Abstract
Hirschsprung's disease (HSCR), or aganglionic megacolon, is a congenital disorder characterized by the absence of enteric ganglia in variable portions of the distal intestine. RET is a well-established susceptibility locus, although existing evidence strongly suggests additional loci contributing to sporadic HSCR. To identify these additional genetic loci, we carried out a genome-wide association study using the Affymetrix 500K marker set. We successfully genotyped 293,836 SNPs in 181 Chinese subjects with sporadic HSCR and 346 ethnically matched control subjects. The SNPs most associated with HSCR were genotyped in an independent set of 190 HSCR and 510 control subjects. Aside from SNPs in RET, the strongest overall associations in plausible candidate genes were found for 2 SNPs located in intron 1 of the neuregulin1 gene (NRG1) on 8p12, with rs16879552 and rs7835688 yielding odds ratios of 1.68 [CI95%:(1.40, 2.00), P = 1.80 × 10−8] and 1.98 [CI95%:(1.59, 2.47), P = 1.12 × 10−9], respectively, for the heterozygous risk genotypes under an additive model. There was also a significant interaction between RET and NRG1 (P = 0.0095), increasing the odds ratio 2.3-fold to 19.53 for the RET rs2435357 risk genotype (TT) in the presence of the NRG1 rs7835688 heterozygote, indicating that NRG1 is a modifier of HSRC penetrance. Our highly significant association findings are backed-up by the important role of NRG1 as regulator of the development of the enteric ganglia precursors. The identification of NRG1 as an additional HSCR susceptibility locus not only opens unique fields of investigation into the mechanisms underlying the HSCR pathology, but also the mechanisms by which a discrete number of loci interact with each other to cause disease.
Keywords: GWA, RET
Hirschsprung's disease (HSCR, aganglionic megacolon) is a developmental disorder characterized by the absence of the enteric ganglia along a variable length of the hindgut, leading to tonic contraction of the affected segment, intestinal obstruction, and massive distension of the bowel. Aganglionosis is attributed to a failure in the time-specific migration of enteric neural crest-derived cells into the intestinal tract. There is significant racial variation in the incidence of the disease, and it is most often found among Asians (2.8 per 10,000 live births) (1, 2). HSCR presents mainly sporadically, although familial aggregation also exists (5–20% of cases) and manifests with low, sex-dependent penetrance and phenotypic variability. HSCR patients can be classified according to the length of the aganglionic segment into short segment (S-HSCR, 80%), long segment (L-HSCR, 15%), and total colonic aganglionosis (TCA, 5%). The male:female ratio (M:F) is ≈4:1 among S-HSCR patients and ≈1:1 among L-HSCR patients. The recurrence risks to siblings vary from 1.5 to 33%, depending on the gender and the length of the aganglionic segment in the proband and the gender of the sibling.
The RET gene, encoding a tyrosine-kinase receptor, is the major HSCR gene and its expression is crucial for the development of the enteric ganglia (3, 4). Mutations in the coding sequence (CDS) of RET account for up to 50% of the familial cases and between 15 and 20% of the sporadic cases, indicating that additional HSCR-causing mutations exist. Other HSCR genes identified so far mainly code for protein members of interrelated signaling pathways involved in the development of enteric ganglia: RET, endothelin receptor B (EDNRB), and the transcriptional regulator Sox10 signaling pathways. Rare mutations in CDS of genes other than RET account for 7% of the cases (5–12). Reduced penetrance and phenotypic variability of mutations in these genes imply that the effect of a mutation is modulated by other loci (modifiers) and that more than one gene is required for the disease manifestation. The current data indicate that RET HSCR-associated single nucleotide polymorphisms (SNPs) could act as modifiers in the RET gene itself, explaining the variability observed between intrafamilial cases of sporadic HSCR patients carrying the same mutation. Also, RET SNPs act as low-penetrance alleles conferring risk. Indeed, common RET SNPs and haplotypes are strongly associated with HSCR (13–16), with the largest contribution to risk made by a functional SNP (rs2435357) lying in an enhancer-like sequence in intron 1 (17). This common mutation has low penetrance, a sex-dependent effect, and explains only a small fraction of the HSCR cases. Noteworthy, the overall frequencies of the HSCR-associated RET SNPs and haplotypes are significantly higher in the Chinese population, not only in patients but also in the general population (17, 18), possibly explaining the higher incidence of HSCR in Asians when compared to Caucasians (1). In addition, population differences in the composition of the RET haplotype at the 3′ end of the gene exist (18, 19).
HSCR is viewed as an oligomeric disorder that, with few exceptions, requires RET and other disease susceptibility alleles for its manifestation. Thus far, a few susceptibility loci for familial aggregation have been identified at the 9q31 (20), 3p21, and 19q12 (21) loci by linkage analysis on non-Chinese individuals. Despite these important findings and the unequivocal implication of RET in HSCR risk, it is likely that additional common genetic variants with a modest effect remain to be discovered to explain the complexity of the disease and, in particular, its sporadic manifestation. Thus, we have conducted a genome-wide association (GWA) study on sporadic HSCR patients, the commonest form of the disease.
