Abstract
The L1 stalk is a mobile domain of the large ribosomal subunit E site that interacts with the elbow of deacylated tRNA during protein synthesis. Here, by using single-molecule FRET, we follow the real-time dynamics of the L1 stalk and observe its movement relative to the body of the large subunit between at least 3 distinct conformational states: open, half-closed, and fully closed. Pretranslocation ribosomes undergo spontaneous fluctuations between the open and fully closed states. In contrast, posttranslocation ribosomes containing peptidyl-tRNA and deacylated tRNA in the classical P/P and E/E states, respectively, are fixed in the half-closed conformation. In ribosomes with a vacant E site, the L1 stalk is observed either in the fully closed or fully open conformation. Several lines of evidence show that the L1 stalk can move independently of intersubunit rotation. Our findings support a model in which the mobility of the L1 stalk facilitates binding, movement, and release of deacylated tRNA by remodeling the structure of the 50S subunit E site between 3 distinct conformations, corresponding to the E/E vacant, P/E hybrid, and classical states.
Keywords: dynamics, single-molecule FRET, translocation
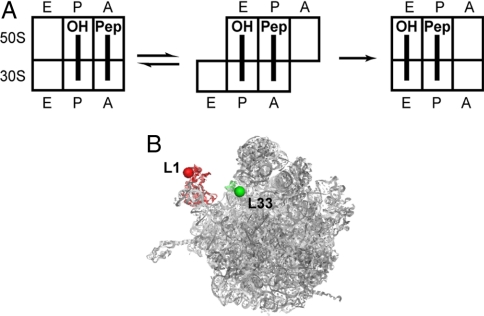
Protein synthesis is a complex multistep process that involves translocation of tRNAs through the ribosome, from the A (aminoacyl) to P (peptidyl) to E (exit) sites, along a path of >100 Å (1). Translocation occurs in separate consecutive steps on the 2 ribosomal subunits. In the first step, after peptide bond formation, the acceptor ends of the tRNAs move from the A and P sites of the 50S subunit to the P and E sites, respectively, whereas the anticodon ends of the peptidyl-tRNA and de acylated tRNA remain in the A and P sites of the 30S subunit, resulting in formation of the A/P and P/E hybrid states (2). Subsequently, the tRNAs move from the A/P and P/E states to the P/P and E/E states, in a step catalyzed by elongation factor (EF) G and GTP (Fig. 1A).
Fig. 1.
Experimental design. (A) Schematic depiction of the movement of deacylated (OH) and peptidyl (Pep) tRNAs through the ribosome from the classical P/P and A/A states (Left) into the hybrid P/E and A/P states (Center) to the classical E/E and P/P states (Right) during one round of translocation. (B) Positions of fluorescent Cy5 and Cy3 dyes attached to cysteines at position 88 of protein L1 (red) and position 29 of protein L33 (green) in the 50S subunit, viewed from the subunit interface side in the crystal structure (15).
The structural dynamics of the ribosome play a central role in translocation. Cryo-EM studies of ribosomes bound with EF-G and a single deacylated tRNA showed an altered conformation, in which the small subunit was rotated counterclockwise with respect to the large subunit, and the unusual orientation of the tRNA suggested that it was bound in the P/E state (3, 4). Combined bulk FRET and chemical probing experiments confirmed that the binding state of the tRNA in the rotated ribosomes is indeed identical to the P/E hybrid state (5, 6). By using single-molecule FRET (smFRET) it was recently demonstrated that pretranslocation ribosomes containing deacylated tRNA in the P site fluctuate spontaneously between 2 rotational conformations corresponding to the classical and hybrid states (7–9). Comparison of cryo-EM and X-ray structures of ribosomal complexes suggests that during translocation intersubunit movement is coupled to additional structural rearrangements within both the small and large subunits (4, 10–12). In particular, movement of a structural feature of the 50S subunit known as the L1 stalk has been implicated in translocation (4, 12, 13).
The L1 stalk, which comprises ribosomal protein L1 and helices 76, 77, and 78 of 23S rRNA, forms the contact with the elbow of tRNA in the E site (14–16). Cryo-EM and X-ray studies show the L1 stalk in at least 3 different orientations relative to the body of the 50S subunit (10). In X-ray structures of ribosomes with a vacant E site or in isolated 50S subunits, the L1 stalk is found in an “open” conformation, extended away from the body of the subunit (11, 17). When deacylated tRNA is bound in the classical E/E state, the stalk moves inward by 30–40 Å, allowing it to contact the elbow of the tRNA (14, 15). In hybrid state complexes, the L1 stalk moves by an additional 15–20 Å relative to its position in the E/E state complex to enable contact with the elbow of P/E tRNA (4, 12).
