Abstract
Recent transcriptome analyses have shown that thousands of noncoding RNAs (ncRNAs) are transcribed from mammalian genomes. Although the number of functionally annotated ncRNAs is still limited, they are known to be frequently retained in the nucleus, where they coordinate regulatory networks of gene expression. Some subnuclear organelles or nuclear bodies include RNA species whose identity and structural roles are largely unknown. We identified 2 abundant overlapping ncRNAs, MENε and MENβ (MENε/β), which are transcribed from the corresponding site in the multiple endocrine neoplasia (MEN) I locus and which localize to nuclear paraspeckles. This finding raises the intriguing possibility that MENε/β are involved in paraspeckle organization, because paraspeckles are, reportedly, RNase-sensitive structures. Successful removal of MENε/β by a refined knockdown method resulted in paraspeckle disintegration. Furthermore, the reassembly of paraspeckles disassembled by transcriptional arrest appeared to be unsuccessful in the absence of MENε/β. RNA interference and immunoprecipitation further revealed that the paraspeckle proteins p54/nrb and PSF selectively associate with and stabilize the longer MENβ, thereby contributing to the organization of the paraspeckle structure. The paraspeckle protein PSP1 is not directly involved in either MENε/β stabilization or paraspeckle organization. We postulate a model for nuclear paraspeckle body organization where specific ncRNAs and RNA-binding proteins cooperate to maintain and, presumably, establish the structure.
Keywords: nuclear bodies, RNA-binding proteins
Recent large-scale transcriptome analyses have revealed large numbers of transcripts that do not have protein-coding potential (1, 2). Many studies have suggested that a number of long noncoding RNAs (ncRNAs) are involved in the regulation of genome organization and/or gene expression in the nucleus. Despite the identification of a handful of functional ncRNAs, including Xist, SRA, Air, and HOTAIR (3–6), the exact functions of the recently identified polyadenylated ncRNAs remain in dispute.
The nucleus consists of many nuclear bodies in addition to nonrandomly arranged chromosomes (7–9). These nuclear bodies are membraneless suborganelles characterized by a distinct set of resident proteins, which provokes the question of how these compartments are assembled and maintained. There are 2 possibilities: First, an unidentified scaffold serves as an organizing center or second, the nuclear bodies are self-organized by transient interactions among their constituents. In addition to protein components, a number of RNA species reside in distinct nuclear structures, including the nucleolus (rRNA and snoRNA), the Cajal body (scaRNA and U-snRNA), and the nuclear stress bodies (satellite III RNAs) (10, 11). However, the structural role of the RNA molecule(s) in these nuclear subcompartments has not been fully investigated.
We hypothesized that some of the newly discovered ncRNAs may be involved in nuclear processes in the context of nuclear bodies, and sought to copurify such ncRNAs with nuclear bodies. The copurified ncRNAs were specifically disrupted in cultured cells by a knockdown method to investigate phenotypic alterations. Here, we describe the identification of MENε/β ncRNAs, which are indispensable for maintenance of the structural integrity of the nuclear body paraspeckle, which is an RNase-sensitive structure (12). We propose a model of paraspeckle organization where MENε/β ncRNA and the paraspeckle-localized RNA-binding proteins cooperate to establish the structure of this nuclear body.
Results
Characterization of a Paraspeckle-Localized Noncoding RNA.
