Abstract
Background
Distal interactions between discrete elements of an enzyme are critical for communication and ultimately for regulation. However, identifying the components of such interactions has remained elusive due to the delicate nature of these contacts. Protein kinases are a prime example of an enzyme with multiple regulatory sites that are spatially separate, yet communicate extensively for tight regulation of activity. Kinase misregulation has been directly linked to a variety of cancers, underscoring the necessity for understanding intramolecular kinase regulation.
Methodology/Principal Findings
A genetic screen was developed to identify suppressor mutations that restored catalytic activity in vivo from two kinase-dead Protein Kinase A mutants in S. cerevisiae. The residues defined by the suppressors provide new insights into kinase regulation. Many of the acquired mutations were distal to the nucleotide binding pocket, highlighting the relationship of spatially dispersed residues in regulation.
Conclusions/Significance
The suppressor residues provide new insights into kinase regulation, including allosteric effects on catalytic elements and altered protein-protein interactions. The suppressor mutations identified in this study also share overlap with mutations identified from an identical screen in the yeast PKA homolog Tpk2, demonstrating functional conservation for some residues. Some mutations were independently isolated several times at the same sites. These sites are in agreement with sites previously identified from multiple cancer data sets as areas where acquired somatic mutations led to cancer progression and drug resistance. This method provides a valuable tool for identifying residues involved in kinase activity and for studying kinase misregulation in disease states.
Introduction
Protein kinases comprise one of the largest gene families and are responsible for regulating cellular processes throughout all stages of growth and development, including gene transcription [1], metabolism [2], cell cycle regulation [3] and apoptosis [4]. Critical regulatory roles performed by kinases are underscored by the occurrence of numerous cancer phenotypes when kinase activity is altered or misregulated [5].
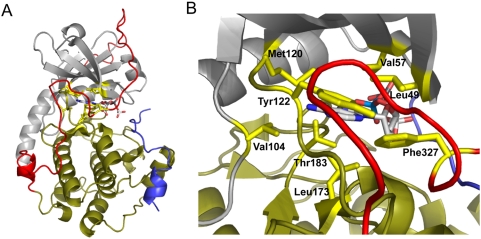
The kinase superfamily shares a conserved core that contains several invariant amino acids that contribute to activity [6]. Of the kinase superfamily, Protein Kinase A (PKA) is perhaps the best characterized. The tetrameric holoenzyme of PKA is composed of two catalytic subunits that possess kinase activity, and two regulatory subunits that are responsive to cAMP [7], [8]. The kinase domain is composed of a small lobe and a large lobe. The active site cleft resides between the lobes and is lined with multiple conserved residues that directly interact with the nucleotide upon binding (Figure 1). The ATP binding pocket is largely preformed but undergoes small structural rearrangements during nucleotide binding, whereas the active site cleft is dynamic and undergoes large structural changes to allow for nucleotide binding and catalysis [9]. In order to achieve tightly regulated catalytic activity, distal regions of the kinase must allosterically communicate with one another in order to synchronize structural changes [10]–[12].
Figure 1. Residues highlighted to interact with the nucleotide base in PKA.
When PKA is in an ATP-bound state, the small lobe (gray) and glycine-rich loop move toward the large lobe (sand) to enclose the nucleotide-binding pocket around ATP. A, A global view of PKA in a nucleotide-bound state. Eight residues have been identified to interact with the nucleotide base: Leu 49, Val 57, Val 104, Met 120, Tyr 122, Leu 173, Thr 183, and Phe 327 (highlighted yellow). The bound nucleotide is represented as a ball-and-stick figure. The C-terminal tail is colored red, and PKI is colored magenta. B, Zoomed view into the nucleotide binding pocket. The nucleotide binding pocket is largely preformed with the exception of Val 57, Met 120, Leu 173, and Phe 327 which undergo structural changes upon nucleotide binding. Structure adapted from PDB accession number 1ATP and modeled using Pymol.