Recently, spontaneous fluctuations in smFRET between fluorescently labeled tRNA and protein L1 have been interpreted as an indication of inward movement of the L1 stalk on movement of deacylated tRNA into the 50S E site during translocation or spontaneous hybrid state formation (13). No significant differences in tRNA-L1 FRET were observed between the classical E/E and hybrid P/E states. However, interpretation of these experiments is ambiguous because of the attachment of the fluorophores to tRNA and protein L1, both of which are mobile structural elements. Thus, movement of the L1 stalk has yet to be observed by smFRET directly and independently of tRNA movement. Moreover, it remains unclear how many conformational states the L1 stalk can adopt, and what their relative stabilities and rates of interconversion are.
Here, we address these questions by using smFRET between 2 fluorophores attached to proteins L1 and L33 of the 50S subunit, allowing direct observation of motions of the L1 stalk between different functional states of the translation elongation pathway. We found that the L1 stalk moves between at least 3 distinct structural states, relative to the body of the 50S subunit. Movement between these states is controlled by the positions and acylation states of the tRNAs bound to the ribosome. These findings bear on the mechanisms by which tRNAs are moved through and released from the ribosome during the elongation cycle of protein synthesis.
Results
We introduced fluorescent labels by conjugation with unique cysteines created by directed mutagenesis, at position 88 of L1 and (to provide a static landmark in the 50S subunit by which to measure movement of the L1 stalk) position 29 of L33 (Fig. 1B). Protein L1 (labeled with Cy5) and protein L33 (labeled with Cy3) were incorporated by in vitro reconstitution with 50S subunits derived from a double-mutant strain containing precise genomic deletions of the genes for both L1 and L33. This dye pair arrangement avoids convoluting the FRET signal due to L1 stalk movement with the motions of intersubunit rotation or tRNA rearrangement. Functional assays showed that at least 50–60% of purified reconstituted, labeled ribosomes 70S:L1(Cy5)/L33(Cy3) were active in in vitro translocation [supporting information (SI) Fig. S1].
Two different pretranslocation complexes were assembled by binding the peptidyl-tRNA analogue N-Ac-Phe-tRNAPhe to the A site of ribosomes containing either deacylated tRNATyr or tRNAfMet in the P site in the presence of the defined mRNAs m301 or m291, respectively (18) (see Materials and Methods). Pretranslocation ribosome complexes were then immobilized in polymer-passivated microscope slide/coverslip chambers via a biotin-derivatized DNA oligonucleotide annealed to the mRNA (7) and were visualized by using total internal reflection microscopy (19). Both complexes were observed to fluctuate between a low FRET (≈0.25) and a high FRET (≈0.55) state (Fig. 2). These FRET transitions were observed in 34% of traces for pretranslocation complexes containing tRNATyr in the P site and in 58% of traces for ribosomes containing tRNAfMet in the P site. The rest either photobleached quickly or did not show fluctuations. Based on the dye locations and previous biochemical and structural characterization of similar complexes, we infer that ribosome populations primarily in a low-FRET regime correspond to an open state of the L1 stalk with populations primarily in the high-FRET regime in a closed conformation of the L1 stalk in the P/E hybrid state. This interpretation is consistent with previous smFRET studies of intersubunit and tRNA movements, which concluded that under similar conditions pretranslocation ribosomes fluctuate spontaneously between the hybrid and classical states (7, 9, 13).
Fig. 2.
Representative FRET time trajectories of L1 stalk dynamics. Traces show fluorescence intensities observed for Cy3 (green) on ribosomal protein L33 and Cy5 (red) on ribosomal protein L1 and the calculated FRET trace in blue. The fitted HaMMy curves are superimposed on the FRET trajectories in black. Fluorescence intensities are only shown for the complex in A. Complexes were formed from labeled 70S ribosomes containing tRNATyr in the P site and N-Ac-Phe-tRNAPhe in the A site (A), tRNAfMet in the P site and N-Ac-Phe-tRNAPhe in the A site (B), or tRNAfMet in the P site with a vacant A site (C).
