Abstract
The binding transition state (TS) is the rate-limiting step for transient molecular interactions. This important step in the molecular recognition process, however, is largely understood only at a qualitative level. To establish a more quantitative picture of the TS structure, we exploit a set of biophysical techniques that have provided major insights in protein folding applications. As a model system representing the large class of “weakly charged” protein–protein interactions, we examine the binding of a variety of human growth hormone (hGH) variants to the human growth hormone receptor (hGHR) and the human prolactin receptor (hPRLR). hGH variants were chosen to probe different features of the TS structure, based on their highly reengineered interfaces. Both Eyring and urea (m value) analyses suggest that the majority of binding surface burial occurs after TS. A comprehensive φ analysis showed that individual hGH interface residues do not contribute energetically to the stability of the TS, but there is a TS “hot spot” in the receptor. Zinc dependence studies that take advantage of an endogenous tetracoordinated interfacial metal binding demonstrate that surfaces of the molecules have attained a high orientational complementarity by the time the TS is reached. The model that best fits these data are that a “knobs-into-holes” process precisely aligns the two molecular interfaces in forming the TS structure. Surprisingly, most of the thermodynamic character of the binding reaction is focused in the fine-tuning process occurring after TS.
Keywords: equilibrium thermodynamics, hot spot, protein recognition, transition state thermodynamics, binding pathways
Organized networks of transient protein–protein interactions play fundamental roles in initiating many types of biological processes. In most cases, proper function depends on the timing and duration of the interaction, which is coordinated with a finely tuned affinity between the binding partners. The energy landscape describing the binding event between two molecules consists of a number of intermediates whose magnitude and location depend on the specific interaction. Nevertheless, highly simplified models of the reaction pathway consist of three principal states (1): Step 1, the unbound reactants; Step 2, the transition state (TS); and Step 3, the bound complex. The molecular details of two of the states, Steps 1 and 3, have been captured in numerous high-resolution structure determinations. However, the molecular recognition processes that connect Steps 1 and 3 go through a poorly understood TS intermediate step, which is characterized by a protein–protein interface that is still partially solvated, containing unoptimized side-chain interactions (2–7).
There are two major classes of protein–protein interactions that describe most protein–protein association processes. One class is regulated primarily by electrostatic forces that provide both a long-range steering function and dominate the overall binding energy. The second class is more predominant and involves molecular contact interfaces having neutral or weakly charged surfaces. The binding energy of weakly charged interfaces is governed chiefly by hydrophobic forces, although charge interactions can still play a major role in defining specificity.
The majority of the detailed experimental work examining the association component of the molecular recognition factors in binding has been performed on systems displaying strong electrostatic enhancement (3–7). In comparison, the partitioning of binding energy within large, weakly charged interfaces appears to be more complex because hydrophobic interfaces are more malleable (8). Studies based on weakly charged interactions by Schreiber and coworkers (3, 9) indicate that individual interface side chains have little or no effect on association rates or energetic consequence on forming the TS. However, a number of fundamental questions about the thermodynamic and structural linkages that actually define the TS in weakly charged interactions are yet to be answered.
To develop a deeper understanding of the thermodynamic and structural nature of the TS complex, we analyzed the thermodynamic, structural, and orientational properties that govern formation of the TS of the Site 1 human growth hormone (hGH)–human growth hormone receptor (hGHR) interaction, a representative large, weakly charged protein–protein interface. To evaluate the thermodynamic profile of the TS in this system, we determined the TS-binding thermodynamics of wild-type (WT) hGH and three related hGH variants. These variants were selected to probe different features of the TS structure based on their modified Site 1 interface. To establish the orientational effects of the TS structure, the thermodynamics of hGH binding to the prolactin receptor (hPRLR) were studied. This system is particularly powerful in this regard because it has an interfacial Zn2+-binding site that can be probed to establish the orientational precision of the TS structure with high accuracy.
Our experimental approach involved the application of a set of biophysical techniques that have been used to characterize protein-folding pathways. Using these techniques, we investigate the amount of surface buried in the TS structure and at what step do desolvation factors contribute most to the thermodynamics of the binding event. We establish how the enthalpic, entropic, and heat capacity components are partitioned between the association and dissociation steps and estimate when the majority of the conformational changes take place, before or after the TS state. A comprehensive φ analysis was performed to identify residues in the binding interface that contribute significantly to formation of the TS and correlate the “hot spot” for TS formation with that determined by traditional alanine scanning for the final bound complex. Finally, we evaluate the orientational requirements of forming the TS complex by using the influence of metal binding on the hGH–hPRL interaction in a manner analogous to a ψ analysis conducted with surface bihistidine-binding sites (10–12).