Identifying distal components involved in kinase activity has remained elusive as a consequence of small structural changes and weak residue interactions. In order to expose these modulators throughout conserved and non-conserved elements of a kinase, a structurally non-biased approach was utilized. A genetic screen was developed to identify suppressor mutations that could restore catalytic activity from a kinase-dead mutant of PKA. Mutations were introduced to generate suppressor libraries, and libraries were screened in an S. cerevisiae strain devoid of native PKA expression. Since PKA activity is essential for survival, a viability screen was used to identify mutations that could suppress a catalytically inactive mutant. Screening revealed previously unidentified residues that are critical for PKA activity. In this report, we identify suppressor mutations that were isolated from two discrete genetic backgrounds. The mutations identified in this study were previously unknown modulators of kinase activity and point to multiple modes for achieving suppression. This genetic screen may be applied as a tool to investigate the relationship between kinase disregulation and various disease states.
Results
The prkaca-L173A and prkaca-F327A alleles cannot support essential PKA function
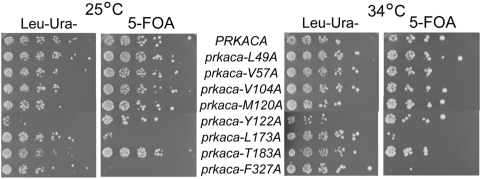
As a basis for the genetic screen, we sought to identify nucleotide binding pocket mutations that would render the kinase catalytically inactive in vivo. Several key residues within the nucleotide binding pocket that interact with the nucleotide base were mutated in the murine isoform of PKA (L49A, V57A, L104A, M120A, Y122A, L173A, T183A and F327A) [9]. The mutants were characterized in vivo using the model organism S. cerevisiae as it affords a genetic background that can be readily manipulated. Since PKA activity is essential for survival in yeast [13], a synthetically lethal strain was created by deleting all three genes encoding PKA (TPK1, TPK2, and TPK3). Plasmid shuffling revealed that viability could be fully maintained in this strain at both permissive and elevated temperatures by expressing the murine isoform of PKA (Figure 2) as previously demonstrated [14]. In order to identify single mutations that could render PKA inactive, each site was individually mutated and transformed in the tpk null strain to evaluate kinase function. The tpk null strain was initially maintained by expression of wild-type TPK1 from a URA3-marked plasmid since it has the highest degree of sequence identity to the murine PKA homolog. Wild-type and mutant PKA proteins were expressed using the endogenous TPK1 promoter in LEU2-marked plasmids. Each PKA mutant was subsequently tested using standard “plasmid shuffle” assays to determine whether essential PKA function was present in the absence of TPK1 expression [15].
Figure 2. prkaca-L173A and prkaca-F327A cannot support viability in a tpk null strain.
Point mutations were introduced at the nucleotide binding sites indicated in Figure 1B . Coexpression of wild-type and mutant alleles of PKA is demonstrated on Leu-Ura- selection plates. Expression of the mutants alone is shown on 5-FOA plates. All mutants tested, except prkaca-L173A and prkaca-F327A, were viable at permissive and elevated temperatures.
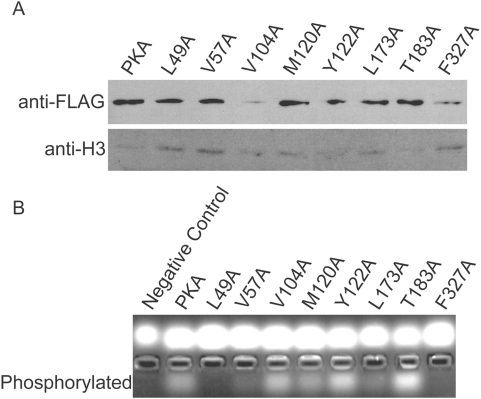
Growth resulting from co-expression of wild-type and mutant plasmids (Leu- Ura-) was compared to that of the mutant plasmid alone (5-fluoroorotic acid, 5-FOA) (Figure 2). When co-expressed with wild-type Tpk1, most PKA mutants grew comparably to the wild-type at both permissive (25°C) and elevated temperatures (34°C). However, a significant growth defect was observed for two mutants: prkaca-L173A and prkaca-F327A. At both temperatures, the prkaca-F327A mutant was completely non-viable and had very low steady-state protein expression (Figure 3A). The prkaca-L173A mutant demonstrated very weak growth only at the highest concentration. Both mutants demonstrated significantly reduced catalytic activity in vitro (Figure 3B). Previous crystallographic analysis reveals that of the eight residues tested, four residues retain their structural positions (L49, L104, Y122 and T183) whereas the other four (V57, M120, L173, and F327) undergo structural changes during nucleotide binding [16]. The dramatic inability of the prkaca-L173A and prkaca-F327A mutants to maintain viability in the tpk null strain may indicate that residues that undergo structural rearrangement may play a more significant role in regulating nucleotide binding for PKA activity.