SmFRET traces for the pretranslocation complexes were then subjected to hidden Markov model (HMM) analysis to determine whether these data correspond to only 2 states or whether additional intermediate states exist (20). For both complexes, only 2 states were observed (Fig. 2 and Fig. S2). The smFRET data were then used to calculate forward and reverse rates for the following “open-closed” structural rearrangement (Scheme 1). For the pretranslocation complex containing tRNATyr in the P site, the closing rate was 1.41 ± 0.20 s−1 with an opening rate of 0.68 ± 0.15 s−1 (Table 1). For the pretranslocation complex containing tRNAfMet in the P site, the rates were virtually reversed with a closing rate of 0.34 ± 0.01 s−1 and an opening rate of 1.22 ± 0.05 s−1 (Table 1). The slower rate of L1 stalk closing and higher rate of L1 stalk opening in pretranslocation ribosomes containing tRNAfMet in the P site are consistent with the known lower propensity of tRNAfMet to move into the hybrid P/E state (5, 21–23).
Scheme 1.
Table 1.
Rates of transitions between open and closed FRET states
| P site tRNA/A site tRNA | Transitions (k−1) | k1, s−1 | Transitions (k−1) | k−1, s−1 |
|---|---|---|---|---|
| tRNATyr/N-Ac-Phe-tRNAPhe | 5484 | 1.41 ± 0.20 | 5350 | 0.68 ± 0.15 |
| tRNAfMet/N-Ac-Phe-tRNAPhe | 10068 | 0.34 ± 0.01 | 9926 | 1.22 ± 0.05 |
| tRNAfMet/vacant | 5863 | 0.41 ± 0.12 | 5738 | 0.33 ± 0.15 |
These data are a result of fitting FRET time trajectories with the HMM algorithm as described above. Each dataset was divided into 3 and separately analyzed. The reported number is the average from each of the 3 datasets together with the standard deviation.
We next tested the effect of A site occupancy on L1 stalk dynamics by using complexes that contained only a single deacylated tRNA bound to the P site. Transitions between just 2 FRET states were again detected by HMM analysis of data from ribosomes containing tRNAfMet in the P site with a vacant A site (Fig. 2C). A closing rate of 0.41 ± 0.12 s−1 and an opening rate of 0.33 ± 0.15 s−1 were observed (Table 1). Thus, although binding of N-Ac-Phe-tRNAPhe to the A site does not affect the rate of L1 stalk closing, it increases the rate of opening by ≈3.7-fold. Interestingly, in our previous studies on intersubunit rotation, binding N-Ac-Phe-tRNAPhe to the A site of ribosomes containing tRNAfMet in the P site also shifted the equilibrium toward the nonrotated, classical state, although in contrast to L1 stalk movement, the rate of forward (classical to hybrid) rotation of the 30S subunit decreased by ≈2-fold and the rate of reverse rotation was unchanged (9).
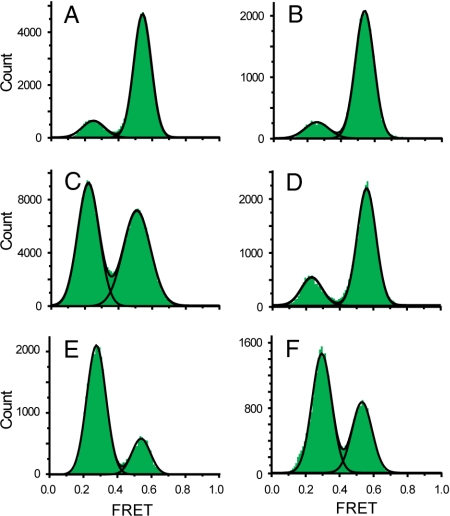
We next tested how EF-G-catalyzed translocation of a pretranslocation complex affects the equilibrium between different conformations of the L1 stalk. FRET histograms built from several hundred traces show that pretranslocation ribosomes containing tRNATyr in the P site and N-Ac-Phe-tRNAPhe in the A site were primarily in the closed (hybrid) state (Fig. 3A) with an associated equilibrium constant of 2.85 (Table S1). Translocation with EF-G and GTP dramatically changed the FRET distribution, shifting the bulk of the complexes from the closed to the open conformation (Fig. 3B). Our previous FRET studies on intersubunit rotation showed that posttranslocation ribosomes are predominantly fixed in the classical (nonrotated) state (9). Deacylated tRNA has been shown to dissociate from the E site after EF-G-dependent translocation (21, 24–26). Our finding that the L1 stalk is found in the open conformation in the majority of posttranslocation ribosomes is consistent with release of the deacylated tRNA from