Based on these analyses, we conclude that the TS structure is characterized by a high orientational complementarity between interacting molecules driven by a “knobs-into-holes” mechanism. Further, the thermodynamic partitioning of both the enthalpic and entropic components of the association and dissociation steps of binding showed that hypermutated interfaces have surprisingly little effect on the thermodynamic factors leading to the TS but differ drastically after TS. This suggests that the fine-tuning process that occurs after TS is the main contributor to magnitude of the enthalpic and entropic components and determines which of these components drives the binding event. Although it is difficult to extend one set of findings into a general mechanism, it appears that for weakly charged interfaces it is the precise matching of surface terrains of the two molecules that defines the competent TS state.
Results
Systems.
Site 1 of the hGH–hGHR interface contains ≈30 residues on each side. Alanine-scanning mutagenesis identified a set of binding hot-spot residues for each molecule (21–23). These hot spots are characterized by a relatively small group of clustered residues that in the context of the complex form the major set of interactions in the interface.
Four individual complexes with hGHR were analyzed: the WT hGH and three hypermutated variants. One variant, hGHv (15 mutations) binds to hGHR with an extremely high affinity (≈10 pM vs. 1 nM for WT hGH) (13). This hGHv variant possesses a different binding mechanism than WT hGH, as characterized by different equilibrium thermodynamic and H/D exchange profiles (8, 14). Thus, it is of interest to determine whether the mechanistic differences manifest themselves during the association or dissociation step of the binding pathway. The two other Site 1 hGH variants, AS1 and AS2, contain a large number of alanine substitutions of residues that participate in Site 1 binding. AS1 has 24 Site 1 mutations, 13 of which are alanine, and AS2 has 21 mutations with 11 alanines (8). Although these “Ala-shaved” variants have a substantially reduced set of possible side-chain interactions, surprisingly they have binding affinities close to the WT hormone (≈1–2 nM) (8). We expect these variants to have significantly different side-chain entropy and desolvation properties than the WT hGH.
TS Thermodynamics Evaluated by Eyring Analysis.
We performed an Eyring analysis for the four hormones binding to hGHR to monitor the association and dissociation rates as a function of temperature to determine the activation thermodynamic components: ΔH‡, ΔS‡, and ΔCp‡. The resulting association and dissociation thermodynamics are presented in supporting information (SI) Fig. S1. Although the three hGH variants have very diverse Site 1 interfaces, they have comparable ΔGon‡ values based on their similar on-rates compared with WT hGH. In addition, the analysis showed that during association all of the hormones had similar ΔHon‡ and ΔSon‡ contributions to binding. In each case, the free-energy barrier to reach the TS is primarily characterized by a larger enthalpic penalty than an entropic one (at 1 M standard state).
In contrast, the dissociation thermodynamics showed significant differences among the four hormones (Fig. S1). For instance, dissociation of WT hGH involves both enthalpic and entropic penalties (ΔHoff‡ = 22 kcal/mol; −TΔSoff‡ = 12 kcal/mol), whereas for hGHv, dissociation is highly enthalpically opposed ΔHoff‡ = 42 kcal/mol), yet favored entropically (−TΔSoff‡ = −18 kcal/mol). The other two variant hormones, AS1 and AS2, have ΔHoff‡ and −TΔSoff† values intermediate to WT hGH and hGHv (Fig. S1).
TS Surface Area Burial.
A denaturant m value analysis was performed to obtain an estimate of the percentage of the denaturant sensitive surface area buried in the final Site 1 interface at the stage where the TS complex is formed (15). The m values for both association and equilibrium free energies of binding were generated by measuring changes in the kinetic and equilibrium binding constants as a function of urea concentration (Fig. S2). The percentage of surface area burial in the TS was calculated from the corresponding ratio between m values obtained for the association and equilibrium free energies. For the WT hGH–hGHR complex, the m value analysis yields a value of massoc/meq = 43 ± 4%, or just under half the net surface area that is found in the final complex.
We similarly analyzed the two Ala-shaved variants, AS1 and AS2 (Fig. S2). Despite the large amount of remodeling where several hundred square angstroms of interface surface area are lost, both AS1 and AS2 possessed very similar fractional TS surface area burial, 49 ± 3% and 43 ± 3%, respectively. As such, the TS surface burial analysis provided a global picture of the extent of intermolecular interactions required. This invariance in surface burial (≈50%) for the different hormone variants implies that the TS surface burial is apparently insensitive to changes in a set of individual side chains, even when these changes are extensive (similar m value data for hGHv binding could not be acquired because it binds too tightly to obtain accurate binding data at low urea concentrations).