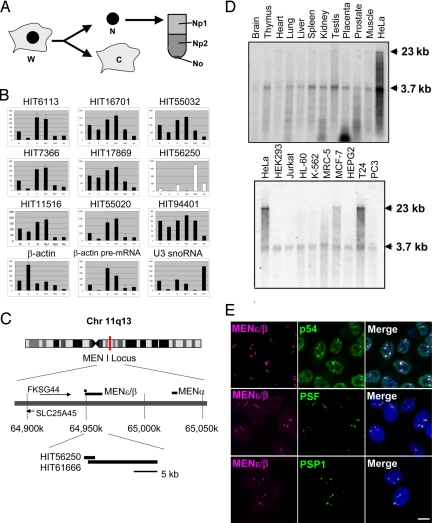
We first assessed the intracellular localization of 9 ncRNAs selected from a human cDNA database (13) (Fig. 1B). Isolated HeLa cell nuclei were fractionated by sucrose step-gradient centrifugation (Fig. 1A). Quantitative RT-PCR (qRT-PCR) of the fractionated ncRNAs revealed that the majority of these ncRNAs were predominantly localized in the low-density nucleoplasmic fraction (Np1, Fig. 1B), which contains various genetic machineries, including spliceosomes and chromosomes. The exception was HIT56250, which was enriched in the Np2 fraction from which only 2–3% of the total RNA was recovered (Fig. 1B). HIT56250 is a partial cDNA clone for a putative ncRNA, transcribed from the multiple endocrine neoplasia I (MEN I) locus on chromosome 11, and overlaps with a longer ncRNA transcript (Fig. 1C). We tentatively designated HIT56250 as MENε (14), rather than by its other synonyms, Tnc (15) or NEAT1 (16), to identify this ncRNA simply by its genomic locus. Northern blot analysis and an RNase protection assay identified the 2 major isoforms of this ncRNA, MENε (3.7 kb) and β (23 kb) [Fig. 1D and supporting information (SI) Fig. S1B]. Both isoforms (MENε/β) have no potential A-to-I editing site and were expressed ubiquitously, although their expression was up-regulated in a few cancer cell lines (Fig. 1D).
Fig. 1.
Identification of paraspeckle-associated noncoding RNAs. (A) Subnuclear fractionation. HeLa cells were first divided into cytoplasmic (C) and nuclear (N) fractions, and the nuclei were then subfractionated into Np1, Np2, and No subfractions by sucrose step-gradient centrifugation. (B) Intracellular localization of 9 representative ncRNAs. The ncRNA levels in the 6 fractions in A (from the left: W, C, N, Np1, Np2, and No) were quantified by qRT-PCR. W represents the total RNA from whole cells, and the RNA level in W was defined as 100%. β-Actin mRNA, β-actin premRNA, and U3 snoRNA are the control RNAs that localized to the cytoplasm (C), nucleoplasm (Np2), and nucleolus (No), respectively. (C) The chromosomal locus of MENε/β. (D) The expression of ncRNA in human tissues (Upper) and cell lines (Lower) by Northern blot hybridization. Arrowheads indicate MENε (3.7 kb) and MENβ (23 kb). (E) MENε/β colocalizes to paraspeckles. FISH was used to detect the nuclear localization of MENε/β (magenta); 3 paraspeckle proteins, PSF, PSP1, and p54 (green), were assayed by immunostaining. Cells were counterstained with DAPI. (Scale bar: 10 μm.)
RNA FISH revealed that the MENε/β signal was localized to discrete puncta, indicative of nuclear bodies (Fig. 1E), which were present in all cell lines examined (Fig. S2). Furthermore, the 2 isoforms colocalized in the same puncta (Fig. S1D). FISH followed by immunofluorescence (FISH-IF) with antibodies against various nuclear body markers demonstrated that the signals of 3 paraspeckle proteins, p54, PSF, and PSP1, overlapped with the MENε/β puncta (Fig. 1E). None of the other marker proteins tested showed any overlaps (Fig. S3). Thus, we confirmed the validity of our biochemical fractionation data and concluded that the MENε/β ncRNAs colocalize to the nuclear body paraspeckle.
Hutchinson et al. (16) recently reported that MENε (NEAT1 in their report) localizes to the periphery of speckle. We reexamined the localization of MENε/β with SC35, a speckle marker, and found that MENε/β did not colocalize to the speckle marker in any of the cell lines examined (Figs. S2B and S3Aq-t). In contrast, MENα, another ncRNA transcribed from the MEN locus, exhibited perfect colocalization with the speckle marker (Fig. S2A), but not with MENε/β (Fig. S3B). Furthermore, our biochemical fractionation data indicate that MENα was predominantly enriched in the Np1 fraction (HIT55020, Fig. 1B), whereas MENε/β were enriched in the Np2 fraction (HIT56250, Fig. 1B), consistent with our results indicating that MENε/β do not localize to nuclear speckles but to paraspeckles.
Specific Knockdown of MENε/β ncRNAs Disintegrates Paraspeckle Structure.