the ribosome after translocation into the E/E state. To saturate the E site of posttranslocation ribosomes with deacylated tRNA bound in the E/E state, we added excess tRNATyr to immobilized complexes containing N-Ac-Phe-tRNAPhe bound to the P site. The distribution of the resulting FRET values were best fitted to 3 Gaussian peaks, in which the majority of complexes were found in a new ≈0.4 FRET state (Fig. 3C). This result suggests that when the L1 stalk interacts with deacylated tRNA bound in the classical E/E state, it adopts an intermediate “half-closed” conformation. We observed a similar distribution of FRET values when complexes containing N-Ac-Phe-tRNAPhe bound to the P site in the presence of m291 mRNA (Fig. 3D) were incubated with an excess of deacylated tRNAfMet (Fig. 3E). Binding of tRNAfMet to the E site, presumably in the E/E state, again stabilized the L1 stalk in the half-closed (≈0.4 FRET) conformation. When ribosomes containing N-Ac-Phe-tRNAPhe bound in the P/P state were treated with puromycin, resulting in deacylation of the peptidyl-tRNA, the L1 stalks of the majority of the ribosomes were converted to the fully closed, hybrid state (≈0.55 FRET) conformation (Fig. 3F), indicating that deacylation of N-Ac-Phe-tRNAPhe resulted in its movement into the hybrid P/E state.
Fig. 3.
FRET histograms for pretranslocation and posttranslocation complexes. (A–C) L1(Cy5)/L33(Cy3)-labeled ribosomes programmed with m301 mRNA, containing (A) tRNATyr in the P site and N-Ac-Phe-tRNAPhe in the A site; (B) as in A with 300 nM EF-G and 250 μM GTP; and (C) as in B with 100 nM tRNATyr added to the imaging buffer. (D–F) FRET histograms for L1(Cy5)/L33(Cy3)-labeled ribosomes programmed with m291 mRNA and containing (D) N-Ac-Phe-tRNAPhe in the P site; (E) N-Ac-Phe-tRNAPhe in the P site with 144 nM tRNAfMet added to the imaging buffer; (F) tRNAPhe in the P site after deacylation of the complex in D with puromycin. (G and H) FRET histograms for S6(Cy5)/L9(Cy3)-labeled ribosomes containing (G) N-Ac-Phe-tRNAPhe in the P site; and (H) N-Ac-Phe-tRNAPhe in the P site with 200 nM tRNAfMet added to the imaging buffer.
In our previous smFRET studies on intersubunit rotation, we showed that S6(Cy5)/L9(Cy3) 70S ribosomes fluctuate between 0.6 and 0.4 FRET states, corresponding to the classical (nonrotated) and hybrid (rotated) states, respectively. Here, we asked whether stabilization of the L1 stalk in its half-closed (E/E state) conformation by filling the E site with deacylated tRNA affects the dynamics of intersubunit rotation, by using our previous S6-L9 construct. An excess of tRNAfMet was added to doubly labeled S6(Cy5)/L9(Cy3) 70S ribosomes containing N-Ac-Phe-tRNAPhe bound in the P/P state. In contrast to the behavior of the L1 stalk, no change in the distribution of intersubunit FRET values was observed, as expected, because the majority of the complexes are predicted to remain in the classical state (compare Fig. 3 G and H). Thus, the L1 stalk is able to move independently of intersubunit rotation.
Movement of the L1 stalk into the half-closed conformation was directly visualized in flow experiments with individual ribosomes (Fig. S3). When tRNAfMet was flowed into sample chambers containing immobilized ribosomes with N-Ac-Phe-tRNAPhe bound in the P/P state in the presence of m291, a FRET transition of the L1 stalk from the open (low FRET) state to the half-closed (0.4 FRET) state was observed (Fig. S3A). Further addition of puromycin induced a subsequent transition from the half-closed state to the fully closed (high FRET) state of the L1 stalk, consistent with movement of the deacylated tRNAPhe into the hybrid P/E state (Fig. S3B).
