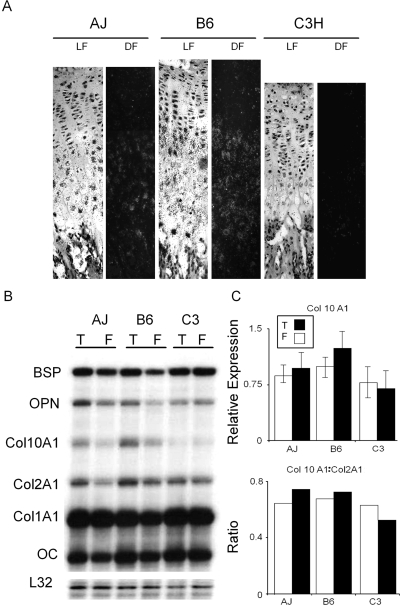
In the final studies, we examined Col10A1 mRNA expression in 28-day postbirth distal femur growth plates using both in situ hybridization (Fig. 7A) and RPA (Figs. 7B and 7C). The in situ hybridization studies provided further evidence that the B6 strain shows the most extended hypertrophic period based on the greater number of cells per column in which cells express Col10A1. In the RPA studies, we also examined mRNA expression of various cartilage and bone mRNAs in the distal and proximal growth plates of the femur and tibia, respectively (Figs. 7B and 7C). Although not statistically significant, the examination of the mRNA expression for various extracellular matrix proteins in the distal growth plates from 28-day postbirth femora and tibias provided further evidence supporting the conclusion that C3H showed the lowest levels of Col10A1 expression.
FIG. 7.
Variation in collagen type X expression in growth plates from three inbred strains of mice. (A) Variation in Col10A1 expression in 28-day postbirth distal femur growth plates. Serial sections from centrally localized regions of the distal femora growth plates depicted in Fig. 6 were hybridized with a Col10A1 antisense probe. Matched dark (DF) and light fields (LF) are presented. Growth plates were defined by the margins with the secondary center and intramedullary bone of the diaphysis. (B) Progression of hypertrophic cartilage maturation based on Col10A1 expression. Growth plates from 28-day postbirth mice were isolated from distal femora and proximal tibia. RNA expression was assessed by RPA and normalized to the L32 mRNA expression found in tibia growth plates (T) and femora growth plates (F). (C) Relative Col10A1 expression. Top panel is the graphical representation of the Col10A1 expression based on analysis of the cpm(s) of the Col10A1 band/cpm(s) of the L32 bands. Bottom panel is the ratio of Col10A1 to Col2a1.
DISCUSSION
The concept that bone has the ability to assemble different sets of traits to achieve its needed mechanical functions is reflective of the network of functional interactions between the genetic variants that affect bone size, material property, and shape.(39) The studies presented here showed that the regain of mechanical functionality after a long bone fracture varied with genetic background. The identification of the types of structural and biological variations that were observed in fracture healing in the well-defined animal models used in these studies therefore will be very informative of the variation expected for human bone healing. The aspects of fracture healing that varied among the three inbred mouse strains can be related to regain in mechanical function included the size of the callus and the timing at which bony bridging occurred in the central region of the callus.
The differences in X-ray density observed by μCT suggested that the osteoid that formed specifically in the central region of the C3H calluses was less mineralized compared with that formed in the A/J and B6 calluses. The comparisons between the μCT, mechanical, and histological findings suggested that the osteoid which formed in the central region of C3H calluses was less organized and the external osteoid that bridged the central regions of the callus formed and mineralized much slower compared with that formed in the A/J and B6 calluses. This biomechanical analysis suggested that the slower rate of regain in mechanical function of C3H calluses resulted from a slower and poorer coordination of the transitional stages between cartilage and bone and thus a slower progression in the mineralization of the primary bone formed within the central region of the callus. This slower transition also seemed to affect the development of the external bone bridging that is needed to restore initial mechanical function to the fractured bone.
The interstrain comparison of fracture healing provided new insight into the cellular basis of the genetic variation in fracture healing. The histological and gene expression profiles showed that there was variation in the recruitment, temporal progression in the differentiation, and activity of the skeletal progenitor cells that formed the bone and cartilage tissues among the inbred strains. The more rapid healing B6 mice had an earlier and more prolonged period of endochondral bone formation. At the tissue level, the B6 calluses underwent a period of mineralization and resorption that overlapped more extensively with primary bone formation. In contrast, the slower healing C3H mice had a much shorter period of chondrogenesis and mineralization with an earlier induction of osteogenesis. In this regard, whereas the temporal patterns of chondrogenic development in the A/J strain appeared to be very similar to that of the C3H mice, the osteogenic transcription factor osterix was expressed later in A/J mice. The later commitment to osteogenesis in both the A/J and B6 strains seemed to lead to a better coordination between endochondral resorption and with the subsequent periods of primary bone formation. Overall, it would seem that variation in the rates of healing resided in the temporal progression of osteogenic and chondrogenic differentiation during endochondral bone formation.
