Abstract
The outer membranes of Gram-negative bacteria, mitochondria, and chloroplasts harbor β-barrel proteins. The signals that allow precursors of such proteins to be targeted to mitochondria were not characterized so far. To better understand the mechanism by which β-barrel precursor proteins are recognized and sorted within eukaryotic cells, we expressed the bacterial β-barrel proteins PhoE, OmpA, Omp85, and OmpC in Saccharomyces cerevisiae and demonstrated that they were imported into mitochondria. A detailed investigation of the import pathway of PhoE revealed that it is shared with mitochondrial β-barrel proteins. PhoE interacts initially with surface import receptors, and its further sorting depends on components of the TOB/SAM complex. The bacterial Omp85 and PhoE integrated into the mitochondrial outer membrane as native-like oligomers. For the latter protein this assembly depended on the C-terminal Phe residue, which is important also for the correct assembly of PhoE into the bacterial outer membrane. Collectively, it appears that mitochondrial β-barrel proteins have not evolved eukaryotic-specific signals to ensure their import into mitochondria. Furthermore, the signal for assembly of β-barrel proteins into the bacterial outer membrane is functional in mitochondria.
Keywords: outer membrane, PhoE, protein import, TOB complex, TOM complex
Mitochondria and chloroplasts contain β-barrel proteins in their outer membranes (1–3). The only other biological membrane known to harbor β-barrel proteins is the outer membrane of Gram-negative bacteria (4). This situation reflects the evolutionary origin of mitochondria and chloroplasts from endosymbionts that belong to the class of Gram-negative bacteria.
Precursors of mitochondrial β-barrel proteins are synthesized on cytosolic ribosomes and recognized initially by the import receptors of the translocase of the mitochondrial outer membrane (TOM complex). They are then translocated across the outer membrane via the general-import pore of the TOM complex (5–8). From the TOM complex, β-barrel precursors are relayed to a specialized hetero-oligomeric protein complex termed either topogenesis of outer-membrane β-barrel proteins (TOB complex) (9) or sorting and assembly machinery (SAM complex) (10).
The major component of the TOB complex is Tob55/Sam50 (9, 11, 12). Its sequence is similar to that of the highly conserved bacterial protein Omp85/YaeT, which mediates the insertion of β-barrel proteins into the bacterial outer membrane (13, 14). Hence, this function has apparently been conserved during evolution of mitochondria from bacteria. Although the mitochondrial machinery for insertion of β-barrel proteins was derived from the bacterial system, some modifications were necessary during evolution to meet the requirements of the organelle. It is believed that the evolvement of 2 further TOB subunits, Mas37/Sam37/Tom37 and Tob38/Sam35/Tom38, is part of such an adaptation process. These latter 2 components are associated with Tob55 at the cytosolic side of the outer membrane (10, 15–17). The role of Mas37 in the biogenesis of mitochondrial β-barrel membrane proteins has not yet been identified. Recent studies pointed to a possible function in the release of β-barrel precursors from the TOB complex (18, 19). Tob38 is tightly bound to Tob55, and both proteins form a functional TOB core complex even in the absence of Mas37. Tob38 is probably required for the stability and assembly of the TOB complex. Furthermore, a recent study suggests that Tob38 recognizes a sorting signal within β-barrel proteins (20).
Like all other mitochondrial outer-membrane proteins, β-barrel precursors do not contain a cleavable N-terminal presequence for their targeting to mitochondria but rather a noncleavable internal signal. The nature of such a signal element has not been characterized so far. Similarly unclear is whether the interaction of the mitochondrial import machinery is specific for β-barrel proteins of the eukaryotic cell or whether this machinery can recognize equivalent structural motifs independent of the origin of the protein. A study addressing this point revealed that PorB of pathogenic Neisseria species can target mitochondria when expressed in eukaryotic cells whereas porins of closely related nonpathogenic bacteria were unable to do so (21). Thus, it is still an open question whether PorB contains unique virulence-related features that make it an exceptional case. Furthermore, very recently, a specific signal that is conserved in eukaryotic β-barrel proteins was suggested to promote the intramitochondrial sorting of these proteins (20). However, because this signal is not conserved in prokaryotic β-barrel proteins, it is particularly interesting to test the fate of the latter proteins when expressed in eukaryotic cells.
To better understand the mechanism by which the eukaryotic cell recognizes β-barrel precursor proteins, we expressed bacterial β-barrel proteins in yeast cells. The bacterial proteins were imported into mitochondria via a pathway shared with mitochondrial β-barrel proteins. Furthermore, these proteins could be assembled in a native-like conformation into the mitochondrial outer membrane. Our results imply that although the machinery that sorts these proteins had to be modified during evolution of mitochondria, such an adaptation of substrate proteins was not required for allowing the machinery to recognize them.
Results
Bacterial β-Barrel Proteins Expressed in Yeast Cells Are Targeted to Mitochondria.