RNAi is reportedly less effective on nuclear RNAs than on cytoplasmic mRNAs, with a few exceptions. We therefore refined a knockdown method in which a phosphothioate-converted antisense chimeric oligonucleotide against an ncRNA is introduced into HeLa cell nuclei by nucleofection (see Materials and Methods). As shown in Fig. S4A, 9 nuclear ncRNAs (shown in Fig. 1B) were efficiently knocked down by their corresponding antisense oligonucleotides. The specificity of each antisense oligonucleotide was verified by comparing the knockdown efficiency of the targeted ncRNA to that of the untargeted ncRNA in each knockdown sample (Fig. S4B).
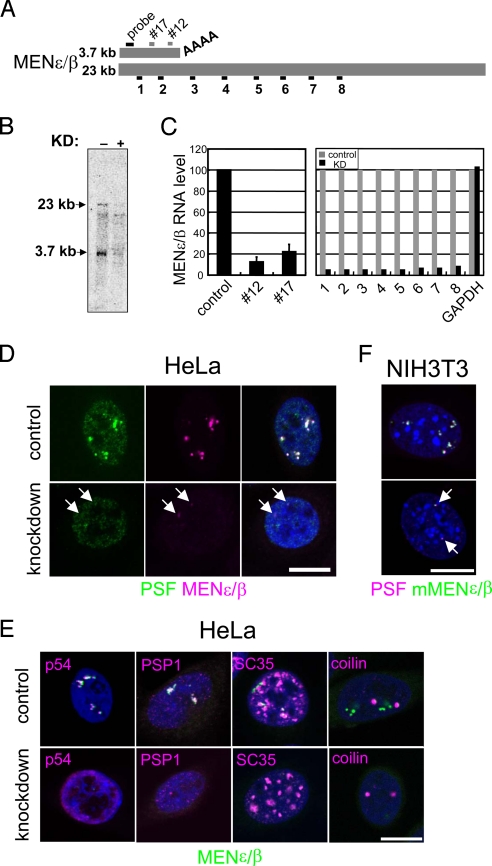
We investigated the function of MENε/β with our nuclear RNA knockdown method. Eight primer pairs for qRT-PCR were designed, along with HIT61666, at ≈2-kb intervals (Fig. 2A). Of the 6 antisense oligonucleotides, 2 (#12 and #17) successfully disrupted both MENε and β isoforms, with an efficiency of up to 80% (Fig. 2C). Alteration of the paraspeckle structure was observed upon MENε/β depletion in HeLa cells, with MENε/β signals diminishing (Fig. 2D and Fig. S5A) as the transcript levels decreased to ≈10% (Fig. 2C Right). Interestingly, the paraspeckle structure, defined by anti-PSF immunostaining, also disappeared, and PSF was localized diffusely throughout the nucleoplasm (Fig. 2D). The other paraspeckle markers, p54 and PSP1, behaved identically (Fig. 2E). MENε/β depletion did not affect the structure of the splicing speckle (SC35) or the Cajal body (coilin) (Fig. 2E), indicating the target specificity of the knockdown phenotype. We occasionally observed dwarfed paraspeckles that were always accompanied by residual MENε/β (Fig. 2D and Fig. S5B). Additionally, knockdown of the mouse MENε/β counterpart led to paraspeckle disintegration (Fig. 2F), indicating a common function for these ncRNAs in 2 different mammalian species.
Fig. 2.
Knockdown of MENε/β ncRNAs leads to disintegration of the paraspeckles. (A) The MENε/β isoforms are shown schematically. The sequences corresponding to the 2 antisense knockdown oligonucleotides (short gray bars) and a Northern blot probe (a black bar) are indicated. Positions of the fragments (1–8) amplified by qRT-PCR are shown below. (B) Northern blot hybridization clearly shows the loss of both isoforms upon knockdown (KD: lane +). (C) MENε/β levels in control and knockdown cells were quantified by qRT-PCR. (Left) Two antisense oligonucleotides, #12 and #17, effectively knocked down MENε/β. Values represent means ± SD. (Right) MENε/β levels in control (with GFP, gray bars) and knocked down (with #12, black bars) cells were quantified with 8 primer pairs and a GAPDH control primer. (D–F) MENε/β were knocked down in HeLa (D and E) and NIH 3T3 (F) cells. Cells were treated with a control (Upper) or with the #12 oligonucleotide (Lower). The signal identities are shown below each image. Arrows indicate remnant paraspeckles (D and F). (Scale bars: 10 μm.)