Results
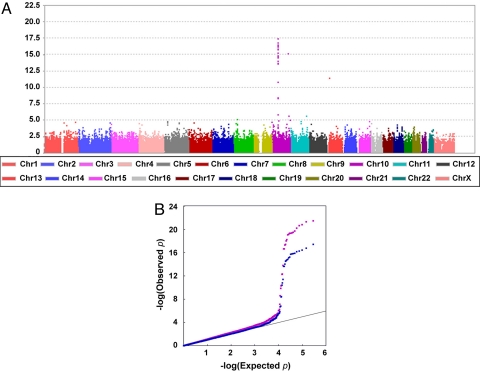
After stringent quality control, the association analysis was conducted on 293,836 SNPs in 181 HSCR patients (10 with TCA, 8 with L-HSCR, 162 with S-HSCR, and 1 undetermined) and 346 controls. A total of 19 HSCR and 62 control samples were excluded after checks for contamination, false identity, non-Chinese ancestry, and relatedness [see supporting information (SI) Text]. Of the SNPs, 293,836 passed the quality-control filters, with an average call rate of 98.69%. The Cochran-Armitage trend test was used to assess the association of the SNPs with disease status after correction for stratification. There was little evidence of stratification in our sample, as noted by the Cochran-Armitage trend test χ2 distribution conforming to the null hypothesis (median λ = 1.14), and most of the initially associated peaks persisted after EIGENSTRAT correction (Fig. 1A), with the mean χ2 dropping to 1 (median λ = 1.01), indicating the effectiveness of the EIGENSTRAT correction. On visual inspection of the plots, additional SNPs apparently showing strong association were filtered out because of gross incompatibilities with the patterns of linkage disequilibrium in the Chinese Han population (CHB) HapMap sample.
Fig. 1.
Results of the Genomewide Association Study. (A) Cochran-Armitage significance test results after EIGENSTRAT correction for population stratification. (B) Q-Q-plot revealing deviation of association from expected, starting from P = 5 × 10−4 (purple before and blue after EIGENSTRAT correction).
In line with previous studies, the strongest association was found at the RET locus in the 10q11 region (rs2742234; P = 3.92 × 10−18) and extended ≈150 Kb downstream, encompassing the GALNACT-2 and RASGEF1A genes. Given the well-known strong effect of the RET intron 1 functional variant (rs2435357) on HSCR susceptibility (17), the extent of the region showing association indicates that even distant SNPs in modest linkage disequilibrium (LD) will show indirect evidence for association. This may be more pronounced in the Chinese population in which LD at the 3′ end of RET extends further than in Caucasians.
On examination of the Q-Q plot (Fig. 1B), we see that those P-values less than 5 × 10−4 have deviated from that expected by chance, and correspond to 195 SNPs (including those removed after visual inspection of the plot). Among these, we prioritized markers according to their biological plausibility. No SNP on or nearby the other previously identified HSCR genes (2) or RET modifiers (20, 21) was identified, although we note that the latter were identified in non-Chinese populations. In addition, this probably reflects the fact that only rare deleterious mutations have been identified in HSCR genes or that the sample size is not large enough.
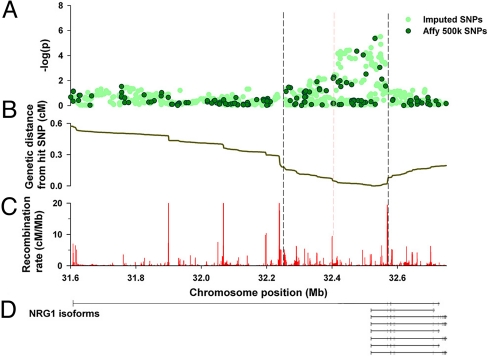
The markers mapping to the promoter and intron 1 regions of the neuregulin gene (NRG1) on 8p12 were our first choice because this gene is known to have functional implications in the enteric nervous system (ENS) (22–24). The most strongly associated marker within the NRG1 region was rs16879552 (P = 7.43 × 10−6), followed by rs7835688 (P = 4.77 × 10−5) just 283 bp downstream (Table 1). As expected, nearby SNPs also displayed association, which extended as far as the recombination hotspots on either side (Fig. 2). To help identify additional putative disease loci, the program IMPUTE was used to generate in silico genotypes for those SNPs in the associated NRG1 region that are not present on the Affymetrix chips. These in silico genotypes were then tested for association, but no more prominent loci were detected by this method (see Fig. 2A). Association peaks of similar magnitude as that of NRG1 were found in other chromosomal regions although none fell in any plausible candidate gene, so we chose not to follow them up at this stage (see Tables S1 and S2).
Table 1.