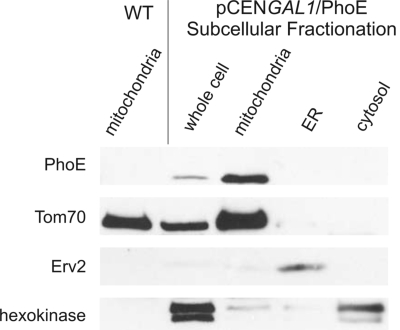
In the present study, we wanted to test whether mitochondrial β-barrel proteins contain specific targeting and sorting signals that are absent in bacterial β-barrel proteins. To address this point, we used the outer-membrane porin PhoE of Escherichia coli that was used extensively as a model protein for studying the biogenesis of bacterial outer-membrane proteins. We constructed a PhoE variant without the signal sequence required for transport across the bacterial inner membrane and produced it in yeast cells by using the strong GAL1 promoter on a low-copy plasmid. We chose high-expression conditions to enhance detection and to challenge the mitochondrial import and sorting systems. Subcellular fractionation of the transformed cells revealed that PhoE was located solely in the mitochondrial fraction (Fig. 1). As a control for the specificity of the antibody against PhoE, we verified that no signal was observed in mitochondria isolated from a nontransformed strain (Fig. 1). The mitochondrial localization of PhoE was confirmed by immunofluorescence microscopy where PhoE colocalized with a mitochondrial marker protein (Fig. S1).
Fig. 1.
Bacterial β-barrel proteins are targeted to mitochondria in yeast cells. Whole-cell lysate of cells expressing PhoE and fractions corresponding to mitochondria, endoplasmic reticulum (ER), and cytosol were analyzed by SDS/PAGE and immunodecoration with antibodies against the bacterial protein, the mitochondrial protein Tom70, the ER protein Erv2, and a control marker protein for the cytosol (hexokinase). Mitochondria isolated from wild-type, untransformed cells were coanalyzed as a control.
We investigated whether other bacterial β-barrel proteins are targeted to mitochondria when expressed in yeast cells. To that end, we examined the outer-membrane proteins OmpA and OmpC of E. coli and Omp85 from Neisseria meningitidis. This group of proteins presents a wide array of β-barrel proteins ranging from the 8-stranded monomer OmpA to the 16-stranded trimers OmpC and PhoE. These 3 additional proteins were constructed without a signal sequence and expressed in yeast cells under the control of a strong promoter. Similarly to PhoE, these proteins were exclusively found in the mitochondrial fractions (Fig. S2). Thus, the property of being targeted to mitochondria when expressed in eukaryotic cells is shared by several bacterial proteins.
PorB from pathogenic Neisseria gonorrhoeae expressed in mammalian cells was reported to dissipate the membrane potential across the mitochondrial inner membrane and to induce apoptosis (21). Therefore, we investigated whether the bacterial β-barrel proteins under study interfere with the biogenesis of mitochondrial β-barrel proteins or with other crucial mitochondrial functions. The levels of outer-membrane β-barrel proteins, such as Tob55, porin, and other mitochondrial proteins, were not affected by the expression of PhoE, Omp85, or OmpC and were only slightly reduced in mitochondria containing OmpA (Fig. S3 A and B). The capacity of isolated mitochondria harboring PhoE to import in vitro the matrix-destined preprotein pSu9 (1–69)-DHFR was similar to that of organelles isolated from control cells (Fig. S3C). Next, the growth rate of cells expressing PhoE was monitored and found to be comparable to that of nontransformed cells under all tested conditions, including growth on a nonfermentable carbon source where yeast cells depend on mitochondrial respiration for energy production (Fig. S3D). Thus, it appears that high-level expression of bacterial β-barrel proteins in yeast does not obstruct major mitochondrial features.
PhoE Is Imported into Mitochondria in a TOM- and TOB-Complex-Dependent Manner.
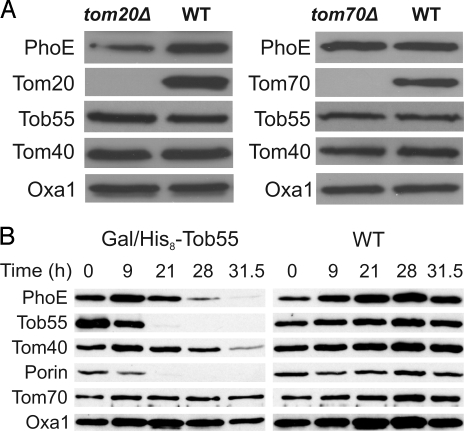
Precursors of the mitochondrial β-barrel proteins like Tom40, porin, and Tob55 are initially recognized at the surface of the organelle by the import receptor Tom20 (5, 6, 8, 18). Therefore, we asked whether the import receptors of the TOM complex play a role in the import of the bacterial β-barrel precursor proteins. To that end, we expressed PhoE in cells lacking either Tom20 or Tom70 and monitored its level in mitochondria. Mitochondria isolated from strains lacking Tom20 had significantly reduced amounts, whereas deletion of Tom70 hardly affected the mitochondrial level of PhoE (Fig. 2A). Hence, similar to their function in recognizing precursors of mitochondrial β-barrel proteins, the import receptor Tom20 plays an important role in the recognition of PhoE precursor, whereas Tom70 has only a minor function in this process.