In this regard it is interesting to note that the vast majority of fractures sustained worldwide are either untreated (e.g., patients in many third world countries) or are treated by cast immobilization, traction, or intramedullary nailing. In each of these instances, the lack of rigidity of the fixation promotes micromotion and supports healing in a manner that involves a periosteal response and an endochondral component of bone formation.(40,41) The pivotal role of endochondral bone formation as the rate-limiting step to the initial regain of mechanical function during fracture healing is supported by previous structural studies that showed that the central region of the callus around the fracture gap is the last region in which the cartilage tissue develops, matures, and undergoes resorption.(25) Such results would be consistent with the structural necessity that cortical bone bridging across the fracture gap must occur to restore the basic mechanical competency to the bone.(25,41)
The primary genetically inherited structural differences among the inbred strains that were recapitulated during fracture healing were the moment of inertia and cross-sectional area. As in prior work, the structural variations observed for the fracture calluses were also accompanied with differences in tissue mineralization.(39) Although contributory factors that would be considered epigenetic to the bone cannot be ruled out, many of these genetically varying traits (e.g., activity, muscle size) arise after postnatal growth when bone size has been established.(14) Although most studies in mice that have characterized these structural features have focused on rates of coupled bone modeling and remodeling,(42) the current data suggested that this feature may be affected by many different biological mechanisms, including variation that resides in the postnatal processes controlling cell lineage selection within skeletal progenitors during endochondral bone formation. This conclusion is consistent with recent findings showing that BMP-2 was necessary for postnatal bone repair and is genetically associated with maintenance of normal bone mass but was not apparently needed for embryological bone formation.(43,44) In the case of the B6 strain, it seems that genetically inherited differences in the mechanisms affecting both the temporal progression of chondrocyte differentiation and chondrocyte cell size may also affect transverse bone growth.
Whereas previous studies have shown that phylogenetic variation in the mechanisms of longitudinal growth reside in part in variable chondrocyte cell size and rates of chondrocyte replication,(31,45) our studies suggested that these mechanisms are also genetically variable within a given species and may also have an effect on transverse growth. In contrast, the greater cross-sectional area of the C3H mice seems to arise because of a substantially longer period of osteogenesis in comparison with either the B6 or the A/J strain. We further speculate that increased cross-sectional area of the callus tissue in C3H mice fails to achieve increased strength as fast as either the A/J or B6 strain, because the greater amount of osteoid that is produced by this strain of mice either needs more time to mineralize or its synthesis is less well coordinated with the shorter period of chondrogenesis and mineralized cartilage resorption.
In summary, these studies are the first to show that genetic background that is related to bone quality affects the rate of fracture healing after traumatic injury. Whereas to date the majority of studies have focused on the genetics of coupled remodeling as the central biological processes that affect bone quality, our studies provide the first direct evidence that genetic variation in bone quality may also be linked to inherited differences in the processes of endochondral bone formation and their relationship to postnatal bone growth.
ACKNOWLEDGMENTS
This study was supported with grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (PO1AR049920, AR44927) to TAE, LCG, and KJJ and the Department of Defense (DAMD17-03-1-0576) to LCG and KJJ. Institutional support was provided by the Department of Orthopaedic Surgery Boston University School of Medicine and by Boston University School of Medicine.
Footnotes
The authors state that they have no conflicts of interest.