Figure 3. The L173A mutant protein is stably expressed but inactive, whereas the F327A mutant has lowered protein expression.
A, Protein expression of each PKA mutant were measured. Of the two mutants that could not complement synthetic lethality in the tpk null strain, F327A also demonstrated very low protein expression. B, The catalytic activity of each PKA mutant was determined. Both L173A and F327A mutants had nearly undetectable catalytic activity.
Intragenic suppression restores catalytic activity to prkaca mutants
Using the nonviable prkaca site-directed mutants identified above, a genetic approach was employed to identify suppressor mutations that could overcome the non-viable phenotype. The prkaca-L173A and prkaca-F327A mutants were both selected for suppressor screening since they provide two unique backgrounds: L173A lies within the conserved kinase core while F327A lies within the non-conserved C-terminal tail. Plasmid libraries were constructed after random mutagenesis of the prkaca-L173A and prkaca-F327A mutant alleles. These were transformed and screened for restoration of viability to the tpk null strain using standard plasmid shuffling. Mutation rates of libraries varied from approximately 1–20 mutations per 1000 base pairs. Approximately 25,000 transformants were screened for each allele, and suppressors were sequenced to identify the acquired mutations. The suppressors were isolated from each background separately.
Suppressor mutations were screened based upon their ability to restore viability to the tpk null strain and were more readily identified from the prkaca-F327A background than from the prkaca-L173A background (Tables 1 and 2). Additionally, suppressors from the pkraca-L173A background required at least two additional amino acid changes in order to suppress the L173A mutation. Due to the variance noted between the two discrete genetic backgrounds during screening, it appears that the modes of suppression are dependent upon the effects caused by the initial mutation.
Table 1. Suppressor mutations identified from the prkaca-L173A background.
| Suppressor | Changes in Suppressors |
| Suppressor 27 | T153S, L160N, S212N |
| Suppressor 28 | T153S, L160N, S212N |
| Suppressor 31 | Q181K, T278S |
| Suppressor 51 | M58V, R190C |
| Suppressor 52 | M58V, R190C |
| Suppressor 53 | M58V, R190C |
| Suppressor 54 | M58V, R190C |
| Suppressor 55 | Wild-type reversion |
| Suppressor 57 | M58V, R190C |
| Suppressor 58 | K28N, I305L, E311V, I315L |
| Suppressor 59 | K28N, I305L, E311V |
| Suppressor 60 | K28N, E311V |
| Suppressor 61 | K21Q, K28N, R308K, E311V |
| Suppressor 62 | Wild-type reversion |
| Suppressor 63 | R190C, R194H, K292Q, I303V |
Table 2. Suppressor mutations identified from the prkaca-F327A background.