Fig. 2.
PhoE is sorted to mitochondria in a TOM- and TOB complex-dependent manner. (A) Mitochondrial import of PhoE depends on the import receptor Tom20. Mitochondria isolated from tom20Δ, tom70Δ, and their corresponding wild-type cells transformed with p2μTPI/PhoE were analyzed by SDS/PAGE and immunodecoration with antibodies against PhoE and the indicated mitochondrial proteins. (B) Cells from wild-type strain or from a strain expressing Tob55 under the control of the GAL10 promoter (Gal/His8-Tob55) were harvested at the indicated time points after a shift to glucose-containing medium (see Fig. S4A). Crude mitochondria were isolated and proteins were analyzed by SDS/PAGE and immunodecoration with antibodies against PhoE and the indicated mitochondrial proteins. Tob55, Tom40, and porin, β-barrel proteins; Tom70, a signal-anchored protein in the outer membrane; Oxa1, an inner membrane protein.
Does the TOB complex mediate the membrane insertion of PhoE? To address this point, we transformed a plasmid encoding PhoE into cells where the essential component Tob55 is under the control of GAL10 promoter (9). The growth of these transformed cells was slowed down ≈32 h after shifting them to glucose-containing medium (Fig. S4A). Mitochondria were isolated from these Tob55-depleted cells at various time points after the shift, and the levels of various mitochondrial proteins were monitored. Clearly, Tob55 was gradually depleted from cells grown on glucose-containing medium. The lower amounts of Tob55 caused a reduction in the detected amounts of the mitochondrial β-barrel proteins porin and Tom40 as well as in the amounts of PhoE (Fig. 2B). The levels of non-β-barrel proteins were unaffected. Of note, the reduction in the amounts of PhoE preceded that of Tom40, suggesting that the depletion of PhoE is not a secondary effect to the reduction in the levels of Tom40. Next, PhoE was expressed in cells lacking Mas37, a peripheral subunit of the TOB complex. The absence of Mas37 resulted in a dramatic reduction in the amount of PhoE detectable in the corresponding mitochondria (Fig. S4B). Collectively, these results demonstrate that PhoE follows a TOM- and TOB-dependent insertion pathway.
Integration of PhoE in a Native-Like Structure into the Mitochondrial Outer Membrane.
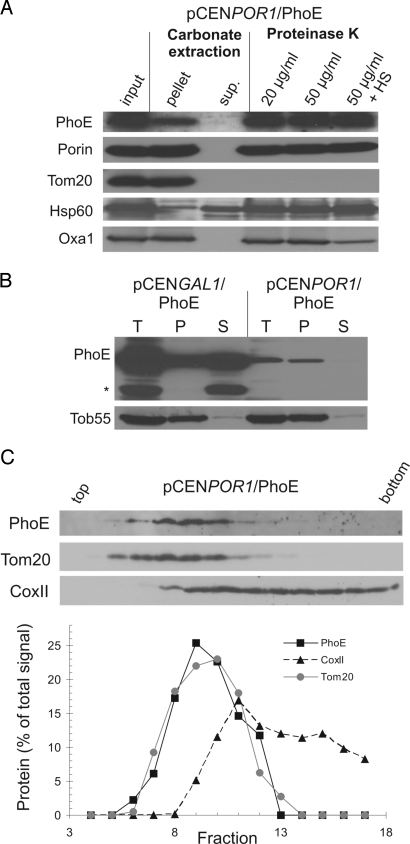
We asked whether PhoE is correctly assembled into the mitochondrial membranes. To that end, the association of PhoE with the mitochondrial membranes was analyzed by alkaline treatment of mitochondria isolated from PhoE-expressing cells. PhoE was recovered mainly in the pellet fraction just like the known membrane proteins porin, Tom20, and OxaI, although a considerable portion was also found in the supernatant (Fig. S5A). The membrane topology of PhoE was further studied by protease treatment of mitochondria because native PhoE is resistant against proteolysis from either side of the bacterial outer membrane (22). PhoE was indeed unaffected by such treatment in intact mitochondria consistent with its uptake into the organelles, but it was partially degraded upon rupturing the outer membrane by hypoosmotic swelling (Fig. S5A). Furthermore, when the membranes were treated with 6 M urea, a common method to analyze the membrane insertion of bacterial outer-membrane proteins (13, 23), a significant fraction of the overexpressed PhoE was extractable (Fig. 3B). Thus, a large portion of PhoE produced at high amounts is present in the mitochondria in a conformation and location where the protein is not embedded within a membrane, and at least some parts of the molecule are exposed to the intermembrane space (IMS).