NRG1 SNPs associated with HSCR
| Chromosome | ID | Position | Strand | Associated allele | Other allele | Before correction |
After correction |
||
|---|---|---|---|---|---|---|---|---|---|
| χ2 | P | χ2 | P | ||||||
| 8p12 | rs16879552 | 32530758 | + | G | A | 19.50 | 1.01 × 10−5 | 20.08 | 7.43 × 10−6 |
| 8p12 | rs7835688 | 32531041 | − | C | G | 25.07 | 5.53 ×10−7 | 16.54 | 4.77 × 10−5 |
P-values obtained before and after correction for stratification are shown. P values represented in this table were obtained using Cochran-Armitage trend test.
Fig. 2.
(A) A −log(p) value for GWA NRG1 SNPs (dark green) and imputed SNPs (light green) under the additive model of association. (B) The cumulative recombination rate (genetic distance) measured away from the most highly associated genotyped SNP. (C) The fine-scale recombination rate across the region. The vertical dashed lines on the plot delineate the main region of association. (D) Schematic representation of the NRG1 isoforms overlapping the associated region. Boxes represent exons and fine lines introns. Notice that exon 1 is in the associated region.
NRG1 SNP Associations Are Confirmed in an Independent Sample and in Combined Analysis.
To validate the finding from the whole-genome scan, we genotyped the NRG1 SNPs in 190 HSCR additional cases (7 with TCA, 29 with L-HSCR, 125 with S-HSCR, and 29 undetermined) and 510 additional controls (see SI Text). In Table 2 we present tests of NRG1 SNPs using logistic regression under an additive model, both for the GWA and follow-up stages. The data strongly suggest that the initial finding was genuine, as both NRG1 SNPs showed strong and significant association with HSCR in the independent sample, with P = 4 × 10−4 for rs16879552 and P = 3.85 × 10−4 for rs7835688. More importantly, when follow-up and genome-wide scan samples were analyzed together, the results showed very strong evidence for association with P = 1.80 × 10−8 for rs16879552 and P = 1.12 × 10−9 for rs7835688, which are significant after a conservative Bonferroni genome-wide correction for multiple testing of 500,000 markers (see Table 2), which would require P < 0.05/500,000, or P < 10−7. Homogeneity between GWA and replication samples was evaluated by testing the interaction between sample (GWA vs. follow-up) and NRG1 (under an additive model using logistic regression). We found no significant difference for either SNP (P = 0.38 for rs16879552 and P = 0.21 for rs7835688).
Table 2.
Summary of the statistics for the NRG1 SNPs association
| NRG1 SNPs | Alleles |
Phase | N |
MAFs |
P | OR (95%CI) | |||
|---|---|---|---|---|---|---|---|---|---|
| Associated | Other | Cases | Controls | Cases | Controls | ||||
| rs16879552 | G | A | |||||||
| 500K | 181 | 343 | 0.5331 | 0.3921 | 1.23 × 10−5 | 1.82 (1.39,2.39) | |||
| Replication | 190 | 507 | 0.4974 | 0.3925 | 4.00 × 10−4 | 1.55 (1.22,1.98) | |||
| Combined | 371 | 850 | 0.5148 | 0.3924 | 1.80 × 10−8 | 1.68 (1.40,2.00) | |||
| rs7835688 | C | G | |||||||
| 500K | 179 | 343 | 0.2682 | 0.1356 | 4.95 × 10−7 | 2.31 (1.67,3.21) | |||
| Replication | 191 | 510 | 0.2330 | 0.1529 | 3.85 × 10−4 | 1.73 (1.28,2.35) | |||
| Combined | 370 | 853 | 0.25 | 0.1460 | 1.12 × 10−9 | 1.98 (1.59,2.47) | |||
N, Number of individuals successfully genotyped; MAF, Minor allele frequency; CI, confidence interval; P values and odd ratios (OR) represented in this table were obtained from fitting an additive model using logistic regression; genotyping rates for rs16879552 were 100% for HSCR patients and 99.13% for controls, and for rs7835688 were 98.90% for HSCR patients and 99.13% for controls.
We then used logistic regression of disease status on the NRG1 SNP genotypes of the combined sample to determine which genetic model best fits the observed data (see Methods). The additive (1 df) model fit the data as well as the unconstrained (2 df) model (difference χ2df = 1 = 0.7, P > 0.4 for rs16879552 and χ2df = 1 = 2.7, P > 0.1 for rs7835688), with an odds ratio (OR) under the additive model for rs16879552 of 1.68 [P = 1.19 × 10−8; CI95%:(1.40, 2.00)] for a single copy of the risk allele (G) and of 1.98 [P = 1.07 × 10−9, CI95% = (1.59, 2.47)] for rs7835688, for a single copy of the risk (C) allele. Neither dominant (χ2df = 1 = 14.9, P < 0.0002 for rs16879552 and χ2df = 1 = 7.9, P < 0.005 for rs7835688) nor recessive (χ2df = 1 = 12.4, P < 0.0005 for rs16879552 and χ2df = 1 = 17.3, P < 4 × 10−5 for rs7835688) models yielded an adequate fit when tested against the unconstrained models.
Combined samples were also used to carry out haplotype analysis of these two NRG1 SNPs. The overall haplotype distribution was significantly different between cases and controls (P = 9.84 × 10−11), and the effect of the haplotype comprising the risk alleles was only slightly higher (P = 9.32 × 10−10) than that of the individual SNPs.