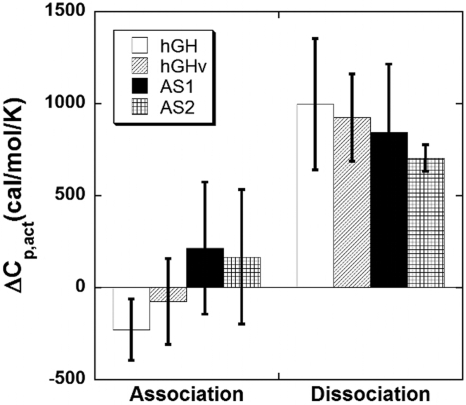
Heat Capacity (ΔCp) Differences in the Association and Dissociation Binding Steps.
Previously, the equilibrium ΔCp values for the hormone–receptor Site 1 interactions were found to be relatively large and negative (−600 to −1,000 cal/mol per K) (8). In addition to suggesting significant surface burial/conformational change upon binding, the large magnitude of the ΔCp values facilitated the observance and dissection of ΔCp into its association and dissociation components. Complementing the m value analysis discussed above, changes in pre- and post-TS surface burial/conformation change were inferred by comparing the ΔCp values between the association and dissociation steps for binding (ΔCp,on‡ and ΔCp,off‡) (Fig. 1). Although error levels were expectedly high from the second derivative, several trends could be established. Within error, ΔCp,on‡ values were similar and close to zero among the four hormones, suggesting that similar overall binding factors (e.g., surface burial) were involved in reaching the TS. This interpretation is supported by the m value analysis discussed above that indicated that the surface burial in the TS complexes of all of the variants was similar. Conversely, ΔCp,off† was large and positive among the four hormones, indicating that the pathways from the TS complex to the bound state possess steps involving the majority of the desolvation and/or conformational changes.
Fig. 1.
TS ΔCp values for hormone–receptor binding, which includes WT hormone, the phage-optimized hGHv, and two alanine-shaved hormones, AS1 and AS2.
The activation free-energy barriers for association and dissociation binding steps as a function of temperature are shown in Fig. S3. Although the ΔGon for the four hormones exhibit minimal changes across the temperature range −20–80 °C, the comparable ΔGoff values are widely distributed across this temperature range. The ΔGoff profiles exhibit a defined point of maximum stability, 35 °C for WT hGH, 6 °C for hGHv, 26 °C for AS1, and 18 °C for AS2. Interestingly, the hGH–hGHR complex appears maximally stable (slowest dissociation rate) at 35 °C, near the physiologically relevant temperature for the hGH–hGHR interaction in vivo, whereas the phage display-generated interfaces (hGHv, AS1, and AS2) possess maximum ΔGoff, max values at lower temperatures. This finding is noteworthy in that it relates protein–protein dissociation energetics with physiological function. The importance of a high barrier against dissociation of a 1:1 complex may stem from the mechanism of hGH signaling that occurs in a stepwise fashion where first a 1:1 hGH–receptor complex is formed followed by binding of a second receptor to form a 1:2 active complex. Although the finding represents a single example, such correlations have been noted for protein folding from thermophillic organisms (16).
φ Analysis: Determination of a TS Hot Spot.
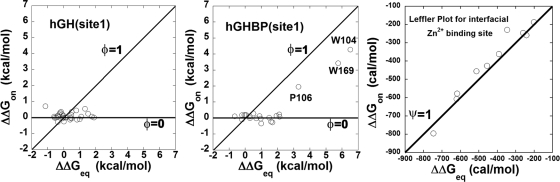
The relative energetic contributions of individual side-chain groups to the TS energy were determined from a mutational φ analysis study performed on 31 WT hGH residues in its Site 1 interface. The corresponding φ values were calculated from differences in the free energy of association vs. the corresponding changes in the equilibrium free energy upon individual WT-to-Ala substitutions φ = ΔΔGon‡/ΔΔGeq (17–19). Residues playing a vital role in forming the TS structure show an accompanying strong correlation between the changes in the ΔΔGon‡ and the ΔΔGeq.
The data for the WT hGH–hGHR Site 1 interaction are presented in a Leffler plot of ΔΔGon relative to ΔΔGeq (20) (Fig. 2), which includes two lines representing φ values of 1 and 0, respectively. The φ values for the hormone cluster tightly around 0 for the entire set of interface mutations (Fig. 2). This indicates that for a given WT-to-Ala mutation, there is very little fluctuation in the binding association rates, even though the comparable effects on ΔΔGeq values can approach ≈2 kcal mol−1 (21–23). This trend suggests that any change in ΔΔGeq stems from effects in the dissociation rates. Additionally, AS1 and AS2, variants of hGHv with multiple alanine substitutions, possess changes in ΔGeq almost exclusively through ΔGoff (8). This suggests that even the simultaneous removal of a large number of side chains in the hormone interface has surprisingly little effect on the influence of individual residues on the TS structure.