Fig. 3.
PhoE expressed at low levels is properly integrated into the mitochondrial outer membrane. (A) Mitochondria isolated from cells expressing PhoE under the control of the POR1 promoter were loaded directly on SDS/PAGE gel (input) or were subjected first to alkaline extraction and then centrifuged to discriminate between membrane proteins (pellet) and soluble proteins in the supernatant (sup.). Additional aliquots of mitochondria were left intact or were treated by hypoosmotic swelling (HS) before their incubation with the indicated amounts of proteinase K. Proteins were analyzed by SDS/PAGE and immunodecorated with antibodies against the indicated proteins. Of note, for unknown reason, the recovery of PhoE in the carbonate extraction experiment was only 75%. Porin, protein embedded in the outer membrane; Tom20, outer-membrane protein exposed to the cytosol; Hsp60, soluble matrix protein; OxaI, an inner-membrane protein exposed to IMS. (B) Mitochondria isolated from PhoE-expressing cells were loaded directly on SDS/PAGE gel (T, Total) or were treated with urea and then centrifuged to separate the nonextractable pellet (P) from the urea-soluble fraction (S). Proteins were analyzed by SDS/PAGE and immunodecoration with antibodies against the indicated proteins. A proteolytic fragment of PhoE is labeled with an asterisk. (C Upper) PhoE is sorted to the mitochondrial outer membrane when expressed at low levels. Mitochondria from cells expressing PhoE under the POR1 promoter were analyzed by sucrose density gradient centrifugation. Proteins from fractions were analyzed by SDS/PAGE and immunodecoration with antibodies against PhoE, Tom20, and the inner-membrane protein CoxII. (C Lower) The bands corresponding to the detected proteins were quantified by densitometry. The relative amount in each fraction is presented as the portion from the total amount of this protein.
Next, we wished to determine in which of the 2 mitochondrial membranes PhoE was located. To that end, the organelles were sonified, and membrane vesicles were separated by sucrose density gradient centrifugation where the 2 mitochondrial membranes can be enriched in distinct fractions. Interestingly, PhoE behaved neither like a typical outer-membrane protein nor as an inner-membrane protein (Fig. S5B). Significant amounts of the protein were found in fractions with high density suggesting that PhoE molecules in these fractions might represent precursors that aggregated in the IMS and/or mistargeted to the IMS surface of the inner membrane. Taken together, it appears that most of the overexpressed PhoE molecules are not properly inserted into the mitochondrial membranes.
Do the high amounts of PhoE cause overloading of the TOB machinery, and therefore, the protein is not efficiently inserted from the IMS into the outer membrane? To address this question, we investigated the membrane integration of PhoE expressed under the control of the promoter of the mitochondrial β-barrel protein POR1. Subcellular fractionation confirmed that mitochondrial targeting of PhoE and the other bacterial proteins, OmpA and Omp85, was retained under these low expression levels (Fig. S6), although detection of OmpC was not possible with the available antibody. Hence, the bacterial β-barrel proteins are targeted to mitochondria independently of their expression level.
The vast majority of the PhoE produced at a low level was in the membrane pellet of the alkaline extraction, and it was protected from proteinase K even when the outer membrane was ruptured (Fig. 3A). Next, we analyzed the membrane integration of PhoE by resistance to extraction with urea. When expressed in reduced amounts, practically all PhoE molecules were not extractable with urea, similar to the endogenous β-barrel protein, Tob55. The submitochondrial location of PhoE expressed at reduced levels was further studied by sucrose density gradient centrifugation and revealed that PhoE migrated like the outer-membrane marker, Tom20 (Fig. 3C). Similarly, when mitochondria harboring OmpA and Omp85 expressed at reduced amounts were analyzed by this method, both proteins were found to comigrate with the outer-membrane marker (Fig. S7). Collectively, these results suggest that low expression levels of bacterial β-barrel proteins allow the precursor molecules to fully integrate into the mitochondrial outer membrane.
The C-Terminal Phenylalanine Residue of PhoE Is Crucial for Its Assembly into Trimeric Structures.