REFERENCES
- 1.Praemer A, Furner S, Rice DP. Park Ridge, IL, USA: American Academy of Orthopaedic Surgeons; 1995. Musculoskeletetal Conditions in the United States. [Google Scholar]
- 2.Consensus Panel NIH. Osteoporosis prevention, diagnosis, and therapy. JAMA. 2001;285:785–795. doi: 10.1001/jama.285.6.785. [DOI] [PubMed] [Google Scholar]
- 3.U.S. Department of Health and Human Services. Rockville, MD, USA: USDHHS/PHS; 2004. Bone Health and Osteoporosis: A Report of the Surgeon General. [Google Scholar]
- 4.Xiong DH, Shen H, Xiao P, Guo YF, Long JR, Zhao LJ, Liu YZ, Deng HY, Li JL, Recker RR, Deng HW. Genome-wide scan identified QTLs underlying femoral neck cross-sectional geometry that are novel studied risk factors of osteoporosis. J Bone Miner Res. 2006;21:424–437. doi: 10.1359/JBMR.051202. [DOI] [PubMed] [Google Scholar]
- 5.Beamer WG, Shultz KL, Ackert-Bicknell CL, Horton LG, Delahunty KM, Coombs HF, III, Donahue LR, Canalis E, Rosen CJ. Genetic dissection of mouse distal chromosome 1 reveals three linked BMD QTLs with sex-dependent regulation of bone phenotypes. J Bone Miner Res. 2007;22:1187–1196. doi: 10.1359/jbmr.070419. [DOI] [PubMed] [Google Scholar]
- 6.Beamer WG, Shultz KL, Donahue LR, Churchill GA, Sen S, Wergedal JR, Baylink DJ, Rosen CJ. Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3HH/HeJ inbred strains of mice. J Bone Miner Res. 2001;16:1195–1206. doi: 10.1359/jbmr.2001.16.7.1195. [DOI] [PubMed] [Google Scholar]
- 7.Jepsen KJ, Pennington DE, Lee YL, Warman M, Nadeau J. Bone brittleness varies with genetic background in A/J and C57BL/6J inbred mice. J Bone Miner Res. 2001;16:1854–1862. doi: 10.1359/jbmr.2001.16.10.1854. [DOI] [PubMed] [Google Scholar]
- 8.Jepsen KJ, Akkus O, Majeska RJ, Nadeau JH. Hierarchical relationship between genetically determined bone traits and whole bone mechanical properties in inbred mice. Mamm Genome. 2003;14:97–104. doi: 10.1007/s00335-002-3045-y. [DOI] [PubMed] [Google Scholar]
- 9.Wergedal JE, Sheng MH, Ackert-Bicknell CL, Beamer WG, Baylink DJ. Genetic variation in femur extrinsic strength in 29 different inbred strains of mice is dependent on variations in femur cross-sectional geometry and bone density. Bone. 2005;36:111–122. doi: 10.1016/j.bone.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Tommasini SM, Morgan TG, van der Meulen MC, Jepsen KJ. Genetic variation in structure-function relationships for the inbred mouse lumbar vertebral body. J Bone Miner Res. 2005;20:817–827. doi: 10.1359/JBMR.041234. [DOI] [PubMed] [Google Scholar]
- 11.Tommasini SM, Nasser P, Schaffler MB, Jepsen KJ. Relationship between bone morphology and bone quality in male tibias: Implications for stress fracture risk. J Bone Miner Res. 2005;20:1372–1380. doi: 10.1359/JBMR.050326. [DOI] [PubMed] [Google Scholar]
- 12.Li CY, Schaffler MB, Wolde-Semait HT, Hernandez CJ, Jepsen KJ. Genetic background influences cortical bone response to ovariectomy. J Bone Miner Res. 2005;20:2150–2158. doi: 10.1359/JBMR.050819. [DOI] [PubMed] [Google Scholar]
- 13.Richman C, Kutilek S, Miyakoshi N, Srivastava AK, Beamer WG, Donahue LR, Rosen CJ, Wergedal JE, Baylink DJ, Mohan S. Postnatal and pubertal skeletal changes contribute predominantly to the differences in peak bone density between C3H/HeJ and C57BL/6J mice. J Bone Miner Res. 2001;16:386–397. doi: 10.1359/jbmr.2001.16.2.386. [DOI] [PubMed] [Google Scholar]
- 14.Price C, Herman BC, Lufkin T, Goldman HM, Jepsen KJ. Genetic variation in bone growth patterns defines adult mouse bone fragility. J Bone Miner Res. 2005;20:1983–1991. doi: 10.1359/JBMR.050707. [DOI] [PubMed] [Google Scholar]
- 15.Beamer WG, Shultz KL, Churchill GA, Frankel WN, Baylink DJ, Rosen CJ, Donahue LR. Quantitative trait loci for bone density in C57BL/6J and CAST/EiJ inbred mice. Mamm Genome. 1999;11:1043–1049. doi: 10.1007/s003359901159. [DOI] [PubMed] [Google Scholar]