Because binding of EF-G was shown to stabilize the hybrid state conformation of the ribosome (4, 9, 21), we next tested the effect of its binding on the dynamics of the L1 stalk. Incubation of ribosomes containing deacylated tRNATyr in the P site with EF-G·GDPNP had virtually no effect on the distribution of FRET values (compare Fig. 4 A and B; Table S1). In this case, stabilization of the hybrid state by EF-G is probably masked by the strong propensity of deacylated tRNATyr to move into the hybrid state. Indeed, even in the absence of EF-G, the L1 stalk of 85% of ribosomes containing tRNATyr in the P site occupied the fully closed, hybrid state conformation. Binding of EF-G to ribosomes containing deacylated tRNAfMet in the P site produced a more marked change, giving an increase in the fraction of high-FRET ribosomes from 50% to 79% with a slight shift in the high FRET peak (Fig. 4 C and D; Table S1). The effect of EF-G binding depended strongly on the presence of P site tRNA; binding of EF-G·GDPNP to vacant ribosomes only marginally affected the FRET distribution (Fig. 4 E and F). These results support the conclusions drawn from FRET studies of intersubunit movement (9) that both the movement of the acceptor stem of deacylated tRNA into the 50S E site and EF-G binding to the ribosome contribute to stabilization of the rotated, hybrid state and (in this work) the closed L1 stalk conformation.
Fig. 4.
FRET histograms for L1(Cy5)/L33(Cy3) labeled ribosomes containing tRNATyr in the P site (A); tRNATyr in the P site with 300 nM EF-G and 255 μM GDPNP (B); tRNAfMet in the P site (C); tRNAfMet in the P site with 300 nM EF-G and 255 μM GDPNP (D); no tRNA (E); and no tRNA with 300 nM EF-G and 255 μM GDPNP (F).
Discussion
The dynamics of the L1 stalk of the ribosome have previously been studied by using changes in FRET between labels attached to protein L1 and tRNA (13); interpretation of these experiments is complicated because both fluorescent probes are connected to mobile elements of the ribosome complex. Our smFRET experiments, in which both fluorescent labels were attached directly to the ribosome, at protein L1 and at protein L33 (which resides in the main body of the 50S subunit; Fig. 1), provide unambiguous observation of the dynamics of the L1 stalk. These data reveal movement of the stalk between at least 3 distinct positions relative to the body of the 50S subunit. We assign the low (≈0.25) FRET state to the open conformation seen in vacant ribosomes (11) and free 50S subunits (17), in which the L1 stalk is maximally rotated away from the body of the subunit (Fig. 5A), presumably facilitating release of deacylated tRNA from the ribosome. The intermediate (≈0.4) FRET state corresponds to the position of the stalk in the classical state most often observed in cryo-EM (27) and X-ray structures (14–16), in which the L1 stalk moves inward to make contact with the elbow of the deacylated tRNA bound in the classical, E/E state (Fig. 5B). The high (≈0.55) FRET state is observed under conditions where the deacylated tRNA is found in the P/E hybrid state, most likely reflecting the furthest inward excursion made by the L1 stalk (Fig. 5C), enabling it to reach the elbow of the P/E tRNA (4, 28, 29). Thus, movement of the L1 stalk results in remodeling of the ribosomal E site, adapting its structure to 3 different functional states of the elongation cycle.
Fig. 5.
Three positions of the L1 stalk, corresponding to 3 functional states of the ribosomal E site. (A) Open state of the L1 stalk with vacant E site; (B) half-closed state of the L1 stalk with deacylated tRNA in the classical E/E state; (C) fully closed state of the L1 stalk with tRNA in the hybrid P/E state. The deacylated tRNA is shown in red; the position of the half-closed E/E state is shown by a dashed green outline.
In previous studies, we observed spontaneous fluctuations between the 2 states of intersubunit rotation associated with translocation, in the absence of EF-G or GTP (9). Here, we show that the L1 stalk of pretranslocation ribosomes fluctuates spontaneously between the fully closed and open conformations. Both modes of movement are associated with tRNA translocation, and indeed, the equilibria between the closed and open states for the various complexes correlate with those of the rotated vs. nonrotated states (Fig. 6A). However, it is not known whether these 2 structural rearrangements are coupled or occur independently. Comparison of forward and reverse rates obtained for both intersubunit rotation and L1 stalk movement bears on this question (Fig. 6). We obtained a sufficient number of time traces in this work to calculate rates of L1 stalk movement for three of our complexes. Forward rates for the 2 processes are similar (correlation coefficient = 0.97; Fig. 6B), indicating that spontaneous closing of the L1 stalk and counterclockwise intersubunit rotation (classical to hybrid state transition) occur at similar rates, consistent with the possibility that these 2 motions might be coupled. In contrast, the rates of opening of the L1 stalk and clockwise rotation (hybrid to classical state transition) are uncorrelated (Fig. 6C); opening of the L1 stalk occurs at consistently faster rates than reverse intersubunit rotation. Coupling of the forward processes seem reasonable, because formation of the contacts between the L1 stalk and the elbow of the P/E hybrid state tRNA occur after intersubunit rotation. Opening of the L1 stalk, however, requires that the L1 stalk break its interactions with tRNA, an event that may depend on the identity of the tRNA. A further indication that movement of the L1 stalk can be uncoupled from intersubunit rotation is that the L1 stalk can be observed in 3 distinct conformations in posttranslocation ribosomes, which are fixed predominantly in the nonrotated, classical state (Fig. 3 C and E).