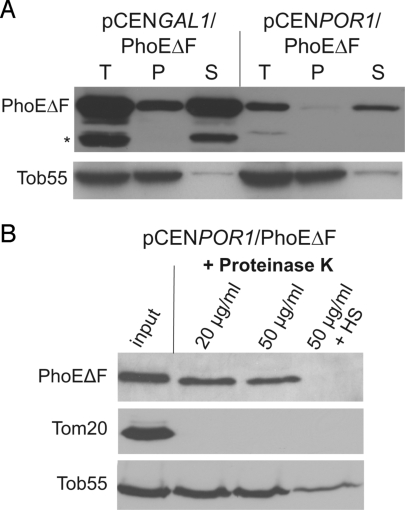
PhoE, like the majority of bacterial β-barrel proteins, contains a C-terminal signature sequence, of which Phe at the ultimate C-terminal position is the most important feature (24). To test whether this signature plays a similar role in the sorting of PhoE in eukaryotic cells, we constructed a PhoE variant where the C-terminal Phe residue was deleted (PhoEΔF). This variant was expressed in yeast cells under the strong GAL1 promoter or the POR1 promoter, and in both cases the mutant protein was targeted to mitochondria with a similar or even slightly higher efficiency as compared with full-length PhoE (Fig. S8 A and B). Is the ultimate Phe residue required for the correct membrane insertion of PhoE? In contrast to the full-length protein, both low- and high-level expression of PhoEΔF resulted in a situation where the vast majority of the protein was extractable by urea (Fig. 4A). Furthermore, at both expression levels, PhoEΔF was sensitive to protease treatment when the outer membrane was not intact (Fig. 4B and Fig. S8C). We next tested whether PhoEΔF is imported into mitochondria in a TOB-dependent manner. As observed for the full-length protein, the absence of Mas37 resulted in a significant reduction in the amount of PhoEΔF molecules in the corresponding mitochondria (Fig. S8D). Hence, PhoEΔF is imported into mitochondria via a pathway shared with the mitochondrial β-barrel proteins. However, its protease sensitivity and extractability by urea suggest that the protein is not assembled correctly into the outer membrane.
Fig. 4.
The C-terminal phenylalanine residue of PhoE is dispensable for targeting to mitochondria but not for proper assembly. (A) Mitochondria isolated from PhoEΔF-expressing cells were loaded directly on SDS/PAGE gel (T, Total) or were treated with urea and then centrifuged to separate the nonextractable pellet (P) from the urea-soluble fraction (S). Proteins were analyzed by SDS/PAGE and immunodecoration with antibodies against the indicated proteins. A proteolytic fragment of PhoE is labeled with asterisk. (B) PhoEΔF is present in mitochondria in a protease-sensitive conformation. Mitochondria isolated from cells expressing low amounts of PhoEΔF were treated and analyzed as described in Fig. 3A.
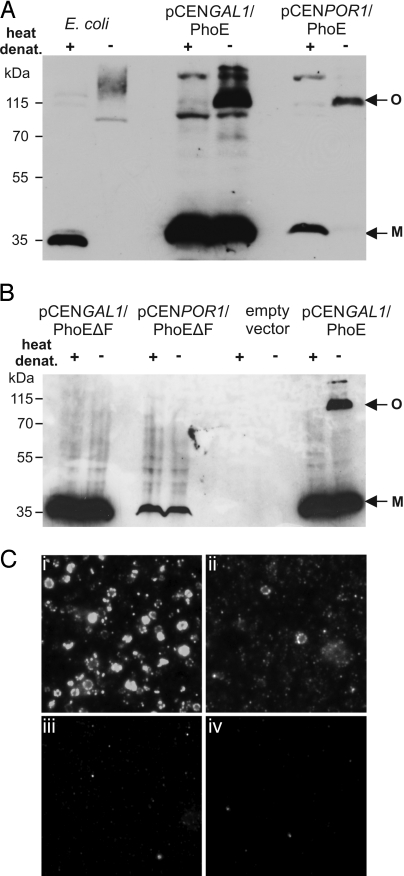
PhoE is known to form homotrimers in the bacterial outer membrane. We monitored whether PhoE expressed in yeast cells is able to form such oligomeric structures. Only a small fraction of the overexpressed mitochondrial PhoE was found in the trimeric structures when mitochondria were analyzed by seminative SDS/PAGE. Most of the PhoE molecules migrated as a monomeric form. In contrast, practically all PhoE molecules were found in the trimeric form when PhoE was expressed under the POR1 promoter (Fig. 5A). The ability to form such assembled trimeric form requires C-terminal Phe residues, because the variant lacking this residue was unable to form such structures independently of its expression level (Fig. 5B).
Fig. 5.
PhoE can form native-like trimeric structures in mitochondria. (A) Full-length PhoE assembles into a trimeric structure in mitochondrial membranes. Cell envelopes from E. coli or mitochondria isolated from cells transformed with plasmid encoding PhoE under either the GAL1 or POR1 promoter were analyzed by seminative PAGE after heat denaturation or without further treatment. The positions of monomeric and oligomeric forms of PhoE are indicated with M and O, respectively. (B) Deletion of the C-terminal Phe residue abolishes PhoE trimer formation in mitochondrial membranes. Mitochondria isolated from cells transformed with the empty plasmid or expressing the PhoEΔF variant from the GAL1 or POR1 promoter were treated and analyzed as in Fig. 5A. Mitochondria harboring high levels of full-length PhoE are coanalyzed as control. (C) Mitochondria derived from cells expressing PhoE (i), PhoE under control of the POR1 promoter (ii), PhoEΔF under the control of the GAL1 promoter (iii), or no PhoE (empty vector control) (iv) were stained with a monoclonal antibody that specifically recognizes native PhoE trimers.