Genetic Properties of NRG1 Susceptibility Alleles.
This study has failed to identify chromosome X-linked genes that could account for the sex bias in severity and incidence of the disease. Sex-specific effects (transmission frequency is higher to affected males than to affected females) have nevertheless been described for the RET 1 enhancer mutation rs2435357 (17). Genotype frequencies were used to estimate the penetrance of the NRG1 SNPs in males and females (see Methods). Although higher penetrance was observed in males when compared to females, the genotypic relative risk of neither NRG1 SNP was significantly different between sexes (P = 0.26 for rs16879552 and P = 0.96 for rs7835688; Table S3). Differences in penetrance between sexes are expected because more males are affected than females. Similarly, the genotypic effect did not differ with the length of the aganglionic segment (Table S4).
We then explored the combined effects of NRG1 SNPs and the RET intron 1 rs2435357 SNP previously described, using all 321 patients and 507 controls from the combined GWA and replication samples. Considering only the RET SNP in this combined sample, and fitting a recessive genetic model (TT vs. CT+CC, where T represents the HSCR risk allele), the risk of HSCR in the presence of TT was very large [9.27; CI95%:(6.75,12.74)] and highly significant (P = 2.96 × 10−49). Table 3 presents the odds ratios for the genotypic combinations of the two risk genes (RET and two NRG1 SNPs), using logistic regression to model the interaction between NRG1 rs7835688 (under an additive model) and RET rs2435357 (under the recessive model described) (see Tables S5 and S6). We see that neither NRG1 SNP conveys significantly increased risk in the presence of the low-risk RET genotype. However, the interaction between RET and NRG1 rs7835688 was significant (P = 0.0095), increasing the odds ratio 2.3-fold to 19.53 for the RET rs2435357 risk genotype (TT) in the presence of the NRG1 heterozygote, as compared to the predicted risk when no interaction was modeled (independent risks of 6.41 for RET × 1.34 for NRG1 = 8.59). In the case of NRG1 rs16879552, the interaction was not significant (P = 0.10), although the odds ratio still increased 1.5-fold for the combined effects of RET and NRG1. Performing a genome-wide scan conditional on the known RET variants may have been desirable, although it would only appreciably influence the obtained P value if the RET variant were associated with the SNP tested. Because of the small number of individual CT or CC, we have limited power to detect an effect of NRG1 in the absence of the RET risk allele.
Table 3.
ORs (relative to doubly low risk genotypes) of individuals with RET rs2435357, under recessive model and NRG1 (rs16879552, rs7835688) genotypes under additive model.
| RET risk | rs16879552 |
rs7835688 |
||||
|---|---|---|---|---|---|---|
| AA | GA | GG | GG | CG | CC | |
| rs2435357 (TT) | 5.89 (3.44, 10.09) | 12.08 (7.64, 19.11) | 24.77 (13.79, 44.51) | 6.41 (4.31, 9.53) | 19.53 (11.85, 32.17) | 59.50 (24.59, 143.94) |
| rs2435357 (CT and CC) | 1 | 1.39 (0.99, 1.95) | 1.93 (0.97, 3.82) | 1 | 1.34 (0.87, 2.06) | 1.80 (0.76, 4.25) |
95%CI in parenthesis.
Combined samples were also used to carry out haplotype analysis of these two NRG1 SNPs. The overall haplotype distribution was significantly different between cases and controls (P = 9.84 × 10−11) and the effect of the haplotype comprising the risk alleles was slightly higher (P = 9.32 × 10−10) than that of the individual SNPs. High D' (0.978) but moderate r2 (0.273) LD values between these two NRG1 markers is a result of these two loci having very different allele frequencies. In addition, the rs16879552 G allele shows population differences in HapMap (25), with frequencies ranging from 0.322 in CHB, 0.817 in YRI, to 0.975 in CEU, while the rs7835688 C allele ranges from 0.125 in YRI, 0.133 in CHB, to 0.415 in CEU (Fig. S1). Nevertheless, despite interpopulation differences, most functional genetic variations are not population-specific (26, 27), as exemplified by the RET rs2435357 mutation. These population differences may further contribute to the population differences in the observed prevalence of HSCR, in a manner similar to that of RET (18, 19). Although little is known about the influence of noncoding regions on bioinformatic analysis performed on the genomic region encompassing these SNPs revealed neither disruption nor creation of binding sites nor regulatory RNAs, despite the relatively high degree of conservation among species in the region. Unless the alleles affect functional sites in the vicinity, the most probable explanation is that these loci are in LD with a functional variant located elsewhere in the gene. Given the pattern of LD encompassing NRG1, if there were a functional variant common to both Chinese and Caucasians, it would most likely be located 5′ to rs16879552, within the region encompassing part of intron 1, exon 1, and the promoter region (see Fig. 2D). We did sequence the NRG1 exon 1 in the patient group and no variant was found (see SI Text).