Fig. 6.
Correlations between equilibria and rates of intersubunit rotation and L1 stalk movement. (A) Correlation between equilibrium constants obtained from smFRET measurements of intersubunit rotation (9) and L1 stalk movement (this work; Table S1). The dashed lines at Keq = 1 divide the plot into 4 quadrants corresponding to the 4 possible combinations of nonrotated and rotated orientations of the subunits and fully closed and open conformations of the L1 stalk. Filled squares correspond to complexes of pretranslocation ribosomes containing deacylated tRNA in the P site (tRNATyr, tRNAPhe, or tRNAfMet). Open circles correspond to posttranslocation ribosomes containing N-Ac-Phe-tRNAPhe in the P site and a vacant E site. Open triangles correspond to vacant ribosomes with or without EF-G·GDPNP bound. (B) Correlation between forward rates: closing of the L1 stalk vs. rotation of subunits from classical to hybrid state. (C) Correlation between reverse rates: opening of the L1 stalk vs. rotation of subunits from hybrid to classical state. Lines represent log-linear fits of the data.
The rates of the observed fluctuation of the L1 stalk determined here (0.3–1.22 s−1 for complexes containing tRNAfMet in the P site) are similar to those previously determined for spontaneous intersubunit movement in the same complexes (0.2–0.5 s−1) and are marginally slower than the rates of L1 stalk fluctuation inferred from FRET measurements between protein L1 and tRNA (0.6–5 s−1) (13) or rates of fluctuation of 2 labeled tRNAs between the hybrid and classical states (1–5 s−1) (7, 8, 30). Such differences are most likely due to variations in experimental conditions and constructs used; for example, the use of reconstituted ribosomes in our experiments, fluorescent labeling of tRNAs in the studies by Kim et al. (2007), Munro et al. (2007), and Fei et al. (2008), and different ionic conditions could all affect the observed frequencies.
Further evidence for the independence of L1 stalk and intersubunit movement comes from the different effects of filling the E site of classical state complexes on these 2 processes. After the movement of the L1 stalk, we were able to enrich for populations of half-closed (≈0.4 FRET) complexes by addition of excess deacylated tRNA to classical state (≈0.25 FRET) complexes containing a vacant E site (Fig. 3 C and E). In contrast, when we monitored intersubunit rotation, addition of excess deacylated tRNAfMet to complexes containing N-Ac-Phe-tRNAPhe bound in the P/P state had no effect on the distribution between the rotated and nonrotated conformations of the ribosome (Fig. 3 G and H). It is noteworthy that decoupling of intersubunit rotation and L1 stalk movement shows that previously observed changes in FRET between S6 and L9 cannot be explained by local conformational changes in the L1 stalk, as suggested by Marshall et al. (31).
Three conformational states of the L1 stalk were not detected in previous smFRET experiments employing energy transfer between fluorophores attached to tRNA and L1 (13). This can probably be explained by the fact that the L1 stalk contacts the deacylated tRNA in both the P/E and E/E states (8, 12, 20), and therefore, fluorophores attached to tRNA and L1 would be expected to have similar intermolecular distances in both states.
The mobility of the L1 stalk enables a dynamic remodeling of the structure of the ribosomal E site during translocation. The closed and half-closed states observed here make possible continuous contact between the head of the L1 stalk and the elbow of tRNA during transitions between classical and hybrid states despite the relatively rigid structure of tRNA, and the open state allows escape of deacylated tRNA from its enclosure by the E site. Although both protein and RNA moieties of the stalk participate in contacts with tRNA, its mobility appears to be based on the inherent flexibility of its RNA stem (10, 14). The evolutionary persistence of RNA as a major structural component of ribosomes may, in part, be a reflection of its dynamic capabilities.