The formation of the oligomeric structure was further studied after solubilizing the organelles with the mild detergent digitonin. Of note, most of the overproduced PhoE was recovered in the pellet fraction when a clarifying spin was performed with this lysate. In contrast, PhoE at lower levels was soluble under these conditions and recovered in the supernatant (Fig. S9A). These observations support the notion that a significant portion of the PhoE molecules is aggregated under high expression levels. The soluble fractions of the lysates were further analyzed by BN/PAGE, and a band corresponding to an oligomeric form of PhoE was detected (Fig. S9B). Of note, because solubilization of bacterial membranes with digitonin is very inefficient, a direct comparison of the migration behavior with that of PhoE derived from E. coli membranes was not possible in this case. Omp85 is also known to form oligomers, and such forms of the purified in vitro refolded E. coli Omp85/YaeT were observed by BN/PAGE (25). Thus, we used the same technique to analyze mitochondria isolated from cells expressing this protein. High molecular-weight oligomers were observed upon solubilization with digitonin (Fig. S9C). Collectively, these results demonstrate that the outer membrane of mitochondria can support the formation of oligomeric structures of bacterial β-barrel proteins.
The formation of correctly folded forms was further monitored by immunofluorescence microscopy employing mitochondria isolated from PhoE-expressing cells and a monoclonal antibody that recognizes specifically native PhoE (26). Specific staining at the surface of intact mitochondria was observed when full-length PhoE was expressed. In agreement with a stronger trimer band in the case of overexpression (Fig. 5A), the signal was stronger for mitochondria from cells overproducing PhoE (Fig. 5 Ci and Cii). Only background staining was observed when mitochondria isolated from cells producing high levels of PhoEΔF were inspected (Fig. 5Ciii). Taken together, PhoE expressed in eukaryotic cells is imported into mitochondria independent of its C-terminal signature sequence. In contrast, like in bacteria, the insertion into the mitochondrial outer membrane in a native-like structure requires this signature motif.
Discussion
Precursors of mitochondrial β-barrel proteins are recognized initially by the import receptors of the TOM complex and are then translocated via the import pore of the translocase through the outer membrane into the IMS. Next, β-barrel precursors are relayed to the TOB complex, which mediates their insertion into the outer membrane (27, 28). Very recently, a sorting signal that facilitates the interaction of eukaryotic β-barrel precursors with the TOB complex was identified. This signal appears to be an intramitochondrial sorting signal and is not involved in targeting of the precursor proteins to the organelle (20). The signals that facilitate the specific targeting of β-barrel precursors to mitochondria are not characterized yet. Studies have failed to identify a linear, well-defined sequence that can function as a targeting sequence. Hence, it can be assumed that the mitochondrial protein-import machinery recognizes β-barrel-specific structural elements (2).
To test this hypothesis, we expressed bacterial β-barrel proteins in yeast cells. These bacterial proteins do not share significant sequence similarity with endogenous mitochondrial β-barrels and should be devoid of any eukaryotic targeting sequence that evolved when mitochondria developed from the endosymbiont. Nevertheless, the proteins were imported into mitochondria suggesting that the ability of a protein to adopt a membrane-embedded, β-barrel conformation could be sufficient to ensure its specific targeting to mitochondria. This proposal is in line with our observation that a partially folded conformation of β-barrel precursors is required for efficient import into mitochondria (8). These findings also imply that mitochondrial β-barrel proteins, which evolved most probably from bacterial β-barrel structures, were not forced to develop specific targeting signals to assure their correct subcellular targeting.
Of note, our results differ from those of Müller et al. (21) who reported that, with the exception of PorB from pathogenic Neisseriae, bacterial porins were unable to be targeted to mitochondria. Differences in the experimental setup of both studies might explain this apparent discrepancy. For example, Müller et al. studied bacterial proteins in mammalian cells, whereas we used proteins expressed in yeast.
Assembly of outer-membrane proteins in bacteria depends on a C-terminal signature motif, which corresponds to the last transmembrane β-strand and is reminiscent of, but not identical to, the recently identified β-sorting signal in mitochondrial proteins (20). Most bacterial outer-membrane proteins contain a Phe residue at their C terminus, which is essential for efficient integration into the outer membrane in vivo but not for the formation of the trimeric structure as appeared from in vitro folding studies (29, 30). The high-level expression of a mutant PhoE lacking the C-terminal Phe in E. coli led to the periplasmic accumulation of the mutant protein in a protease-sensitive monomeric conformation (24). Furthermore, we demonstrated recently that the bacterial C-terminal signature sequence interacts directly with Omp85 (25). To assess whether the recognition of substrates is conserved between the TOB and the Omp85/YaeT complexes, we investigated whether deletion of the C-terminal Phe residue of PhoE affects assembly of the protein into the mitochondrial outer membrane. The PhoEΔF variant could not form the native-like trimers in the outer membrane. Collectively, our results demonstrate that the ancestral bacterial signal is still recognized and correctly processed by the mitochondrial assembly machinery.