- 16.Parsons CA, Mroczkowski HJ, McGuigan FE, Albagha OM, Manolagas S, Reid DM, Ralston SH, Shmookler Reis RJ. Interspecies synteny mapping identifies a quantitative trait locus for bone mineral density on human chromosome Xp22. Hum Mol Genet. 2005;14:3141–3148. doi: 10.1093/hmg/ddi346. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Gu W, Masinde G, Hamilton-Ulland M, Rundle CH, Mohan S, Baylink DJ. Genetic variation in bone-regenerative capacity among inbred strains of mice. Bone. 2001;29:134–140. doi: 10.1016/s8756-3282(01)00497-5. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Gu W, Masinde G, Hamilton-Ulland M, Xu S, Mohan S, Baylink DJ. Genetic control of the rate of wound healing in mice. Heredity. 2001;86:668–674. doi: 10.1046/j.1365-2540.2001.00879.x. [DOI] [PubMed] [Google Scholar]
- 19.Masinde GL, Li X, Gu W, Davidson H, Mohan S, Baylink DJ. Identification of wound healing/regeneration quantitative trait loci (QTL) at multiple time points that explain seventy percent of variance in (MRL/MpJ and SJL/J) mice F2 population. Genome Res. 2001;11:2027–2033. doi: 10.1101/gr.203701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBrearty BA, Clark LD, Zhang XM, Blankenhorn EP, Heber-Katz E. Genetic analysis of a mammalian wound-healing trait. Proc Natl Acad Sci USA. 1998;95:11792–11797. doi: 10.1073/pnas.95.20.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson C, Alpern E, Miclau T, Helms JA. Does adult fracture repair recapitulate embryonic skeletal formation. Mech Dev. 1999;87:57–66. doi: 10.1016/s0925-4773(99)00142-2. [DOI] [PubMed] [Google Scholar]
- 22.Vortkamp A, Pathi S, Peretti GM, Caruso EM, Zaleske DJ, Tabin CJ. Recapitulation of signals regulating embryonic bone formation during postnatal growth and in fracture repair. Mech Dev. 1998;71:65–76. doi: 10.1016/s0925-4773(97)00203-7. [DOI] [PubMed] [Google Scholar]
- 23.Gerstenfeld LC, Cho TJ, Kon T, Aizawa T, Tsay A, Fitch J, Barnes GL, Graves DT, Einhorn TA. Impaired fracture healing in the absence of TNF-alpha signaling: The role of TNF-alpha in endochondral cartilage resorption. J Bone Miner Res. 2003;18:1584–1592. doi: 10.1359/jbmr.2003.18.9.1584. [DOI] [PubMed] [Google Scholar]
- 24.Gerstenfeld LC, Cullinane DM, Barnes GL, Graves DT, Einhorn TA. Fracture healing as a post-natal developmental process: Molecular, spatial, and temporal aspects of its regulation. J Cell Biochem. 2003;88:873–884. doi: 10.1002/jcb.10435. [DOI] [PubMed] [Google Scholar]
- 25.Gerstenfeld LC, Alkhiary YM, Krall EA, Nicholls FH, Stapleton SN, Fitch JL, Bauer M, Kayal R, Graves DT, Jepsen KJ, Einhorn TA. Three dimensional reconstruction of fracture callus morphogenesis demonstrates asymmetry in callus development. J Histochem Cytochem. 2006;54:1215–1228. doi: 10.1369/jhc.6A6959.2006. [DOI] [PubMed] [Google Scholar]
- 26.Seeman E, Delmas PD. Bone quality–the material and structural basis of bone strength and fragility. N Engl J Med. 2006;354:2250–2261. doi: 10.1056/NEJMra053077. [DOI] [PubMed] [Google Scholar]
- 27.Bonnarens F, Einhorn TA. Production of a standard closed fracture in laboratory animal bone. J Orthop Res. 1984;2:97–101. doi: 10.1002/jor.1100020115. [DOI] [PubMed] [Google Scholar]
- 28.Gerstenfeld LC, Wronski TJ, Hollinger JO, Einhorn TA. Perspective: The application of histomorphometric methods to the study of bone repair. J Bone Miner Res. 2005;20:1715–1722. doi: 10.1359/JBMR.050702. [DOI] [PubMed] [Google Scholar]
- 29.Hunziker EB, Herrmann W, Schenk RK. Improved cartilage fixation by ruthenium hexammine trichloride (RHT): A prerequisite for morphometry in growth cartilage. J Ultrastruct Res. 1982;81:1–12. doi: 10.1016/s0022-5320(82)90036-3. [DOI] [PubMed] [Google Scholar]