Materials and Methods
Materials and Sample Preparation.
tRNAfMet was purchased from MP Biomedicals; GTP, GDPNP, puromycin, tRNAPhe, and tRNATyr were purchased from Sigma. Defined mRNA m291 and m301 were transcribed in vitro and further purified as described in ref. 32. The biotin-labeled DNA primer (5′ biotin-CTTTATCTTCAGAAGAAAAACC-3′) was synthesized by Integrated DNA Technologies. NeutrAvidin used for sample immobilization at a final concentration of 0.2 mg/mL was purchased from Pierce. N-Ac-Phe-tRNAPhe and EF-G with a 6-histidine tag were prepared and purified as described (2, 32, 33). The components of the oxygen-scavenging system (glucose oxidase from Aspergillus niger, glucose and 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid) were purchased from Sigma. Catalase from beef liver was from Roche.
Construction of the double-mutant strain ΔL1ΔL33, containing precise genomic deletions for proteins L1 and L33, was done by using the method of Church and coworkers (33) as previously described for deletion of the C-terminal tails of proteins S9 and S13 (34). First, to create the L1 deletion, the chromosomal DNA from Escherichia coli strain CSH142 corresponding to gene rplA, which codes for protein L1, and 1 kb of flanking sequence was cloned into plasmid pKO3 (33) by PCR to create pLH30. Quickchange (Strategene) mutagenesis was used to delete the gene from the pLH30 plasmid to create pLH33, leaving only the 1 kb of flanking sequence; this construct was confirmed by DNA sequencing. pLH33 contains a temperature-sensitive origin of replication and a CamR gene from the parental pKO3 plasmid. CSH142 transformed with pLH33 was subjected to selection by growth on chloramphenicol plates at 42 °C for bacteria in which pLH33 has integrated into the chromosome by a recombination event. The chromosome of the resulting strain, LH104, contains a wild-type copy of rplA, a mutant version of the gene where the rplA gene has been deleted, and the rest of the pKO3 plasmid in between the 2 genes, as confirmed by PCR. The integration event can occur on the chromosome either upstream or downstream of the wild-type rplA gene at equal frequencies. A third gene contained in pLH33 from the pKO3 parental plasmid, sacB, is a counterselective marker that is lethal to E. coli when grown on sucrose and used to select for a recombination event that removes pLH33 from the chromosome. Surviving colonies are screened by PCR for the deletion of the L1 gene. In the same way, the gene rpmG, coding for protein L33, was deleted from CSH142 to generate strain ΔL33. Precise deletions of rplA and rpmG from strains ΔL1 and ΔL33, respectively, were confirmed by DNA sequencing. P1 transduction was then used to move the L33 deletion into the ΔL1 strain to create the double-mutant strain ΔL1ΔL33.
Tight-couple 70S ribosomes were prepared from wild-type E. coli MRE600 and ΔL1ΔL33 strains, respectively, as described in ref. 35. Mutant variants of ribosomal proteins L33 (T29C) and L1 (A88C) were created by site-directed mutagenesis, expressed, purified, and labeled separately with maleimide derivatives of Cy3 (donor) or Cy5 (acceptor) dyes (Amersham Biosciences) as described in ref. 9. Labeled protein L1 and L33 were incorporated into 70S ribosome subunits by partial reconstitution by incubation for 15 min at 4 °C in a buffer containing 20 mM Hepes·KOH (pH 7.5), 4 mM MgCl2, 330 mM NH4Cl, 6 mM β-mercaptoethanol, and 0.1% Nikkol. Unbound proteins were removed by using YM-100 Centricons (Millipore). To purify reconstituted ribosomes from possible contamination with endogenous tRNA the 70S ribosomes were dissociated in 20 mM Hepes·KOH (pH 7.5), 1 mM MgCl2, 100 mM NH4Cl, 6 mM β-mercaptoethanol, and 0.1% Nikkol and washed with the same buffer by using YM-100 Centricons (Millipore). Then buffer was exchanged for association buffer containing 20 mM Hepes·KOH (pH 7.5), 20 mM MgCl2, 100 mM NH4Cl, 6 mM β-mercaptoethanol, 0.1% Nikkol, and 2 mM spermidine. Ribosomal subunits were activated for 10 min at 42 °C and reassociated for 10 min at 37 °C. Finally, association buffer was exchanged for storage buffer containing 20 mM Hepes·KOH (pH 7.5), 6 mM MgCl2, 150 mM NH4Cl, 6 mM β-mercaptoethanol, 2 mM spermidine, and 0.1 mM spermine by using YM-100 Centricons (Millipore). Preparation of S6(Cy5)/L9(Cy3) 70S ribosomes was done as described in ref. 9.