Discussion
This study, in addition to further demonstrating the unequivocal role of RET in HSRC in the Chinese population, provides evidence that variations in NRG1 increase the risk of disease conferred by RET, modifying the penetrance of the disease. The latter is shown by the strong interaction between these 2 HSCR susceptibility genes whereby NRG1 risk genotypes increase the chances of disease more for those individuals bearing RET risk alleles than for those individuals with the non-RET risk genotype. Although the mechanisms whereby these 2 genes lead to HSCR are yet unknown, our findings are supported by the strong biological plausibility described below.
The genetic complexity observed in HSCR can be conceptually understood in the light of the molecular and cellular events that take place during the development of the ENS. The human gut is formed from the association of the endoderm, which gives rise to the mucosa lining, and the splanchnic mesenchyme, which differentiates into the muscle layers. Concomitantly, neural crest cells (NCCs), derived from the vagal region of the neural tube, migrate, proliferate, and differentiate into neurons and glia and coalesce into ganglion plexuses in the myenteric region (28) forming the enteric nervous system (ENS). During these processes, the NCCs have to adapt to a constantly changing intestinal environment that may strongly influence their fate (28). HSCR may result from severe mutations in a major gene encoding a crucial molecule involved in the development of the ENS (whose penetrance may be modulated by other alleles) and/or from the accumulation of less severe, more common, mutations in several gene members of the same signaling network. In this context, variations in NRG1 could affect the expression of the already defective major gene, RET, and interfere with the ENS development.
NRG1 and its receptors (the ErbB family of tyrosine kinase receptors) are among the molecular regulators of the NCC's development. The NRG1/ErbB system promotes neuronal survival amid other biological functions. Several lines of evidence suggest that NRG1 signaling plays a role in the development and maintenance of the ENS: (i) ErbB2/ErbB3 are expressed in mouse vagal neural crest cells entering the developing gut (24, 29, 30) and in adult intestinal epithelia of both humans and mice (24, 29, 31, 32); (ii) NRG1 is expressed in mice intestinal mucosa (33) and enteric ganglia (34); and (iii) postnatal colonic aganglionosis in mice is because of the loss of ErbB2 signaling in the colonic epithelial cells through which NRG1 induces the production of survival factors required for the postnatal maintenance of the ENS (23).
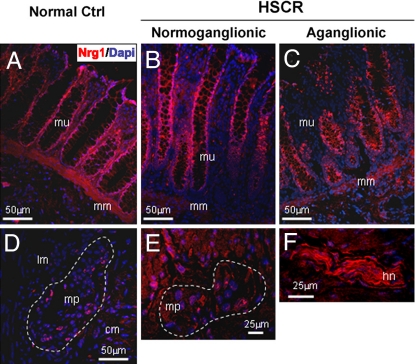
To verify the expression of the NRG1 protein in the human colon, we performed immunofluorescence and immunohistochemistry analysis on normoganglionic and aganglionic tissue sections of a HSCR patient's colon, as well as in colon tissue of a non-HSCR infant (see SI Text). NRG1 is mainly expressed in the enteric ganglia and in the intestinal mucosa of both ganglionic and aganglionic gut (Fig. 3). Expression of the ErbB2 receptor was also observed by immunohistochemistry in the enteric ganglia and colonic mucosa (data not shown).
Fig. 3.
Expression of NRG1 in the intestinal mucosa of (A) a non-HSCR infant, and in the (B) normoganglionic (C) and aganglionic mucosa of a HSCR-affected infant. NRG1 expression in the (D) enteric plexuses of a non-HSCR infant, (E) HSCR infant, and (F) in the hypertrophic nerve trunk. The abnormal nerve bundles characteristic of the aganglionic bowel are extrinsic cholinergic fibers of sacral origin. cm, circular muscle; hn, hypertrophic nerve; lm, longitudinal muscle; mm, muscularis mucosa; mp, myenteric plexuses; mu, intestinal mucosa.
Most of the HSCR genes identified so far code for protein members of interrelated signaling pathways (RET, EDNRB, and that of the transcriptional regulator Sox10) (5–8), involved in the development of enteric ganglia (35–37). Importantly, the expression of the NRG1 receptor ErbB3 by enteric NCCs' precursors is regulated by Sox10, which is essential for NCCs' survival and fate decision processes. Notably, NRG1 signaling mediates these interactions in a Sox10-dependent manner (38). At the molecular level, Sox10 interacts with Ret and Ednrb (11, 12, 39) genes and this would be in line with the genetic interaction with NRG1 and RET reported in this study. In humans, Sox10 mutations are associated with the Waardenburg-Hirschsprung's syndrome (WS4) (8), phenotypes involving myelin deficiencies and sensory neuropathies (40), and with the neurological variant of Waardenburg-Shah syndrome, which includes peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, Waardenburg syndrome, and Hirschsprung disease (41). Remarkably, NRG1 is crucial for the development of both myelinating and nonmyelinating Schwann cells (NCC derivates), and the myelination process, and has been shown to cause peripheral sensory neuropathies in mice (42). It is tempting to speculate that miss-expression of NRG1 could affect the Sox10-mediated maintenance of ENS progenitors and contribute to aganglionosis. In addition, similarly to NRG1, Sox10 is important for defining and maintaining the identity of glial cells in later development stages (43), including the mature human ENS, and abnormal Sox10 expression has been observed in the aganglionic bowel of HSCR patients (44). Whether the latter could be linked to the role proposed above for NRG1 in adult gut and whether they altogether could account for the abnormalities of the extra-cellular matrix characteristic of the aganglionic bowel needs further investigation. Biological interaction between RET and NRG1 signaling has been reported and linked to the survival and maintenance of the peripheral nervous system, where injury-induced expression of the RET ligand GDNF by nonmyelinating Schwann cells is ErbB-dependent.