We report that the expression levels of the β-barrel protein influence the fraction of molecules that can be properly integrated into the outer membrane. At high expression levels, all of the protein was found in mitochondria, thus, it appears that the TOM complex can process such elevated amounts of β-barrel precursors. In contrast, the TOB complex is probably the limiting factor in the biogenesis pathway as overexpressed precursor proteins accumulated in the IMS. Interestingly, even under these conditions, the import into the organelle depended on the TOB complex. Based on these findings, the function of the TOB complex can be divided into 2 distinct steps. First, the complex is involved in the translocation of β-barrel precursors via the TOM complex across the outer membrane. It is not entirely clear yet whether a direct interaction between the precursor and the TOB complex is required at this stage. If there is a direct interaction, it does not involve the intramitochondrial sorting signal of the β-barrel precursor or its bacterial substitute because PhoEΔF and mutant mitochondrial proteins with a deficient signal (20) were still efficiently imported into the mitochondria. Perhaps, the TOB complex, like the TOM complex, recognizes a high β-sheet content at this stage. In the second step, the TOB complex mediates the membrane integration of the β-barrel precursors. At this stage, an interaction between the intramitochondrial sorting signal of the precursor and the Tob38 component of the TOB complex is required. This sorting signal is present in the last predicted β-strand of the mitochondrial proteins and was not recognized in bacterial outer-membrane proteins (20). Nevertheless, we found that the assembly of bacterial β-barrel proteins into a native-like oligomeric structure at the mitochondrial outer membrane depended on their ancestral recognition motif.
Collectively, the current study demonstrates that no eukaryotic-specific signals that are essential for the intracellular mitochondrial targeting of these proteins have evolved within mitochondrial β-barrel proteins. Moreover, it shows that bacterial outer-membrane proteins can be properly assembled into the mitochondrial outer membrane. These findings support the notion that β-barrel protein assembly is conserved between bacteria and eukaryotic cell organelles of endosymbiont origin.
Materials and Methods
SI Text includes a description of immunofluorescence microscopy, in vitro import assay, and drop-dilution assay.
Yeast Strains and Growth Methods.
The wild-type strain YPH499 was used. The GAL10-TOB55 and mas37Δ strains were described in refs. 9 and 18, respectively. The tom20 null strain YTJB64 and its corresponding parental strain YTJB4 were used (31). The tom70 deletion strain was obtained from Research Genetics.
Biochemical Procedures.
Mitochondria were isolated from yeast cells by differential centrifugation, as described in ref. 32. Seminative gel electrophoresis and urea extraction were performed basically as described in refs. 13 and 33. The separation of mitochondrial membranes was based on published procedures (34). Mitochondria were subjected to hypoosmotic shock in swelling buffer, adjusted to a final sucrose concentration of 0.45 M, and disrupted by treatment with a Branson 250 sonifier. The homogenate was clarified by centrifugation (35,000 × g for 30 min at 2 °C). Vesicles were sedimented by ultracentrifugation (200,000 × g for 90 min at 2 °C) onto a 2 M sucrose cushion and overlaid with a sucrose step gradient ranging from 1.4 to 1 M. Gradients were centrifuged in a swing-out rotor (230,000 × g for 16 h at 4 °C), and fractions were analyzed by SDS/PAGE.
Recombinant DNA Techniques.
The sequences encoding PhoE, OmpA, OmpC of E. coli, and Omp85 from N. meningitidis lacking their signal sequences were cloned by PCR amplification from corresponding plasmids encoding full-length proteins. PhoEΔF was generated in a similar way; however, the codon for Phe-330 was omitted from the 3′ primer of the PCR. The PCR products were inserted into the yeast expression vectors pRS426-TPIp-URA3 or pYX113-GAL1p-URA3. To construct low-expression plasmids, the GAL1 promoter in pYX113 was replaced by the S. cerevisiae POR1 promoter.
Fluorescence Microscopy.
Mitochondria were fixed with 2% formaldehyde in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM Mops/KOH, pH 7.2) and immobilized on poly-l-lysine-coated glass cover slips. After removing the fixative by 3 washes with PBS, the cover slips were blocked with 3% BSA in PBS. The mitochondria were labeled with monoclonal antibody PP4–1, which specifically recognizes PhoE trimers (26), followed by labeling with Alexa 488-conjugated goat anti-mouse antibodies. Control samples were treated similarly but without the first antibody. Cover slips were mounted in PBS-buffered glycerol and viewed in a Zeiss Axioskop 2 plus microscope.
Supplementary Material
Acknowledgments.
We thank W. Neupert and M. Ott for helpful discussions; K. Rehn and J. Arenas for technical support; E. Kracker for help in performing some experiments; and K.S. Dimmer and G. Dodt for help with the fluorescence microscopy. This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 446-A30, and the Netherlands Research Council for Earth and Life Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807830106/DCSupplemental.