| Suppressor | Changes in Suppressors |
| Suppressor 1 | Y229S |
| Suppressor 3 | G186C |
| Suppressor 4 | T51P |
| Suppressor 5 | F327S, L173V |
| Suppressor 7 | E17R |
| Suppressor 12 | M120T, L40S |
| Suppressor 14 | K285P |
| Suppressor 32 | V310A |
| Suppressor 38 | H62L, Q149H |
| Suppressor 39 | T37K, D75Q, Q149H, K189N, R190H, V226A, Q242H |
| Suppressor 40 | Q35L, A38T, E140Y, Q149H, E170D, G178V, I180L, R190H, C199S, G214K |
| Suppressor 41 | K28E, L40S, R93S |
| Suppressor 42 | Q35L, K295E, I305L |
| Suppressor 43 | L40S, K189T, T299P, Q307H, F318I |
| Suppressor 44 | L40S, K47T, Y164H |
| Suppressor 46 | L40S, K63N, L95M |
| Suppressor 47 | M71I, V251I, F257I |
| Suppressor 48 | L40S, R308K, I315L |
| Suppressor 49 | V251F |
| Suppressor 66 | F108I, Y215C |
| Suppressor 67 | L40S, L157M |
| Suppressor 68 | L40P |
| Suppressor 69 | L40S |
| Suppressor 70 | T37I, L40P, G282A |
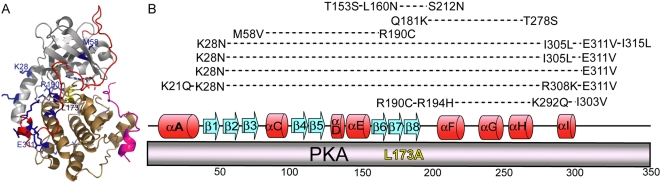
Suppression of the prkaca-L173A mutant
The prkaca-L173A suppressors were first investigated (Figure 4). The original L173A mutation is buried within the nucleotide binding pocket of PKA and directly interacts with the nucleotide base. Notably, some suppressors were independently isolated several times. Of these, the most commonly isolated suppressor was one that had acquired mutations at sites M58V and R190C. Both residue sites are conserved among the PKA family. Met58 is positioned within the β-sheets of the small lobe and is partially buried between the small lobe and the C-terminal tail in the nucleotide-bound state. Arg190 is also partially buried and points away from the nucleotide binding pocket near the hinge region flanking the magnesium-positioning DFG loop. Both residues are members of an intramolecular interaction network that provides crosstalk between the hydrophobic lining and the gly-rich and DFG loops, respectively, of AGC kinases [10]. The high frequency of isolating this suppressor pair may be due to the simultaneous effects on multiple known regulatory components of PKA.
Figure 4. Suppressors identified from the prkaca-L173A background.
A, The original L173A mutation is highlighted yellow and the suppressor mutations are shown in blue. PKA features are highlighted as follows: small lobe, gray; large lobe, sand; C-terminal tail,red; PKI: magenta. Some frequently isolated suppressor mutations are labeled. Mutations are shown using PDB file 1ATP and modeled using PyMol. B, Multiple mutations isolated from the same suppressor are connected by a dashed line and generally aligned with secondary structural elements of PKA. Secondary structural elements are represented where alpha-helices are red and beta-sheets are blue.
Another commonly isolated suppressor from the prkaca-L173A background acquired a pair of mutations: K28N and E311V. Interestingly, both residues reside outside of the conserved kinase core. Lys28 is a solvent-exposed residue in the non-conserved A-helix of the small lobe. The A-helix regulates kinase activity by acting as a communication bridge between the small and large lobes, and also partially anchors the C-helix for nucleotide positioning. Glu311 is positioned in the C-terminal tail and acts to help anchor the tail to the large lobe as it undergoes structural rearrangement upon nucleotide binding to act as a regulator for access to the active site. Kinase activity may have been restored to the prkaca-L173A background by coupling effects stemming from non-conserved elements to alter the activity of the conserved kinase core.