- 30.Hunziker EB, Schenk RK. Physiological mechanisms adopted by chondrocytes in regulating longitudinal bone growth in rats. J Physiol. 1989;414:55–71. doi: 10.1113/jphysiol.1989.sp017676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barreto C, Wilsman NJ. Hypertrophic chondrocyte volume and growth rates in avian growth plates. Res Vet Sci. 1994;56:53–61. doi: 10.1016/0034-5288(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 32.Otsu N. A threshold selection method from gray-scale histogram. IEEE Trans Syst Man Cybernet. 1978;8:62–66. [Google Scholar]
- 33.Burstein AH, Frankel VH. A standard test for laboratory animal bone. J Biomech. 1971;4:155–158. doi: 10.1016/0021-9290(71)90026-1. [DOI] [PubMed] [Google Scholar]
- 34.Hausman MR, Schaffler MB, Majeska RJ. Prevention of fracture healing in rats by an inhibitor of angiogenesis. Bone. 2001;29:560–564. doi: 10.1016/s8756-3282(01)00608-1. [DOI] [PubMed] [Google Scholar]
- 35.Kon T, Cho TJ, Aizawa T, Yamazaki M, Nooh N, Graves D, Gerstenfeld LC, Einhorn TA. Expression of osteoprotegerin, receptor activator of NF-kappaB ligand (osteoprotegerin ligand) and related proinflammatory cytokines during fracture healing. J Bone Miner Res. 2001;16:1004–1014. doi: 10.1359/jbmr.2001.16.6.1004. [DOI] [PubMed] [Google Scholar]
- 36.Cho TJ, Gerstenfeld LC, Einhorn TA. Differential temporal expression of members of the transforming growth factor beta superfamily during murine fracture healing. J Bone Miner Res. 2002;17:513–520. doi: 10.1359/jbmr.2002.17.3.513. [DOI] [PubMed] [Google Scholar]
- 37.Wang K, Vishwanath P, Eichler GS, Al-Sebaei MO, Edgar CM, Einhorn TA, Smith TF, Gerstenfeld LC. Analysis of fracture healing by large-scale transcriptional profile identified temporal relationships between metalloproteinase and ADAMTS mRNA expression. Matrix Biol. 2006;25:271–281. doi: 10.1016/j.matbio.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Nakashima K, de Crombrugghe B. Transcriptional mechanisms in osteoblast differentiation and bone formation. Trends Genet. 2003;19:458–466. doi: 10.1016/S0168-9525(03)00176-8. [DOI] [PubMed] [Google Scholar]
- 39.Jepsen KJ, Hu B, Tommasini SM, Courtland HW, Price C, Terranova CJ, Nadeau JH. Genetic randomization reveals functional relationships among morphologic and tissue-quality traits that contribute to bone strength and fragility. Mamm Genome. 2007;18:492–507. doi: 10.1007/s00335-007-9017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips AM. Overview of the fracture healing cascade. Injury. 2005;36S:55–57. doi: 10.1016/j.injury.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 41.Nork SE. Fractures of the shaft of the femur. In: Bucholz RW, Heckman JD, Court-Brown C, editors. Rockwood and Green's Fractures in Adults. 6th ed. Philadelphia, PA, USA: Lippincott Williams and Wilkins; 2006. pp. 1845–1914. [Google Scholar]
- 42.Linkhart TA, Linkhart SG, Kodama Y, Farley JR, Dimai HP, Wright KR, Wergedal JE, Sheng M, Beamer WG, Donahue LR, Rosen CJ, Baylink DJ. Osteoclast formation in bone marrow cultures from two inbred strains of mice with different bone densities. J Bone Miner Res. 1999;14:39–46. doi: 10.1359/jbmr.1999.14.1.39. [DOI] [PubMed] [Google Scholar]
- 43.Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, Einhorn T, Tabin CJ, Rosen V. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006;38:1424–1429. doi: 10.1038/ng1916. [DOI] [PubMed] [Google Scholar]
- 44.Xiong DH, Shen H, Zhao LJ, Xiao P, Yang TL, Guo Y, Wang W, Guo YF, Liu YJ, Recker RR, Deng HW. Robust and comprehensive analysis of 20 osteoporosis candidate genes by very high-density single-nucleotide polymorphism screen among 405 white nuclear families identified significant association and gene-gene interaction. J Bone Miner Res. 2006;21:1678–1695. doi: 10.1359/JBMR.060808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breur GJ, Van Enkevort BA, Farnum CE, Wilsman NJ. Linear relationship between the volume of hypertrophic chondrocytes and the rate of longitudinal bone growth in growth plates. J Orthop Res. 1991;9:348–359. doi: 10.1002/jor.1100090306. [DOI] [PubMed] [Google Scholar]