This study provides strong evidence for a role of NRG1 in HSCR pathology and further supports the biological evidence of NRG1 as a player in the signaling network implicated in ENS development and maintenance which is also in line with its role as a modifier of the RET gene.
Given the genetic heterogeneity of the disease, establishing a genotype-phenotype relationship poses an enormous challenge, as it is likely that the effect of NRG1 on the phenotype may be dependent on other genes in addition to that already demonstrated for RET. If the causal variant has an effect on gene-regulation and this is to be detected, one would expect either different levels of total NRG1 expression or differences in the type or relative levels of NRG1 isoforms (46).
It would be legitimate to ask whether there are differences in the frequencies of the RET HSCR-associated alleles between L-HSCR and S-HSCR patients, but a much larger sample would be needed to draw significant conclusions.
We have shown that the GWA approach could be used to disentangle the genetic components underlying those less common diseases that result from a combination of a discreet number of loci.
Methods
Subjects.
The overall study was approved by the Institutional Review Board of The University of Hong Kong together with the Hospital Authority (IRB: UW 06–349 T/1374). Blood samples were drawn from all participants after obtaining informed consent (parental consent in newborns and children below age 7).
HSCR Patients.
A total of 390 sporadic HSCR patients had prospectively been collected throughout Hong Kong and Mainland China. HSCR diagnosis was based on histological examination of either biopsy or surgical resection material for absence of enteric plexuses. Patients were initially grouped into genome-wide scan (200 patients) and replication series (190 patients). The overall male-to-female ratio was ≈5:1 (Table S7).
Controls.
For the genome-wide scan, we included 306 individuals (106 males, 200 females) of southern Chinese origin participating in a GWA study aimed at the discovery of genetic factors for the quantitative trait of disk degeneration (DD, see details in SI Text) that is currently ongoing in our institution.
In addition, we derived a synthetic control set, referred to as “pseudocontrols,” by implementing the principle of Haplotype-based Haplotype Relative Risk (47) on parental genotypes of 57 HSCR patients whose parents were also genotyped. For each SNP locus, each pair of parental nontransmitted alleles (except those with Mendelian error) was treated as the genotype of a pseudocontrol. To further increase the power, Affymetrix-generated genotypes for the Chinese Han population (n = 45) were retrieved from the International HapMap Project. This resulted in the inclusion of 102 additional controls.
For replication, we used DNA from 510 (350 males and 160 females) individuals of southern Chinese origin with no diagnosis of HSCR (volunteer blood donors).
Whole-Genome Genotyping.
The whole-genome scan was performed using the Affymetrix GeneChip Human Mapping 500K Array Set (250K NspI and StyI array), which assays 493,840 SNPs across the genome. Genotypes were subsequently called from cell intensity data by the stand-alone command-line BRLMM (Bayesian Robust Linear Model with Mahalanobis distance) program.
Data Analyses.
PLINK (48) was used for data management and manipulation, quality-control statistics (see SI Text) and haplotypic tests. To account for spurious associations resulting from ancestral differences of individual SNPs between cases and controls, we applied a principal components-based analysis to model the differences and correct for the variation in frequency using EIGENSTRAT (49). The Cochran-Armitage trend test was used to assess the levels of association of the SNPs with HSCR. To capture putative disease-susceptibility SNPs in our region of interest not included on the Affymetrix chips, in silico genotypes were also generated based on HapMap Phase II CHB-known haplotypes using IMPUTE (50). Logistic regression was used for model fitting of RET intron 1 and NRG1 SNPs in the combined GWA and follow-up datasets, including assessment of sex effects on NRG1, interaction between RET and NRG1, and interaction between stage (GWA vs. targeted follow-up) and NRG1, with P-values given based on the difference of deviance (−2 log-likelihood), which follow the χ2 distribution with one degree of freedom. Haploview 4.0 (51) was used for visualization of whole genome association data and LD patterns.
Supplementary Material
Acknowledgments.
We thank all subjects who participated in the study. We also thank the Genome Research Centre of the University of Hong Kong, and in particular to Dr. Agnes Chan and Prof. Kathryn S. E. Cheah for their assistance. This work was supported by research Grants HKU 765407M (to M.-M.G.-B.) and HKU 775907M (to P.K.T.) from the Hong Kong Research Grants Council, the University Grants Committee of Hong Kong Grant AoE/M-04/04 and National Institutes of Health Grant EY-12562 (to S.S.C. and P.C.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809630105/DCSupplemental.