All ribosome samples were prepared for visualization in a buffer containing 20 mM Hepes·KOH (pH 7.5), 6 mM MgCl2, 150 mM NH4Cl, 6 mM β-mercaptoethanol, 2 mM spermidine, and 0.1 mM spermine. Imaging buffer was identical except for addition of an oxygen-scavenging system consisting of 0.8 mg/mL glucose oxidase, 0.625% dextrose, ≈1.5 mM 6-hydroxy-2,5,7,8-tetramethyl-chromane-2-carboxylic acid (Trolox), and 0.03 mg/mL catalase to prevent photobleaching during data acquisition. All ribosome samples were assembled as described in ref. 9 or in the text. m301 (18) was used for complexes containing tRNATyr in the P site. For all other complexes, m291 (32) was used.
Data Acquisition and Analysis.
Experiments were performed at room temperature (23 °C) by using total internal reflection fluorescence (TIRF) microscopy as described in detail in ref. 9. Data were acquired by using in-house software, processed by using IDL, and analyzed by using Matlab and Origin. All data were acquired with 100 ms time binning. Time trajectories used in histograms were truncated to remove photobleaching and blinking events and were smoothed with a 5-point window. Trajectories analyzed by using HaMMy (20) were also truncated to remove photobleaching and blinking events and fit to 3 FRET states.
Supplementary Material
Acknowledgments.
We thank John Paul Donohue for help with figure preparations and Andrei Korostelev and Laura Lancaster for discussions. This work was supported by National Institutes of Health Grant GM-17129 and National Science Foundation (NSF) Grant MCB-0212689 (to H.F.N.), NSF Physics Frontiers Grant 0822613 through the Center for the Physics of Living Cells (to T.H), American Cancer Society Postdoctoral Fellowship PF-07-123-01-GMC (to P.V.C.), and a NATO-NSF postdoctoral fellowship (D.N.E.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813180106/DCSupplemental.
References
- 1.Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Cate JH. Translocation of tRNA during protein synthesis. FEBS Lett. 2002;514:11–16. doi: 10.1016/s0014-5793(02)02327-x. [DOI] [PubMed] [Google Scholar]
- 2.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 3.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 4.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 5.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 6.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 7.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12(6):674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 12.Gao H, et al. Study of the structural dynamics of the E coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 13.Fei J, Kosuri P, MacDougall DD, Gonzalez RL. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 15.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 16.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 17.Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 18.Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 19.Zhuang X, et al. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 20.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 24.Semenkov YP, Rodnina MV, Wintermeyer W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc Natl Acad Sci USA. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spirin AS. Testing the classical two-tRNA-site model for the ribosomal elongation cycle. FEBS Lett. 1984;165:280–284. doi: 10.1016/0014-5793(84)80186-6. [DOI] [PubMed] [Google Scholar]
- 26.Robertson JM, Wintermeyer W. Mechanism of ribosomal translocation. tRNA binds transiently to an exit site before leaving the ribosome during translocation. J Mol Biol. 1987;196:525–540. doi: 10.1016/0022-2836(87)90030-1. [DOI] [PubMed] [Google Scholar]
- 27.Agrawal RK, et al. Effect of buffer conditions on the position of tRNA on the 70 S ribosome as visualized by cryoelectron microscopy. J Biol Chem. 1999;274:8723–8729. doi: 10.1074/jbc.274.13.8723. [DOI] [PubMed] [Google Scholar]
- 28.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julian P, et al. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci USA. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HD, Puglisi J, Chu S. Fluctuations of tRNAs between classical and hybrid states. Biophys J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci USA. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 33.Link AJ, Phillips D, Church GM. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: Application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoang L, Fredrick K, Noller HF. Creating ribosomes with an all-RNA 30S subunit P site. Proc Natl Acad Sci USA. 2004;101:12439–12443. doi: 10.1073/pnas.0405227101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hickerson R, Majumdar ZK, Baucom A, Clegg RM, Noller HF. Measurement of internal movements within the 30 S ribosomal subunit using Forster resonance energy transfer. J Mol Biol. 2005;354:459–472. doi: 10.1016/j.jmb.2005.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.