References
- 1.Gabriel K, Buchanan SK, Lithgow T. The alpha and beta: Protein translocation across mitochondrial and plastid outer membranes. Trends Biochem Sci. 2001;26:36–40. doi: 10.1016/s0968-0004(00)01684-4. [DOI] [PubMed] [Google Scholar]
- 2.Rapaport D. How to find the right organelle-targeting signals in mitochondrial outer membrane proteins. EMBO Rep. 2003;4:948–952. doi: 10.1038/sj.embor.embor937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schleiff E, et al. Prediction of the plant β-barrel proteome: A case study of the chloroplast outer envelope. Protein Sci. 2003;12:748–759. doi: 10.1110/ps.0237503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wimley WC. The versatile beta-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 5.Krimmer T, et al. Biogenesis of the major mitochondrial outer membrane protein porin involves a complex import pathway via receptors and the general import pore. J Cell Biol. 2001;152:289–300. doi: 10.1083/jcb.152.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Model K, et al. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat Struct Biol. 2001;8:361–370. doi: 10.1038/86253. [DOI] [PubMed] [Google Scholar]
- 7.Rapaport D. Biogenesis of the mitochondrial TOM complex. Trends Biochem Sci. 2002;27:191–197. doi: 10.1016/s0968-0004(02)02065-0. [DOI] [PubMed] [Google Scholar]
- 8.Rapaport D, Neupert W. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J Cell Biol. 1999;146:321–331. doi: 10.1083/jcb.146.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paschen SA, et al. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- 10.Wiedemann N, et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 11.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozjak V, et al. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- 13.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 14.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J Cell Biol. 2004;166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milenkovic D, et al. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J Biol Chem. 2004;279:22781–22785. doi: 10.1074/jbc.C400120200. [DOI] [PubMed] [Google Scholar]
- 17.Waizenegger T, et al. Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep. 2004;5:704–709. doi: 10.1038/sj.embor.7400183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Habib SJ, Waizenegger T, Lech M, Neupert W, Rapaport D. Assembly of the TOB complex of mitochondria. JBiol Chem. 2005;280:6434–6440. doi: 10.1074/jbc.M411510200. [DOI] [PubMed] [Google Scholar]
- 19.Chan NC, Lithgow T. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol Biol Cell. 2008;19:126–136. doi: 10.1091/mbc.E07-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutik S, et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 21.Müller A, et al. VDAC and the bacterial porin PorB of Neisseria gonorrhoeae share mitochondrial import pathways. EMBO J. 2002;21:1916–1929. doi: 10.1093/emboj/21.8.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tommassen J, Lugtenberg B. Amino terminus of outer membrane PhoE protein: Localization by use of a bla-phoE hybrid gene. J Bacteriol. 1984;157:327–329. doi: 10.1128/jb.157.1.327-329.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collin S, Guilvout I, Chami M, Pugsley AP. YaeT-independent multimerization and outer membrane association of secretin PulD. Mol Microbiol. 2007;64:1350–1357. doi: 10.1111/j.1365-2958.2007.05743.x. [DOI] [PubMed] [Google Scholar]
- 24.Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 25.Robert V, et al. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Gelder P, Tommassen J. Demonstration of a folded monomeric form of porin PhoE of Escherichia coli in vivo. J Bacteriol. 1996;178:5320–5322. doi: 10.1128/jb.178.17.5320-5322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson AE, Jensen RE. Barreling through the membrane. Nat Struct Mol Biol. 2004;11:113–114. doi: 10.1038/nsmb0204-113. [DOI] [PubMed] [Google Scholar]
- 28.Paschen SA, Neupert W, Rapaport D. Biogenesis of beta-barrel membrane proteins of mitochondria. Trends Biochem Sci. 2005;30:575–582. doi: 10.1016/j.tibs.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 29.de Cock H, Struyve M, Kleerebezem M, van der Krift T, Tommassen J. Role of the carboxy-terminal phenylalanine in the biogenesis of outer membrane protein PhoE of Escherichia coli K-12. J Mol Biol. 1997;269:473–478. doi: 10.1006/jmbi.1997.1069. [DOI] [PubMed] [Google Scholar]
- 30.Jansen C, Heutink M, Tommassen J, de Cock H. The assembly pathway of outer membrane protein PhoE of Escherichia coli. Eur J Biochem. 2000;267:3792–3800. doi: 10.1046/j.1432-1327.2000.01417.x. [DOI] [PubMed] [Google Scholar]
- 31.Lithgow T, Junne T, Wachter C, Schatz G. Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J Biol Chem. 1994;269:15325–15330. [PubMed] [Google Scholar]
- 32.Daum G, Gasser S, Schatz G. Import of proteins into mitochondria: Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J Biol Chem. 1982;257:13075–13080. [PubMed] [Google Scholar]
- 33.Bos MP, Robert V, Tommassen J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 2007;8:1149–1154. doi: 10.1038/sj.embor.7401092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahedi RP, et al. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol Biol Cell. 2006;17:1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.