References
- 1.Chakravarti A, Lyonnet S. In: The Metabolic and Molecular Basis of Inherited Diseases. Scriver CR, Sly WS, Valle D, Beaudet AL, editors. New York: McGraw–Hill; 2001. pp. 6231–6255. [Google Scholar]
- 2.Amiel J, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. doi: 10.1136/jmg.2007.053959. [DOI] [PubMed] [Google Scholar]
- 3.Lyonnet S, et al. A gene for Hirschsprung disease maps to the proximal long arm of chromosome 10. Nat Genet. 1993;4:346–350. doi: 10.1038/ng0893-346. [DOI] [PubMed] [Google Scholar]
- 4.Schuchardt A, D'Agati V, Larsson-Blomberg L, Costantini F, Pachnis V. RET-deficient mice: an animal model for Hirschsprung's disease and renal agenesis. J Intern Med. 1995;238:327–332. doi: 10.1111/j.1365-2796.1995.tb01206.x. [DOI] [PubMed] [Google Scholar]
- 5.Puffenberger EG, et al. Identity-by-descent and association mapping of a recessive gene for Hirschsprung disease on human chromosome 13q22. Hum Mol Genet. 1994;3:1217–1225. doi: 10.1093/hmg/3.8.1217. [DOI] [PubMed] [Google Scholar]
- 6.Angrist M, Bolk S, Halushka M, Lapchak PA, Chakravarti A. Germline mutations in glial cell line-derived neurotrophic factor (GDNF) and RET in a Hirschsprung disease patient. Nat Genet. 1996;14:341–344. doi: 10.1038/ng1196-341. [DOI] [PubMed] [Google Scholar]
- 7.Hofstra RM, et al. A loss-of-function mutation in the endothelin-converting enzyme 1 (ECE-1) associated with Hirschsprung disease, cardiac defects, and autonomic dysfunction. Am J Hum Genet. 1999;64:304–308. doi: 10.1086/302184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pingault V, et al. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 9.Carrasquillo MM, et al. Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat Genet. 2002;32:237–244. doi: 10.1038/ng998. [DOI] [PubMed] [Google Scholar]
- 10.McCallion AS, Stames E, Conlon RA, Chakravarti A. Phenotype variation in two-locus mouse models of Hirschsprung disease: tissue-specific interaction between Ret and Ednrb. Proc Natl Acad Sci USA. 2003;100:1826–1831. doi: 10.1073/pnas.0337540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang D, Epstein JA. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum Mol Genet. 2003;12:937–945. doi: 10.1093/hmg/ddg107. [DOI] [PubMed] [Google Scholar]
- 12.Cantrell VA, et al. Interactions between Sox10 and EdnrB modulate penetrance and severity of aganglionosis in the Sox10Dom mouse model of Hirschsprung disease. Hum Mol Genet. 2004;13:2289–2301. doi: 10.1093/hmg/ddh243. [DOI] [PubMed] [Google Scholar]
- 13.Borrego S, et al. RET genotypes comprising specific haplotypes of polymorphic variants predispose to isolated Hirschsprung disease. J Med Genet. 2000;37:572–578. doi: 10.1136/jmg.37.8.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sancandi M, et al. Single nucleotide polymorphic alleles in the 5′ region of the RET proto-oncogene define a risk haplotype in Hirschsprung's disease. J Med Genet. 2003;40:714–718. doi: 10.1136/jmg.40.9.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burzynski GM, et al. Localizing a putative mutation as the major contributor to the development of sporadic Hirschsprung disease to the RET genomic sequence between the promoter region and exon 2. Eur J Hum Genet. 2004;12:604–612. doi: 10.1038/sj.ejhg.5201199. [DOI] [PubMed] [Google Scholar]
- 16.Fitze G, Schreiber M, Kuhlisch E, Schackert HK, Roesner D. Association of RET protooncogene codon 45 polymorphism with Hirschsprung disease. Am J Hum Genet. 1999;65:1469–1473. doi: 10.1086/302618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emison ES, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Barcelo MM, et al. Chinese patients with sporadic Hirschsprung's disease are predominantly represented by a single RET haplotype. J Med Genet. 2003;40:e122. doi: 10.1136/jmg.40.11.e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Barcelo M, et al. TTF-1 and RET promoter SNPs: regulation of RET transcription in Hirschsprung's disease. Hum Mol Genet. 2005;14:191–204. doi: 10.1093/hmg/ddi015. [DOI] [PubMed] [Google Scholar]
- 20.Bolk S, et al. A human model for multigenic inheritance: phenotypic expression in Hirschsprung disease requires both the RET gene and a new 9q31 locus. Proc Natl Acad Sci USA. 2000;97:268–273. doi: 10.1073/pnas.97.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabriel SB, et al. Segregation at three loci explains familial and population risk in Hirschsprung disease. Nat Genet. 2002;31:89–93. doi: 10.1038/ng868. [DOI] [PubMed] [Google Scholar]