Fig. 2.
Leffler plots. (Left and Center) Kinetic response for an alanine scan of both the hGH (Left) and hGHR (Center) interfaces. Kinetic and equilibrium data were analyzed from previous reports (21–23). The interface residues included in the analysis are described in Material and Methods. (Right) Zn2+ binding to the interfacial binding site. The solid line is a slope of 1, indicative of a φ or ψ value of unity (native-like interaction).
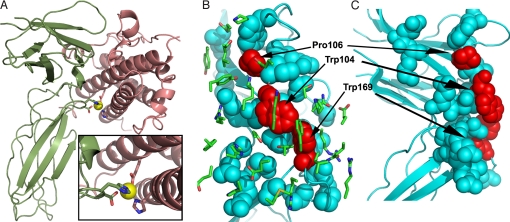
The corresponding φ analysis of the hGHR interface is presented in Fig. 2. Generally, there is a tight clustering of the majority of the hGHR interface residues around ΔΔGon‡ ≈0, which is similar to the WT hGH case. However, three residues show significant variation in the ΔΔGon‡. These are W104, P106, and W169 (see Fig. 3) with corresponding φ values of 0.65, 0.59, and 0.59, respectively. It is interesting to note that these same three residues also make up the receptor's interaction hot spot (21–23).
Fig. 3.
hGH–hGHR complex structures. (A) Ribbon structure of the hGH(salmon)–hPRLR(green) complex highlighting the interface Zn2+-binding site. The four Zn2+ ligands include hGH residues His-18 and Glu-174 and hPRLR residues Asp-187 and His-188. (B) View of the hGHR interface residues (blue and red spheres) illustrating the location of the transition state hot-spot residues (Trp-104, Pro-106, and Trp-169; shaded red). The Trp residues represent the knobs that fit into the hGH interface hole. The Site 1 hGH interface residues (green sticks) are displayed in the foreground. (C) Side view of the hGHR interface displayed in B. Figures were generated with PyMOl (35).
Together, these data suggest a conceptual model of the TS complex where only a small subset of residues (W104, P106, and W169) contributes to the TS hot spot. This finding leads to interesting interpretations about the nature of this complex on several levels. First, the TS hot spot is asymmetric with respect to the binding partners, individual residues possessing significant energetic contributions reside on hGHR, whereas hGH residues are energetically neutral toward TS formation even if they significantly contribute to the equilibrium binding affinity. This contrasts with what is observed from equilibrium alanine-scanning results for WT hGH–hGHR, which display a more symmetric layout with hot-spot residues residing on both sides of the binding interface (22, 23).
Orientation Requirements at the Transition State.
To evaluate the orientational specificity of the TS structure, we exploited a native zinc-binding site that bridges the interface between hGH and hPRLR (hGH binds to the hPRLR extracellular domain (ECD) by using virtually the same binding epitope as it does binding to its cognate receptor (hGHR) (23, 24). In this interface, Zn2+ is bound by residues His-18 and Glu-174 of hGH and Asp-187 and His-188 of hPRLR (see Fig. 3); however, neither the hormone nor receptor binds Zn2+ in their unbound states with a Kd tighter than 1 μM (25). To form the Zn2+-binding site, proper orientation is required for the side-chain ligands involved, and hence binding requires that the proteins themselves be aligned in a native, or near-native geometry. Although the Zn2 + site is endogenous, association is still possible in the absence of the ion. In this regard, ion binding is unlike zinc finger motifs where metals are required for folding. Rather, this situation is analogous to the ψ analysis approach using exogenous surface bihistidine ion-binding sites, which has been used to reveal folding pathways (10–12).
To determine the effect of Zn2+ binding on the TS structure, we performed a ψ analysis by monitoring the association rate and equilibrium constant as a function of Zn2+concentration (11). Fig. 2 is the Leffler plot of the free-energy change for association and free-energy equilibrium change for different Zn2+ concentrations. A clear trend is observed exhibiting a unity correlation between ΔΔGon and ΔΔGeq as the Zn2+ concentration is altered between 5 nM and 5 μM. Over the measured range of 200–800 cal mol−1, the slope of this correlation 1.05 ± 0.08. This strong correlation indicates that the ion-binding site has native-like affinity in the TS. Therefore, the side-chain ligands forming Zn2+-binding site have adopted their native arrangement in the TS. Ala substitutions at Asp-187 and His-188 of hGHR show φ values of close to 0, suggesting that the TS structure is not dependent on any individual Zn2+ ligand itself, but on the presence of an ion-binding site that is formed by a specific hormone/receptor orientation.