Suppression of the prkaca-F327A mutant
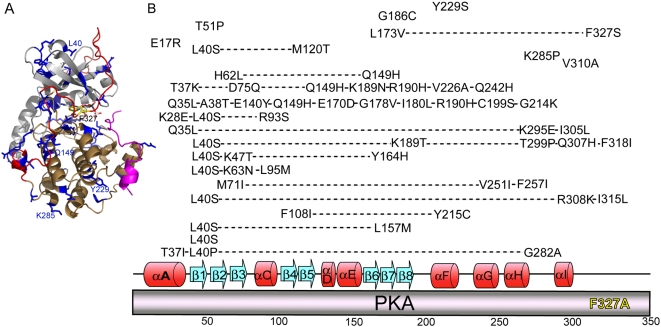
In order to determine how suppression could be achieved from a key catalytic residue outside of the conserved kinase core, suppression of the prkaca-F327A mutant was subsequently examined. The prkaca-F327A background demonstrates the significance of a non-conserved element on kinase activity. The F327A mutation may prevent the C-terminal tail from fully undergoing the structural transition after a nucleotide is bound, deeming it both inactive and structurally unstable (Figure 3). Viable suppressors were isolated from the prkaca-F327A background (Table 2 and Figure 5). Of note, suppressors were isolated at a much higher rate as compared to the prkaca-L173A background and could be isolated from libraries with low mutation rates.
Figure 5. Suppressor mutations from the prkaca-F327A background.
A, Highlighted suppressor mutations identified from the prkaca-F327A background. The original F327A mutation is highlighted in yellow, and suppressor mutations are shown in blue. PKA features are highlighted as follows: small lobe, gray; large lobe, sand; C-terminal tail,red; PKI: magenta. Some frequently highlighted mutations are labeled. The image was made using PDB file 1ATP and modeled using PyMol. B, Mutations isolated from the same suppressor are connected by a dashed line. Single suppressor mutations are listed above the corresponding secondary structural elements of PKA. Secondary structural elements are represented where alpha-helices are red and beta-sheets are blue.
As seen in the prkaca-L173A background, some suppressors were identified multiple times. The most frequently isolated mutation occurred at Leu40. Leu40 is part of the conserved kinase core and is the first residue of β-strand 1 in the small lobe. This conserved residue also belongs to an interaction network connecting the hydrophobic lining of AGC kinases with the glycine-rich loop [10]. The acquired L40S/P mutation likely augments propagated signaling from the hydrophobic lining of the catalytic domain to the glycine-rich loop.
Another frequently isolated suppressor was Q149H. This site is located on the E-helix of the large lobe and interacts with the proximal end of the C-terminal tail. In a nucleotide-bound state, Gln149 becomes completely buried by the C-terminal tail. The Q149H suppressor may promote packing interactions within the large lobe to support anchoring of the C-terminal tail as it undergoes the structural transition during nucleotide binding. Additionally, Q149 is involved in allosteric regulation of the DFG loop during transitions between active and inactive states [12]. The compensatory Q149H mutation may ease the allosteric regulation to allow for suppression of the original F327A mutation.
Discussion
Many of the acquired suppressor mutations would not have necessarily been predicted to significantly impact kinase activity. As such, it was not obvious in many cases how suppression was achieved. Of note, many suppressor mutations are structurally distal to the original L173A mutation. All of the prkaca-L173A suppressors acquired at least two additional mutations, perhaps indicating that overcoming a momentous deficiency stemming from loss of a key catalytic residue within the kinase core required multiple changes throughout the kinase.
In contrast to suppressors identified from the prkaca-L173A background, it is interesting to note that many of the acquired mutations from the prkaca-F327A background are at sites that are solvent-exposed. It is possible that these mutations affect substrate or other protein-protein interactions, leading to enhanced or altered affinities for inhibitor interactions or protein complex formation to ultimately suppress the original prkaca-F327A mutation. Additionally, multiple prkaca-F327A suppressors acquired a proline mutation. Proline inherently decreases localized mobility, thereby inhibiting conformational malleability. In this instance, buried Pro mutations may disallow conformational changes to impact allosteric regulation for catalysis, but surface Pro mutations may alter interactions involving substrate binding or protein-protein interactions.