Paraspeckle disintegration caused by MENε/β depletion could be due either to degradation of the paraspeckle proteins or to alteration of the interactions among these proteins. To differentiate between these 2 possibilities, we compared the amount of paraspeckle proteins in the control and the knocked-down cells. Throughout the experiments, successful knockdown was sustained, as judged by the disappearance of both MENε/β and PSF signals in the knocked-down cells (Fig. S5B). In contrast to the change in MENε/β levels (Fig. S5C), no significant changes in the protein levels of 3-paraspeckle markers were observed (Fig. S5D). Thus, the disintegration of paraspeckles upon MENε/β knockdown was not due to degradation of the resident proteins.
Nuclear bodies are highly dynamic structures, because their resident proteins relocate depending on the physiological status of the cell (8, 9). MENε/β knockdown facilitated the relocation of paraspeckle proteins, turning the distinct puncta of the paraspeckles into “microspeckles.” To examine the localization of the paraspeckle proteins, we used a stable HeLa cell line expressing a fluorescent PSP1 fusion protein (PSP1-Venus). The localization of this reporter protein was faithful to that of the endogenous protein (Fig. S5E). Upon MENε/β knockdown, the PSP1-Venus protein relocated to a previously unrecognized nucleoplasmic space, exclusive of the speckle proteins (Fig. S5F).
Distinct Relocation of MENε/β ncRNAs and Paraspeckle Proteins upon Actinomycin D Treatment.
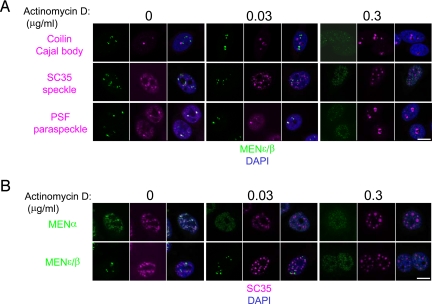
Upon transcriptional inhibition, the paraspeckle proteins relocate to the perinucleolar region and form distinct cap structures (17). We examined whether the MENε/β ncRNAs concomitantly relocate to the perinucleolar cap with the paraspeckle proteins or dissociate from the paraspeckle proteins upon transcriptional inhibition. We also monitored the relocation of coilin and SC35, both of which are known to be sensitive to transcriptional inhibition (18, 19). We determined the minimum dose of the transcription inhibitor actinomycin D required to relocate nuclear body marker proteins. A low dose of actinomycin D (0.03 μg/mL, 4 h) was sufficient to cause the Cajal body marker coilin to form a perinucleolar cap structure. SC35 was also affected at the same dose; speckles increased in size and rounded up, and the number of subspeckles decreased (Fig. 3A). In contrast, the low dose of actinomycin D had little effect on PSF with respect to perinucleolar cap formation: MENε/β ncRNAs and PSF remained in paraspeckles (Fig. 3A). With a high dose of actinomycin D (0.3 μg/mL, 4 h), PSF formed a marked cap structure, whereas MENε/β were diffusely distributed throughout the nucleoplasm, resulting in the disappearance of paraspeckles (Fig. 3A). Thus, MENε/β dissociated from the paraspeckle when the paraspeckle proteins relocated to the perinucleolar region. The paraspeckles were less susceptible to actinomycin D than the Cajal body or speckles.
Fig. 3.
MENε/β dissociated from the paraspeckles upon transcriptional inhibition. (A) HeLa cells were treated with actinomycin D at the concentrations indicated for 4 h. The relocation of marker proteins for each nuclear body was examined by using the MENε/β probe together with the appropriate antibodies [anti-coilin for the Cajal body (Top); anti-SC35 for speckles (Middle); anti-PSF for paraspeckles (Bottom)]. RNA signals (green) were enhanced to view the scattered microspeckles. (B) Actinomycin D treatment and FISH-IF were performed as in A. The MENα probe was used (Upper) for comparison with MENε/β (Lower). (Scale bars: 10 μm.)
When similar experiments were conducted with different combinations of RNA probes and antibodies, we observed the redistribution of the MENα ncRNA coincident with the relocation of SC35 at low doses of actinomycin D (Fig. 3B). The redistribution of the ncRNAs coincided with the relocation of their corresponding nuclear bodies. Actinomycin D treatment reduced the MENε/β level to ≈60% of control levels (Fig. S6). This reduction in MENε/β level may be due to transcriptional inhibition and dissociation of MENε/β from the paraspeckle proteins, the latter event causing destabilization of the MENε/β ncRNAs. Taken together, these results allow us to conclude that MENε/β ncRNAs are indispensable to the maintenance of paraspeckle integrity.