Discussion
The hGH–hGHR Site 1 interface is a classic example of a weakly charged interaction. This class of interactions is believed to be driven primarily by hydrophobic forces that provide an attractive force over a small but significant region of the effective binding surface (8, 23). A common feature of weakly charged interactions is that changes in binding affinity are due principally to effects involving the off-rates of the binding reaction (23). Even in highly mutated systems, on-rates remain remarkably constant (8). This trend has led to the interpretation that TS structures are not very sensitive to individual or groups of mutations and thus, can probably be characterized by a set of relatively nonspecific interactions that loosely align the molecules that, nevertheless, establish a productive orientation that can proceed to the final bound complex. The object of this work was to provide new types of experimental data to help refine this conceptual model and in particular, to better establish the relationships existing between measured thermodynamic parameters and the structural properties of the TS complex.
Orientation Factors in Formation of the TS Structure.
Based on Zn2+ binding only influencing the on-rate for hGH– hPRLR association, we conclude that the binding TS contains a native-like Zn2+-binding site. Structural data indicate that Zn2+ binding requires the specific alignment of four ligands, two from the hormone (His-18 and Glu-174) and two from the hGHR ECD (Asp-187 and His-188) (24). These data provide strong evidence that at the TS for binding, the two partners are correctly aligned with high angular precision and minimal distance separation (Fig. 3).
The Zn2+ data are in contrast with the other structural probes (φ values, ΔCp,on† urea m value), which point to a less specific complex at the level of individual side-chain interactions. Hence, the critical element of the TS for binding is the precise orientation of the two partners. This presents somewhat of a conundrum. How can the neutral interface be so well aligned in the absence of specific interactions compared with the case for electrostatic interfaces? To explain this we propose a knobs-into-holes mechanism. The structure of the Site 1 interface reveals that W104 and W106 in hGHR insert as knobs into holes formed by a group of side chains from helix 4 and helix 1 of hGH (Fig. 3). The φ values for alanine mutations in the hormones indicated that increasing the size of the holes has little effect on on-rates; whereas removing either the knobs by the Trp to Ala mutations or altering the scaffold orienting the knobs by the Pro to Ala mutation at position 106 has a major influence on the on-rate and the corresponding φ value.
Our conclusions are largely consistent with models proposed by Northrup and Erickson (26), and Janin (1) and the studies of Schreiber and coworkers (5). They concluded that in electrostatically driven barnase–barstar binding a high degree of orientation is reached at the TS (1, 3), although a less specific “diffusive” TS was observed for the association of two nonelectrostatically guided systems, TEM1-BLIP and IFNα2–IFNAR2 (9).
Thermodynamic Partitioning Between Association and Dissociation Steps.
The ΔGon‡ thermodynamic values established for Site 1 binding are virtually identical between WT hGH and the three highly mutated hGH variants. Even though these molecules possess drastically different Site 1 interfaces, they bury approximately the same amount of surface area (≈50%) in their TS structures compared with the final stable complex. It is expected that at the atomic level the interfaces differ significantly in their enthalpic and entropic propensities; however, Eyring analysis indicates that their activation enthalpies and entropies are very similar (Fig. S1). This trend holds even for variants that bind through different thermodynamic mechanisms (8).
In contrast, the partitioning of the ΔHoff‡ and ΔSoff‡ components in the ΔGoff‡ step of the reaction pathway is quite different between WT hGH and the three variants. This difference is particularly striking comparing the dissociation steps of WT hGH and hGHv. Whereas for WT hGH both ΔHoff‡ and −TΔSoff‡ disfavor dissociation (12.8 and 8.7 kcal/mol, respectively), for hGHv binding ΔHoff‡ is highly unfavorable (43 kcal/mol), counterbalanced by a favorable −TΔSoff‡ (−17.8 kcal/mol). These differences indicate that the energy landscapes characterizing the dissociation processes (or post-TS-binding pathway) for these two hormones differ in significant ways, as might be expected based on previous thermodynamic and H/D studies (8, 14).
Interpretation of the Measured ΔCp Values in the Association and Dissociation Steps of Binding.
It has been proposed that ΔCp values can be correlated with the amount of hydrophobic surface area desolvated in the subsequent protein–protein interface (27, 28). Based on a compilation of published thermodynamic data the average equilibrium ΔCp value for a dataset of “typical” protein–protein interactions is −333 ± 202 cal mol−1 K−1 (29). The ΔCp values obtained for the binding of hGHR to WT hGH and the three variants are considerably larger, averaging ∼–750 cal mol−1 K−1 (8). This large ΔCp value allows the TS ΔCp values to be clearly assigned as the magnitude of ΔCp increases the degree of curvature in the Eyring plots.