A noteworthy example of a proline suppressor mutation was identified in the F327A/K285P suppressor. Further structural and biochemical characterization was performed on this suppressor to elucidate the mechanism of suppression [17]. It was found that F327 and the dynamic part of the C-tail flanking this site play an essential role in regulating kinase activity by participating in ATP binding, coordinating the positioning of the Glycine-rich Loop for catalysis and by stabilizing the linker joining the two lobes of the kinase core. Thus, the F327A mutation caused a loss of catalytic function by distorting the abovementioned elements. However, biochemical studies revealed that the double mutant remained catalytically defective, yet kinase function was restored in vivo. Suppression was achieved by reducing the inhibitory interactions with either the RII-subunits or the yeast homolog Bcy1p. Even though the mutations are distal to one another, crystallographic studies revealed that K285 provides a key contact point for interactions with the regulatory subunits. The side chain of Lys285, located in the αH-αI Loop of the large lobe, is typically solvent exposed and often disordered in crystal structures where the C-subunit is not bound to a regulatory subunit. Therefore, its functional importance was not previously appreciated. The suppressor mutation prevented the regulatory subunit from binding to the kinase domain, thereby allowing the kinase to remain constitutively active. Further, the single mutants K285P or F327A had a modest effect on inhibition by RIIα, however, a synergistic negative effect was observed for the double mutant. This suggests an allosteric connection of the αH-αI Loop to the active site. Detailed analysis of this suppressor provides an example of how distal sites of PKA are functionally integrated in order to regulate PKA activity.
Functional differences between PKA and Tpk2
In this study, several key residues within the nucleotide-binding site of PKA were tested in vivo for their necessity in kinase function in an S. cerevisiae strain devoid of native PKA expression. Despite site conservation among the kinase superfamily, only two sites, L173 and F327, demonstrated a loss of viability when mutated to Ala. The biological consequences of these mutations were previously tested in vivo using the PKA homolog Tpk2 [18]. Interestingly, although viability was lost in the tpk2-T183V mutant, absolutely no effects were seen for the same mutant in PKA. Likewise, prkaca-L173A and prkaca-F327A caused loss of viability in the tpk null strain, but no consequences were evident for the corresponding Tpk2 mutants. This demonstrates that although the nucleotide binding pocket is conserved among the kinase superfamily, their functional roles may not be conserved.
Some identical suppressor mutations were identified from screening against both of the prkaca backgrounds as well as the yeast homolog tpk2-T183V [18]. One example of this is the acquisition of mutations at site R190. This residue lies within the conserved kinase core and forms a socket with R93 at the end of the C-helix. This socket interacts with W30 and F26 of the A-helix to anchor and position the C-helix for catalytic activity. This socket also provides support to the A-helix to maintain conformational rigidity between the large and small lobes. Identification of acquired mutations at R190 from two distinct genetic backgrounds of PKA as well as the yeast homolog Tpk2 underscores the significance of the interactions between the A- and C-helices on kinase function.
There is a clear link between cancer progression and a variety of conserved, somatic mutations that accumulate over time in kinases. A recent study analyzing multiple cancer data sets has identified four residues within the catalytic core of multiple kinases that play a dominant role in tumor progression [19]. In PKA, these positions are L49, R190, M120 and G126. Our genetic screening against three unique backgrounds using two PKA homologs yielded acquisition of suppressor mutations at positions R190 and M120 for several suppressors. This not only validates the genetic screen as a method for identifying residues that modulate catalytic activity, but also reconfirms the significance of these residues in drug resistance and cancer.
Genetic screening as a tool for studying kinase regulation
Although crystal structures have shed tremendous light on catalytic components of kinases, the more subtle regulatory interactions distributed throughout the kinase have been difficult to distinguish. These results suggest that viability-based genetic screening can identify suppressors that indirectly reverse the functional defectiveness derived from the original mutation by compensatory effects distributed throughout the kinase. Due to the significance of kinases in a variety of diseases, understanding kinase regulation is critical. This method provides a platform for in vivo identification of previously unsuspected sites that significantly impact PKA regulation. The genetic screen presented in this study may act as a tool to broaden our understanding of kinase regulation and can be applied to study kinase misregulation in disease states.