Paraspeckle Reassembly Requires MENε/β ncRNAs.
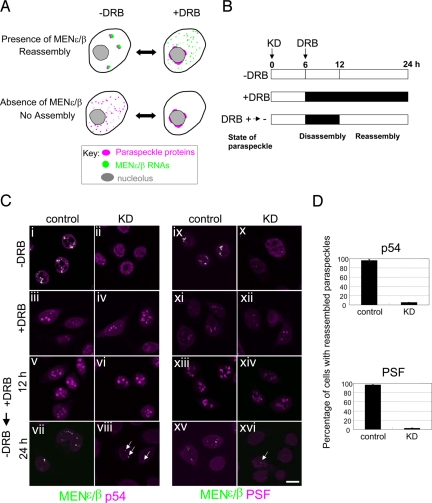
To clarify the role of MENε/β in paraspeckle formation, we took advantage of the reversibility of cap formation by treatment with 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (12). The paraspeckle is first disassembled by DRB and then reassembled by removing DRB. If MENε/β ncRNAs are essential to the reassembly step, then the paraspeckle would not reassemble in cells depleted of these ncRNAs (Fig. 4A). Six hours after the start of MENε/β knockdown, DRB was administered to cell cultures for 18 h (Fig. 4B). Perinucleolar cap formation by DRB was observed in both control and MENε/β-depleted cells (Fig. 4Ciii, iv, xi, and xii), consistent with a previous report that cap formation is independent of RNA (12), while no cap formation occurred in control preparations (Fig. 4Ci, ii, ix, and x). The MENε/β level was reduced to ≈20% by DRB and knockdown treatment (Fig. S6). After perinucleolar cap was formed by DRB (Fig. 4Cv, vi, xiii, and xiv), DRB was removed from the culture medium at 12 h, distinct paraspeckles reassembled only in MENε/β-expressing cells (Fig. 4Cvii and xv). In MENε/β-depleted cells, dwarfed paraspeckles with residual MENε/β were observed (arrows in Fig. 4Cviii and xvi). More than 95% of MENε/β-expressing cells and <6% of MENε/β-depleted cells reassembled paraspeckles (Fig. 4D). To examine whether MENε/β RNAs alone are sufficient to form paraspeckles, we ectopically expressed human MENε, as well as a 13-kb MENβ lacking the downstream 10 kb, the longest clone obtained thus far, in the NIH 3T3 cells. The ectopic transcripts localized to paraspeckles; however, the RNAs were incapable of reassembling paraspeckles when endogenous MENε/β RNAs were knocked down (Fig. S7). This observation suggests a functional difference between MENε and MENβ in mediating paraspeckle formation and integrity. RNA motif(s) in the missing 3′ region of MENβ may be crucial to paraspeckle formation through interactions with paraspeckle proteins. Taken together, our data strongly suggest that MENβ is involved in reorganization of the disassembled paraspeckle.
Fig. 4.
MENε/β ncRNAs are indispensable for paraspeckle reassembly. (A) Schematic diagram of how paraspeckle dynamics depend on MENε/β. (B) Experimental protocol for paraspeckle reassembly. The time course (0–24 h) is shown along the bar, with the black bar representing the duration of DRB administration. The time when MENε/β knockdown commenced (KD) was defined as 0 h. In the third protocol (+DRB → −DRB), DRB was administered (to disassemble the paraspeckle) from 6 to 12 h after KD. Removal of DRB enabled reassembly of the paraspeckle. (C) The behavior of MENε/β and paraspeckle marker proteins (p54 and PSF) were monitored during the processes shown in B. Arrows indicate the paraspeckle remnants. (Scale bar: 10 μm.) (D) Percentage of cells with reassembled paraspeckles. Values represent the means± SD (n = 365–497 cells) from 2 independent experiments.
MENβ ncRNA–Paraspeckle Protein Interactions Are Prerequisite for the Integrity of the Paraspeckle.