The ability to dissect ΔCp into its association and dissociation components reveals unique insights into the binding mechanism. For WT hGH–hGHR Site 1 binding, the measured ΔCp value for the association step was ΔCp,on‡ = −75 cal mol−1 K−1, whereas the dissociation step was an order of magnitude larger, ΔCp,off‡ = 870 cal/mol−1 K−1. This partitioning suggests that the major contribution to effects such as surface burial, conformational change, and solvent reordering occur after the TS. Given our evidence that the two constituents already have a near-native orientation in the TS and the m value data indicate that ≈45% of the contact interface between the proteins is buried at that point, it is hard to reconcile why ΔCp,on‡ is so small compared with its dissociation step counterpart. This would suggest that the effects contained in the equilibrium ΔCp are most affected in the fine-tuning, post-TS stages of binding and not on the initial association process. Functionally, this may be advantageous because any conformational change related to receptor signaling would only occur after a productive hormone–receptor complex was formed, thereby acting as a mechanism to prevent errant signaling.
Model of the Energy Landscape for Binding of Weakly Charged Molecular Interactions.
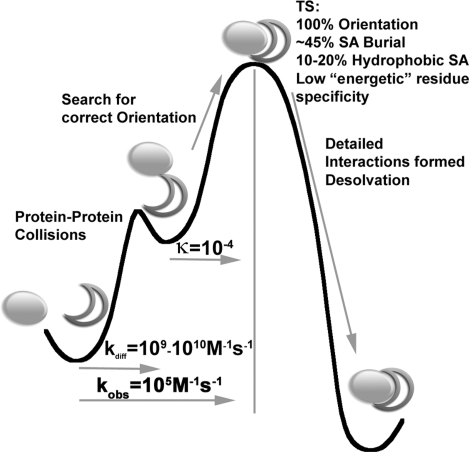
The thermodynamic data described above allow for the generation of a more refined model for the TS structure for weakly charged interactions (Fig. 4). We realize that “weakly charged” is a subjective term and there will be exceptions to any general rules, but the hormone–receptor systems used as a model here have over the years proven to be broadly representative of this class of interactions (8, 23). This model includes the following components: (i) Comparing the on-rates with diffusion controlled interactions, only ≈1 of 104 to 105 collisions are productive. (ii) Denaturant m value analysis indicates a ≈45–50% surface burial at the TS. (iii) φ value analysis determined that no single residue on the hormones alters on-rates, whereas three receptor side chains (W104, W169, and P106) alter it significantly. The two tryptophans form a dominant topographical feature (P106 orients W104) that matches the overall surface terrain of the hormones. A fully comprehensive scan of the hGH interface demonstrated that binding could be improved by changes that optimized the holes for the Trp knobs to fit into (30). (iv) The Zn2+ dependence of kon vs. Keq for hGH–hPRLR binding is a very stringent test that indicates that the molecules are precisely aligned in the TS structure. (v) Eyring analyses show that ΔH, ΔS, and ΔCp values for all hormone–receptor combinations leading to the TS are similar but vary significantly in the dissociation step. (vi) Most of the action happens after the TS. ΔH‡, ΔS‡, and ΔCp‡ are much larger and varied after the TS. It is very clear that in this case the major contributions to the measured equilibrium ΔH and ΔCp components come from the fine-tuning process, not global effects. Taken together with the other data, this suggests that although the precise alignment achieved in the TS is critical, it is only the starting point. The final fine-tuning of the structure after the TS contributes significantly to the thermodynamic character of the interaction.
Fig. 4.
Free-energy diagram of the hGH–receptor binding pathway. The first barrier results in the unbound proteins collide to form an encounter complex after which only a certain fraction (as indicated by the transmission coefficient, T, proceed to the rate-limiting step, the TS. φ, urea, and ψ results presented here suggest that this point along the pathway has very specific orientation requirements. After this point, the post-TS pathway involves the formation of detailed interactions (including those that energetically distinguish the different hGH variants).
Materials and Methods
Protein Expression and Purification.
All proteins were expressed in the periplasm of Escherichia coli BL21 cells as described in ref. 31. All hormones included the G120R mutation to limit hormone receptor binding to a 1:1 stoichiometry. The hGH variants, hGHv, SG1, and SG2 have been described (8, 13, 32). hPRLR was expressed at 20 °C and purified as described in ref. 33.
Surface Plasmon Resonance (SPR).
All SPR experiments were performed on a Biacore 2000 using a CM5 sensor chip. hGHR (29–238) with an engineered cysteine (S237C), allowed site-specific, C-terminal, thiol coupling to the chip surface as described in ref. 8. All urea solutions for m value analysis were prepared in HBS buffer and used within 24 h. Zinc buffers were made with zinc chloride (Puratronic grade, Alfa Aesar) and ranged in concentrations from 5 nM to 5 μM in 10 mM Tris·HCl, 150 mM NaCl, 0.005% Tween 20 (pH 7.4). Additional details of the SPR chip preparation and run conditions are described in SI Materials and Methods.