Materials and Methods
Strains and Media
The yeast strain used in this study was derived from the Saccharomyces Genome Deletion Project [20]. The strain, LPY 06292 has the following genotype: MATα his3-1 leu2-0 lys2-0 met15-0 ura3-0 tpk1Δ::KANMX tpk2Δ::KANMX tpk3Δ::KANMX. To survive, it carries a CEN, URA3-marked plasmid bearing wild-type TPK1 (pLP2024). Where indicated, wild-type and PKA mutants were co-expressed in a CEN, LEU2-marked vector [21]. Growth was at 30°C unless otherwise specified and standard techniques for yeast manipulation were performed [15]. Cultures were grown in either yeast extract/ peptone/ dextrose medium (YPD), Leu- Ura- drop-out medium, or Leu- 5-FOA medium as indicated that was prepared as described [22].
Generation of PKA pocket mutations and screening for restored PKA function
Point mutations were introduced into sequences encoding the nucleotide-binding pocket of Mus musculus PKA-Cα (Gene symbol: Prkaca) using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) using the following forward primers and their reverse complements, respectively:
Leu49Ala:
5′-AGAATCAAGACCGCGGGCACCGGATCC,
5′-GGATCCGGTGCCCGCGGTCTTGAATTCT;
Val57Ala:
5′-TCCTTTGGCCGAGCGATGCTGGTGAAG,
5′-CTTCACCAGCATCGCTCGGCCAAAGGA;
Val104Ala:
5′-TTCCCGTTCCTGGCCAAACTTGAATTC,
5′-GAATTCAAGTTTGGCCAGGAACGGGAA;
Met120Ala:
5′-CTGTACATGGTGGCCGAGTATGTAGCT,
5′-AGCTATATACTCGGCCACCATGTACAG;
Tyr122Ala:
5′-ATGGTCATCGAGGCTGTGGCCGGTGGC,
5′-GCCACCGGCCACAGCCTCCATGACCAT;
Leu173Ala:
5′-CCCGAGAATCTTGCGATCGACCAGCAG,
5′-CTGCTGGTCGATCGCAAGATTCTCGGG;
Thr183Ala:
5′-TATATTCAGGTGGCCGACTTCGGTTTT,
5′-AAAACCGAAGTCGGCCACCTGAATATA;
Phe327Ala:
5′-GACACTAGTAACGCTGACGACTATGAG,
5′-CTCATAGTCGTCAGCGTTACTAGTGTC
Intragenic suppressor libraries of either the prkaca-L173 or prkaca-F327A mutants were generated as previously described [18]. Coding sequence templates of prkaca bearing the mutations encoding either L173A or F327A were used as target templates for PCR amplification in pRS315 plasmids bearing 2000 bp upstream and 1000 bp downstream flanking sequences from the closest PKA-Cα homolog in S. cerevisiae, TPK1.
Phenotypic analysis of strains with point mutations in PKA
Cultures were grown in Leu- Ura- medium. Cell densities were normalized to OD600 = 1.0 and were 5-fold serially diluted in sterilized water. Dilutions were plated onto selective medium and grown at the indicated temperatures for 6 days.
Protein expression, immunodetection, purification of FLAG-labeled PKA catalytic subunit, and catalytic kinase activity assays
Immunodetection, protein purification, and catalytic activity assays were performed as previously described [18].