All 3 paraspeckle proteins have tandem RNA recognition motifs that are required for PSP1 and PSF to localize to the paraspeckle (12, 20), and p54 forms heterodimers with PSF and PSP1 (12, 21). Moreover, a drastic decrease in the MENε/β level coincides with their dissociation from paraspeckles (Fig. S6). Using a pair of siRNAs for each paraspeckle protein, we examined the influence of these RNAis on the MENε/β level. Depletion of either p54 or PSF decreased the longer MENβ by ≈20% (Fig. 5A, lanes 3–8), whereas the shorter MENε was unaffected (Fig. 5A, lanes 1 and 2). These data suggest that MENβ is stabilized by p54 and PSF. Notably, PSP1 depletion did not affect either isoform (Fig. 5A, PSP1), suggesting that PSP1 involvement in paraspeckle organization differs from p54 and PSF.
Fig. 5.
MENε/β ncRNAs and the paraspeckle marker proteins cooperate to organize the paraspeckle structure. (A) The MENε/β levels were quantified by qRT-PCR upon RNA interference (RNAi) of each paraspeckle protein. The relative abundance (100% in control cells treated with a control oligonucleotide) is shown in the graph. Two siRNAs were used for each paraspeckle protein. Numbers below the graphs correspond to the primer pairs for qPCR, as in Fig. 2A. Values represent means ± SD. *, P < 0.01. (B) The effect of RNAi on the paraspeckle structure was monitored by FISH-IF. Cells were treated with the siRNA indicated above each panel for 48 h and were then probed with MENε/β probe (green) in combination with an antibody to one of the three paraspeckle proteins (magenta: p54, PSF, or PSP1, top to bottom). Arrows point to the remnant MENε/β puncta with paraspeckle proteins. Open triangles point out imperfect MENε/β puncta lacking at least 1 paraspeckle protein. (Scale bar: 10 μm.) (C) Immunoprecipitation (IP) of MENε/β with antibodies against paraspeckle proteins. Flag-tagged p54 was used instead of endogenous p54 for p54-IP. Numbers below the graphs correspond to those in Fig. 2A. Values represent means ± SD. *, P < 0.01. (D) The paraspeckle protein p54 directly interacts with MENβ ncRNA. Intact HeLa cells transfected either with Flag-p54 or a control plasmid (FLAG-pcDNA) were irradiated with 254-nm UV light to induce cross-links between RNA and interacting proteins in vivo (+UV). Cell extracts were prepared under strong denaturing conditions and subsequently subjected to IP with the αFlag antibody. The relative IP efficiencies (the practical IP efficiency detected by primer #4 was defined as 100%) are shown, because IP efficiency in each of 3 independent experiments was variable (1.23%, 2.70%, and 11.5%). The large fluctuation in IP efficiency may be due to the efficiency of UV cross-linking. Values represent means ± SD. *, P < 0.05. (E) Possible interaction between MENε/β ncRNAs and 3 known paraspeckle proteins that underlie paraspeckle organization. The protein–protein interactions are depicted according to previous reports (12, 21).
Our FISH-IF data clearly differentiated the influence of PSP1 RNAi on paraspeckle structure from those of p54 or PSF RNAis. In control siRNA-treated cells, the paraspeckle structure appeared to be intact (Fig. 5Bi–iii). In p54 siRNA-treated cells, MENε/β and the other paraspeckle proteins were redistributed throughout the nucleoplasm, resulting in paraspeckle disintegration (Fig. 5Biv–vi). The PSF siRNA result mirrored that of p54 depletion (Fig. 5Bvii–ix). In contrast, PSP1 depletion had little influence on paraspeckle structure (Fig. 5Bx–xii). We verified PSP1 depletion by IF (Fig. 5Bxii) and by Western blot analysis (Fig. S8A). The RNAi results were also confirmed by FISH-IF using a different set of siRNAs (Fig. S8B). The above results imply a physical interaction between MENε/β and p54 as well as PSF. Quantification of MENε/β coimmunoprecipitated with p54 or PSF revealed that only MENβ formed a complex with PSF and Flag-tagged p54 (Fig. 5C). Furthermore, immunoprecipitation of in vivo UV cross-linked ribonucleoprotein complexes after isolation of the complexes under denaturing conditions revealed a direct interaction between Flag-p54 and MENβ (Fig. 5D). These observations support the hypothesis that paraspeckle integrity depends on MENβ RNA–paraspeckle protein (p54 and presumably PSF) interactions as well as secondary recruitment of MENε RNA and PSP1 (Fig. 5E).