TS Thermodynamics.
TS thermodynamics for protein–protein association and dissociation (ΔGà, ΔHà, ΔSà, and ΔCpà) were calculated by determining the on- and off-rates as a function of temperature (15–40 °C) through an Eyring analysis. The equations used to determine the TS thermodynamics are presented in SI Materials and Methods.
Denaturant Dependence Analysis.
The m values relate how urea affects the energetics (e.g., ΔGon, ΔGfolding, ΔGeq) of a particular process (e.g., folding or binding). This value has also been used to estimate the amount of surface area that is buried upon the particular process. To determine the m values and the amount of surface area that is buried at the protein–protein TS, the effect of urea (0–0.8M) on ΔGassoc and ΔGeq was determined:
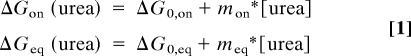 |
where ΔG0,on and ΔG0,eq are association and equilibrium free energies at zero urea and mon and meq are the association and equilibrium m values, respectively. To determine the percentage surface area that is buried at the TS, the ratio of the association and equilibrium m values are determined:
Mutational Analysis of Binding Kinetics.
Data examining the kinetics (association and dissociation) and equilibrium dissociation constant for hGH–hGHR binding have been determined (21, 34). A complete listing of the residues included are presented in SI Materials and Methods. To analyze the contribution of the residue to formation of the protein–protein TS, the ΔΔGon = −RT ln (kon,mut/kon,WT) upon mutation must be compared with the ΔΔGeq upon mutation:
Therefore, φ values were determined by examining:
Supplementary Material
Acknowledgments.
This work was supported by the National Institutes of Health Grants DK61062 (to A.A.K.) and GM55694 (to T.R.S.). J.R.H. was supported by a postdoctoral fellowship from the American Heart Association.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809800106/DCSupplemental.
References
- 1.Janin J. The kinetics of protein–protein recognition. Proteins. 1997;28:153–161. doi: 10.1002/(sici)1097-0134(199706)28:2<153::aid-prot4>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 2.Camacho CJ, Weng Z, Vajda S, DeLisi C. Free energy landscapes of encounter complexes in protein–protein association. Biophys J. 1999;76:1166–1178. doi: 10.1016/S0006-3495(99)77281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frisch C, Fersht AR, Schreiber G. Experimental assignment of the structure of the transition state for the association of barnase and barstar. J Mol Biol. 2001;308:69–77. doi: 10.1006/jmbi.2001.4577. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber G, Fersht AR. Energetics of protein–protein interactions: Analysis of the barnase–barstar interface by single mutations and double mutant cycles. J Mol Biol. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 5.Vijayakumar M, et al. Electrostatic enhancement of diffusion-controlled protein–protein association: Comparison of theory and experiment on barnase and barstar. J Mol Biol. 1998;278:1015–1024. doi: 10.1006/jmbi.1998.1747. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber G, Fersht AR. Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry. 1993;32:5145–5150. doi: 10.1021/bi00070a025. [DOI] [PubMed] [Google Scholar]
- 7.Schreiber G, Fersht AR. Rapid, electrostatically assisted association of proteins. Nat Struct Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 8.Kouadio JL, Horn JR, Pal G, Kossiakoff AA. Shotgun alanine scanning shows that growth hormone can bind productively to its receptor through a drastically minimized interface. J Biol Chem. 2005;280:25524–25532. doi: 10.1074/jbc.M502167200. [DOI] [PubMed] [Google Scholar]
- 9.Harel M, Cohen M, Schreiber G. On the dynamic nature of the transition state for protein–protein association as determined by double-mutant cycle analysis and simulation. J Mol Biol. 2007;371:180–196. doi: 10.1016/j.jmb.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 10.Krantz BA, Dothager RS, Sosnick TR. Discerning the structure and energy of multiple transition states in protein folding using ψ analysis. J Mol Biol. 2004;337:463–475. doi: 10.1016/j.jmb.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 11.Krantz BA, Sosnick TR. Engineered metal binding sites map the heterogeneous folding landscape of a coiled coil. Nat Struct Biol. 2001;8:1042–1047. doi: 10.1038/nsb723. [DOI] [PubMed] [Google Scholar]