- 22.Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- 23.Crone SA, Negro A, Trumpp A, Giovannini M, Lee KF. Colonic epithelial expression of ErbB2 is required for postnatal maintenance of the enteric nervous system. Neuron. 2003;37:29–40. doi: 10.1016/s0896-6273(02)01128-5. [DOI] [PubMed] [Google Scholar]
- 24.Britsch S. The neuregulin-I/ErbB signaling system in development and disease. Adv Anat Embryol Cell Biol. 2007;190:1–65. [PubMed] [Google Scholar]
- 25.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinds DA, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 28.Newgreen D, Young HM. Enteric nervous system: development and developmental disturbances–Part 2. Pediatr Dev Pathol. 2002;5:329–349. doi: 10.1007/s10024-002-0002-4. [DOI] [PubMed] [Google Scholar]
- 29.Britsch S, et al. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paratore C, Eichenberger C, Suter U, Sommer L. Sox10 haploinsufficiency affects maintenance of progenitor cells in a mouse model of Hirschsprung disease. Hum Mol Genet. 2002;11:3075–3085. doi: 10.1093/hmg/11.24.3075. [DOI] [PubMed] [Google Scholar]
- 31.Press MF, Cordon-Cardo C, Slamon DJ. Expression of the HER-2/neu proto-oncogene in normal human adult and fetal tissues. Oncogene. 1990;5:953–962. [PubMed] [Google Scholar]
- 32.Prigent SA, et al. Expression of the c-erbB-3 protein in normal human adult and fetal tissues. Oncogene. 1992;7:1273–1278. [PubMed] [Google Scholar]
- 33.Meyer D, Birchmeier C. Distinct isoforms of neuregulin are expressed in mesenchymal and neuronal cells during mouse development. Proc Natl Acad Sci USA. 1994;91:1064–1068. doi: 10.1073/pnas.91.3.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orr-Urtreger A, et al. Neural expression and chromosomal mapping of Neu differentiation factor to 8p12–p21. Proc Natl Acad Sci USA. 1993;90:1867–1871. doi: 10.1073/pnas.90.5.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pachnis V, Durbec P, Taraviras S, Grigoriou M, Natarajan D. III. Role Of the RET signal transduction pathway in development of the mammalian enteric nervous system. Am J Physiol. 1998;275:G183–G186. doi: 10.1152/ajpgi.1998.275.2.G183. [DOI] [PubMed] [Google Scholar]
- 36.Natarajan D, Marcos-Gutierrez C, Pachnis V, de Graaff E. Requirement of signalling by receptor tyrosine kinase RET for the directed migration of enteric nervous system progenitor cells during mammalian embryogenesis. Development. 2002;129:5151–5160. doi: 10.1242/dev.129.22.5151. [DOI] [PubMed] [Google Scholar]
- 37.Barlow A, de Graaff E, Pachnis V. Enteric nervous system progenitors are coordinately controlled by the G protein-coupled receptor EDNRB and the receptor tyrosine kinase RET. Neuron. 2003;40:905–916. doi: 10.1016/s0896-6273(03)00730-x. [DOI] [PubMed] [Google Scholar]
- 38.Paratore C, Goerich DE, Suter U, Wegner M, Sommer L. Survival and glial fate acquisition of neural crest cells are regulated by an interplay between the transcription factor Sox10 and extrinsic combinatorial signaling. Development. 2001;128:3949–3961. doi: 10.1242/dev.128.20.3949. [DOI] [PubMed] [Google Scholar]
- 39.Zhu L, et al. Spatiotemporal regulation of endothelin receptor-B by SOX10 in neural crest-derived enteric neuron precursors. Nat Genet. 2004;36:732–737. doi: 10.1038/ng1371. [DOI] [PubMed] [Google Scholar]
- 40.Pingault V, et al. Peripheral neuropathy with hypomyelination, chronic intestinal pseudo-obstruction and deafness: a developmental “neural crest syndrome” related to a SOX10 mutation. Ann Neurol. 2000;48:671–676. [PubMed] [Google Scholar]
- 41.Inoue K, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, et al. Disruption of ErbB receptor signaling in adult non-myelinating Schwann cells causes progressive sensory loss. Nat Neurosci. 2003;6:1186–1193. doi: 10.1038/nn1139. [DOI] [PubMed] [Google Scholar]
- 43.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sham MH, Lui VC, Fu M, Chen B, Tam PK. SOX10 is abnormally expressed in aganglionic bowel of Hirschsprung's disease infants. Gut. 2001;49:220–226. doi: 10.1136/gut.49.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, et al. Disruption of ErbB receptor signaling in adult non-myelinating Schwann cells causes progressive sensory loss. Nat Neurosci. 2003;6:1186–1193. doi: 10.1038/nn1139. [DOI] [PubMed] [Google Scholar]
- 46.Kwan T, et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 47.Sham PC, Curtis D. An extended transmission/disequilibrium test (TDT) for multi-allele marker loci. Ann Hum Genet. 1995;59:323–336. doi: 10.1111/j.1469-1809.1995.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 48.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 50.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 51.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.