Discussion
We have identified the ncRNA MENε/β as a potent organizer of paraspeckles. MENε/β may be identical to the previously predicted RNA constituent of paraspeckles (12). In experiments, paraspeckles lacking MENε/β were never observed, implying that this ncRNA plays a principal role in initiating paraspeckle assembly. Although the precise mechanism by which MENε/β RNAs and the paraspeckle proteins interact remains to be determined, we postulate that MENβ RNA–paraspeckle protein interactions are crucial to paraspeckle integrity (Fig. 5E).
Furthermore, the function of the paraspeckle also remains enigmatic. Recently, Prasanth et al. (22) proposed a role for the paraspeckle in the regulation of gene expression through the nuclear retention of CTN-RNA. The paraspeckle may also serve as a repository for proteins. Given the unique MENε/β localization, RNA knockdown offers many advantages over protein depletion for investigating this nuclear body. This advantage may not be confined to the paraspeckle but may be applicable to other nuclear bodies such as nuclear stress bodies (11). Further investigation of the MENε/β knockdown phenotype should reveal the physiological role of the paraspeckle.
To our knowledge, no vertebrate nuclear ncRNA has yet been proven to be an integral part of a nuclear subcompartment or an “architectural RNA” (Fig. 5E). Other than in mammals, there are a few RNAs known to be potentially involved in the organization of cellular architecture. In Xenopus, the mitotic spindle has been shown to be an RNase-sensitive ribonucleoprotein complex (23), and RNAs function in maintaining the integrity of the cytokeratin network (24). Moreover, in Drosophila, the nuclear ncRNA hsr-ω is involved in the formation of a subnuclear structure (25). Global analysis of mRNA localization during embryogenesis provides ample evidence for the structural role of mRNAs (26). It is therefore of great interest to determine how conserved RNA–protein interactions and the ability of RNAs to organize cellular architectures are during evolution.
Materials and Methods
Reagents and Cell Biological Protocols.
All chemicals used were purchased from Nacalai Tesque, unless otherwise stated. See SI Text and Tables S1–S5 for additional information.
Transfection of Antisense Oligonucleotides.
The antisense chimeric oligonucleotides (IDT) used for knockdown experiments were phosphothioate-converted at their backbone to increase their stability. Five terminal nucleotides from the 5′ and 3′ ends were substituted by 2′-O-methoxyribonucleotides. Trypsinized HeLa cells (1 × 106 cells) were suspended in 100 μL of Solution R of the Cell Line Nucleofector Kit R (Amaxa Biosystems) and then mixed with oligonucleotides (4 μM final concentration). Transfection was conducted in an electroporation cuvette by using the Nucleofector instrument (Amaxa Biosystems). The transfected cells were transferred to fresh DMEM plus 10% FBS, incubated at 37 °C and 5% CO2 for 24 h, and cells were harvested for RNA preparation. The chimeric oligonucleotides used are provided in Table S4.
Immunoprecipitation of Ribonucleoprotein Complex.
HeLa cell lysates were prepared as described previously (27). In brief, 1 × 107 cells were trypsinized and centrifuged at 1,000 × g for 3 min at 4 °C. The cells were washed in cold PBS and centrifuged, and the cell pellet was resuspended and incubated in 1 mL of buffer A [10 mM Pipes (pH 6.8), 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, and 0.2 mg/ml PMSF] for 5 min on ice. Pellet A, obtained by centrifugation of the solution at 1,000 × g for 5 min, was resuspended and incubated in 1 mL of buffer B [10 mM Pipes (pH 6.8), 250 mM ammonium sulfate, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.2 mg/ml PMSF] for 5 min at 4 °C. Supernatant B, obtained by centrifugation at 1,000 × g for 5 min, was precleared and used for IP using an αPSF antibody (Sigma), αFlag (M2 Sigma) for Flag-p54, or control IgG. For IP of Flag-p54, cells were transiently transfected with a Flag-p54 construct, and the lysate was prepared after an incubation of 48 h. Antibodies were incubated with protein-G Sepharose beads (Pierce) for 1 h, followed by washing 5 times in buffer B (27). Supernatant B (5%) was stored to prepare input RNAs. The remaining supernatants were mixed with antibody-bead conjugates and rotated for 3 h at 4 °C, and the beads were washed by using an automatic bead washer (Thermo).