- 12.Sosnick TR, Krantz BA, Dothager RS, Baxa M. Characterizing the protein folding transition state using ψ analysis. Chem Rev. 2006;106:1862–1876. doi: 10.1021/cr040431q. [DOI] [PubMed] [Google Scholar]
- 13.Lowman HB, Wells JA. Affinity maturation of human growth hormone by monovalent phage display. J Mol Biol. 1993;234:564–578. doi: 10.1006/jmbi.1993.1612. [DOI] [PubMed] [Google Scholar]
- 14.Horn JR, et al. The role of protein dynamics in increasing binding affinity for an engineered protein–protein interaction established by H/D exchange mass spectrometry. Biochemistry. 2006;45:8488–8498. doi: 10.1021/bi0604328. [DOI] [PubMed] [Google Scholar]
- 15.Dwyer JJ, Dwyer MA, Kossiakoff AA. High affinity RNase S-peptide variants obtained by phage display have a novel“hot spot” of binding energy. Biochemistry. 2001;40:13491–13500. doi: 10.1021/bi011703b. [DOI] [PubMed] [Google Scholar]
- 16.Jaswal SS, Truhlar SM, Dill KA, Agard DA. Comprehensive analysis of protein folding activation thermodynamics reveals a universal behavior violated by kinetically stable proteases. J Mol Biol. 2005;347:355–366. doi: 10.1016/j.jmb.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 17.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 18.Matthews CR. Effects of point mutations on the folding of globular proteins. Methods Enzymol. 1987;154:498–511. doi: 10.1016/0076-6879(87)54092-7. [DOI] [PubMed] [Google Scholar]
- 19.Goldenberg DP. Mutational analysis of protein folding and stability. In: Creighton TE, editor. Protein Folding. New York: Freeman; 1992. pp. 353–403. [Google Scholar]
- 20.Leffler JE. Parameters for the description of transition states. Science. 1953;107:340–341. doi: 10.1126/science.117.3039.340. [DOI] [PubMed] [Google Scholar]
- 21.Cunningham BC, Wells JA. Comparison of a structural and a functional epitope. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 22.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 23.Clackson T, Ultsch MH, Wells JA, de Vos AM. Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity. J Mol Biol. 1998;277:1111–1128. doi: 10.1006/jmbi.1998.1669. [DOI] [PubMed] [Google Scholar]
- 24.Somers W, Ultsch M, De Vos AM, Kossiakoff AA. The X-ray structure of a growth hormone-prolactin receptor complex. Nature. 1994;372:478–481. doi: 10.1038/372478a0. [DOI] [PubMed] [Google Scholar]
- 25.Cunningham BC, Mulkerrin MG, Wells JA. Dimerization of human growth hormone by zinc. Science. 1991;253:545–548. doi: 10.1126/science.1907025. [DOI] [PubMed] [Google Scholar]
- 26.Northrup SH, Erickson HP. Kinetics of protein–protein association explained by Brownian dynamics computer simulation. Proc Natl Acad Sci USA. 1992;89:3338–3342. doi: 10.1073/pnas.89.8.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 28.Murphy KP, Freire E. Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv Prot Chem. 1992;43:313–361. doi: 10.1016/s0065-3233(08)60556-2. [DOI] [PubMed] [Google Scholar]
- 29.Stites WE. Protein–protein interactions: Interface structure, binding, thermodynamics, and mutational analysis. Chem Rev. 1997;97:1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 30.Pal G, Kouadio JLK, Artis DR, Kossiakoff AA, Sidhu SS. Comprehensive and quantitative mapping of energy landscapes for protein–protein interactions by rapid combinatorial scanning. J Biol Chem. 2006;281:22378–22385. doi: 10.1074/jbc.M603826200. [DOI] [PubMed] [Google Scholar]
- 31.Fuh G, et al. The human growth hormone receptor: Secretion from Escherichia coli and disulfide bonding pattern of the extracellular binding domain. J Biol Chem. 1990;265:3111–3115. [PubMed] [Google Scholar]
- 32.Pal G, Kossiakoff AA, Sidhu SS. The functional binding epitope of a high affinity variant of human growth hormone mapped by shotgun alanine-scanning mutagenesis: Insights into the mechanisms responsible for improved affinity. J Mol Biol. 2003;332:195–204. doi: 10.1016/s0022-2836(03)00898-2. [DOI] [PubMed] [Google Scholar]
- 33.Cunningham BC, Bass S, Fuh G, Wells JA. Zinc mediation of the binding of human growth hormone to the human prolactin receptor. Science. 1990;250:1709–1712. doi: 10.1126/science.2270485. [DOI] [PubMed] [Google Scholar]
- 34.Bernat B, Sun M, Dwyer M, Feldkamp M, Kossiakoff AA. Dissecting the binding energy epitope of a high-affinity variant of human growth hormone: Cooperative and additive effects from combining mutations from independently selected phage display mutagenesis libraries. Biochemistry. 2004;43:6076–6084. doi: 10.1021/bi036069b. [DOI] [PubMed] [Google Scholar]
- 35.DeLano WL. San Carlos, CA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.