Acknowledgments
The authors thank Natarajan Kannan, Olivier Lichtarge, and members of the Pillus lab for their helpful discussions and insights; Robert Levenson for assistance on this work; and Anne Ritter for proofreading of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work has been supported by the Minority Access to Science, Engineering and Mathematics Predoctoral Fellowship (National Science Foundation, 9978892), Cellular and Molecular Genetics Training Grant (National Institutes of Health, GM07240), and American Heart Association Predoctoral Fellowship (0315003Y) for E.J.K.; funding from the National Institutes of Health for the labs of L.P., G.G.(GM071862) and S.S.T.(GM19301); and funding from the Howard Hughes Medical Institute for S.S.T. The funders had no role in research design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Walsh DA, Perkins JP, Krebs EG. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem. 1968;243:3763–3765. [PubMed] [Google Scholar]
- 3.Hirai H, Kawanishi N, Iwasawa Y. Recent advances in the development of selective small molecule inhibitors for cyclin-dependent kinases. Curr Top Med Chem. 2005;5:167–179. doi: 10.2174/1568026053507688. [DOI] [PubMed] [Google Scholar]
- 4.Heyninck K, Beyaert R. A novel link between Lck, Bak expression and chemosensitivity. Oncogene. 2005 doi: 10.1038/sj.onc.1209157. [DOI] [PubMed] [Google Scholar]
- 5.Lengyel E, Sawada K, Salgia R. Tyrosine kinase mutations in human cancer. Curr Mol Med. 2007;7:77–84. doi: 10.2174/156652407779940486. [DOI] [PubMed] [Google Scholar]
- 6.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. Faseb J. 1995;9:576–596. [PubMed] [Google Scholar]
- 7.Knighton DR, Zheng JH, Ten Eyck LF, Xuong NH, Taylor SS, et al. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 8.Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIalpha) subunits of PKA. Science. 2005;307:690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 9.Taylor SS, Yang J, Wu J, Haste NM, Radzio-Andzelm E, et al. PKA: a portrait of protein kinase dynamics. Biochim Biophys Acta. 2004;1697:259–269. doi: 10.1016/j.bbapap.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 10.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc Natl Acad Sci U S A. 2008;105:506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Ten Eyck LF, Xuong NH, Taylor SS. Crystal structure of a cAMP-dependent protein kinase mutant at 1.26A: new insights into the catalytic mechanism. J Mol Biol. 2004;336:473–487. doi: 10.1016/j.jmb.2003.11.044. [DOI] [PubMed] [Google Scholar]
- 12.Ten Eyck LF, Taylor SS, Kornev AP. Conserved spatial patterns across the protein kinase family. Biochim Biophys Acta. 2008;1784:238–243. doi: 10.1016/j.bbapap.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 14.Zoller MJ, Yonemoto W, Taylor SS, Johnson KE. Mammalian cAMP-dependent protein kinase functionally replaces its homolog in yeast. Gene. 1991;99:171–179. doi: 10.1016/0378-1119(91)90124-t. [DOI] [PubMed] [Google Scholar]
- 15.Amberg DC, Burke DJ, Strathern JN. 2005. Methods in Yeast Genetics: A Cold Springs Harbor Laboratory Course Manual (Cold Springs Harbor Laboratory Press)
- 16.Akamine P, Madhusudan, Wu J, Xuong NH, Ten Eyck LF, et al. Dynamic features of cAMP-dependent protein kinase revealed by apoenzyme crystal structure. J Mol Biol. 2003;327:159–171. doi: 10.1016/s0022-2836(02)01446-8. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Kennedy EJ, Wu J, Deal MS, Brown S, et al. Contribution of Non-Catalytic Core Residues to Activity and Regulation in Protein Kinase A. J Biol Chem. 2009 Epub ahead of print. [Google Scholar]
- 18.Kennedy EJ, Pillus L, Ghosh G. Identification of functionally distinct regions that mediate biological activity of the Protein Kinase A homolog Tpk2. J Biol Chem. 2008;283:1084–1093. doi: 10.1074/jbc.M704028200. [DOI] [PubMed] [Google Scholar]
- 19.Torkamani A, Schork NJ. Prediction of cancer driver mutations in protein kinases. Cancer Res. 2008;68:1675–1682. doi: 10.1158/0008-5472.CAN-07-5283. [DOI] [PubMed] [Google Scholar]
- 20.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 21.Sikorski RS, Michaud WA, Tugendreich S, Hieter P. Allele shuffling: conjugational transfer, plasmid shuffling and suppressor analysis in Saccharomyces cerevisiae. Gene. 1995;155:51–59. doi: 10.1016/0378-1119(94)00915-f. [DOI] [PubMed] [Google Scholar]
- 22.Sherman F. Getting started with yeast. Methods in Enzymology. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]