In Vivo Cross-Linking.
For each interaction tested, 5 × 106 HeLa cells transfected either with Flag-p54 or the control plasmid were trypsinized and collected by centrifugation, washed twice with cold PBS, and resuspended in 400 μL of PBS in 6-well plates. Cells were irradiated (or not irradiated as a negative control) on ice with 254-nm UV light and collected in 1.5 mL of microfuge tubes. Cell pellets were resuspended with vortexing in 200 μL of lysis buffer [2% SDS, 50 mM Tris-Cl (pH 8), 1 mM EDTA, 1 mM DTT] and boiled at 95 °C for 5 min. After dilution with 4 volumes of collection buffer (28), each sample was gently sonicated 3 times and centrifuged for 90 min at 4 °C. The supernatants were directly subjected to immunoprecipitation as described above.
Supplementary Material
Acknowledgments.
We thank S. Nakagawa and S. Ishida for instruction in FISH analysis, Y. Kurihara (Yokohama National University) for antibodies, M. Kinjo and I. Nagao for use of the microscope facility, and K. Watanabe, T. Kawaguchi, A. Tanigawa, and members of the T.H. laboratory for valuable discussions. This work was supported by a grant from the New Energy and Industrial Technology Development Organization, a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and the Astellas Foundation for Research on Metabolic Disorders (to T.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807899106/DCSupplemental.
References
- 1.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 3.Heard E, Chaumeil J, Masui O, Okamoto I. Mammalian X-chromosome inactivation: an epigenetics paradigm. Cold Spring Harb Symp Quant Biol. 2004;69:89–102. doi: 10.1101/sqb.2004.69.89. [DOI] [PubMed] [Google Scholar]
- 4.Lanz RB, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 5.Braidotti G, et al. The Air noncoding RNA: An imprinted cis-silencing transcript. Cold Spring Harb Symp Quant Biol. 2004;69:55–66. doi: 10.1101/sqb.2004.69.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- 8.Misteli T. Concepts in nuclear architecture. BioEssays. 2005;27:477–487. doi: 10.1002/bies.20226. [DOI] [PubMed] [Google Scholar]
- 9.Misteli T. Protein dynamics: Implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 10.Matera AG, Tern RM, Tern MP. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 11.Biamonti G. Nuclear stress bodies: A heterochromatin affair? Nat Rev Mol Cell Biol. 2004;5:493–498. doi: 10.1038/nrm1405. [DOI] [PubMed] [Google Scholar]
- 12.Fox AH, Bond CS, Lamond AI. p54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki YTF, et al. Identification and characterization of human non-coding RNAs with tissue-specific expression. Biochem Biophys Res Commun. 2007;357:991–996. doi: 10.1016/j.bbrc.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 14.Guru SC, et al. A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res. 1997;7:725–735. doi: 10.1101/gr.7.7.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geirsson A, Lynch RJ, Paliwal I, Bothwell AL, Hammond GL. Human trophoblast noncoding RNA suppresses CIITA promoter III activity in murine B-lymphocytes. Biochem Biophys Res Commun. 2003;301:718–724. doi: 10.1016/s0006-291x(03)00028-7. [DOI] [PubMed] [Google Scholar]
- 16.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox AH, et al. Paraspeckles: A novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 18.Cioce M, Boulon S, Matera AG, Lamond AI. UV-induced fragmentation of Cajal bodies. J Cell Biol. 2006;175:401–413. doi: 10.1083/jcb.200604099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shav-Tal Y, et al. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dye BT, Patton JG. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp Cell Res. 2001;263:131–144. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- 21.Peng R, et al. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA. 2002;8:1334–1347. doi: 10.1017/s1355838202022070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Blower MD, Nachury M, Heald R, Weis KA. Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 24.Kloc M, et al. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- 25.Prasanth KV, Rajendra TK, Lai AK, Lakhotia SC. Omega speckles—A novel subclass of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci. 2000;113:3485–3497. doi: 10.1242/jcs.113.19.3485. [DOI] [PubMed] [Google Scholar]
- 26.Lecuyer E, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook HL, Mischo HE, Steitz JA. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host AU-rich element-containing mRNA levels in virally transformed T cells. Mol Cell Biol. 2004;24:4522–4533. doi: 10.1128/MCB.24.10.4522-4533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.