Abstract
Activation-induced cytidine deaminase (AID) is a single-stranded (ss) DNA-specific cytidine deaminase that initiates Ig heavy chain (IgH) class switch recombination (CSR) and Ig somatic hypermutation (SHM) by deaminating cytidines within, respectively, IgH switch (S) regions and Ig variable region (V) exons. AID that is phosphorylated on serine residue 38 interacts with replication protein A (RPA), a ssDNA binding protein, to promote deamination of transcribed double-stranded DNA in vitro, which, along with other evidence, suggests that AID may similarly gain access to transcribed S regions and V exons in vivo. However, the physiological role of AID phosphorylation at serine residue 38 (S38), and even the requirement for the S38 residue, with respect to CSR or SHM has been debated. To address this issue, we used gene targeting to generate an endogenous mouse AID locus that produces AID in which S38 is substituted with alanine (AIDS38A), a mutant form of AID that retains similar catalytic activity on ssDNA as WT AID (AIDWT). B cells homozygous for the AIDS38A mutation show substantially impaired CSR and SHM, correlating with inability of AIDS38A to interact with endogenous RPA. Moreover, mice haploinsufficient for AIDS38A have even more severely impaired CSR when compared with mice haploinsufficient for AIDWT, with CSR levels reduced to nearly background levels. These results unequivocally demonstrate that integrity of the AID S38 phosphorylation site is required for normal CSR and SHM in mice and strongly support a role for AID phosphorylation at S38 and RPA interaction in regulating CSR and SHM.
Keywords: activation-induced deaminase, protein kinase A, R-loop
In response to antigenic stimulation, mature B cells undergo Ig heavy chain (IgH) class switch recombination (CSR) (1, 2). CSR is a B-cell–specific recombination/deletion process that replaces the initially expressed Cμ IgH constant region (CH) exons with one of several sets of downstream IgH CH exons (Cγ, Cε, or Cα). Thereby, CSR allows a B cell to switch from expression of IgM to expression of a secondary IgH isotype (IgG, IgE, or IgA) with a different effector function. Each set of CH exons is preceded by a repetitive switch (S) region 1–12 kb in length. CSR involves introduction of DNA double-strand breaks (DSBs) into the donor Sμ and into a downstream acceptor S region, followed by their joining and the accompanying deletion of intervening sequences. Mature B cells further diversify their antibody repertoire by undergoing somatic hypermutation (SHM), a process in which point mutations, and occasional deletions and insertions are introduced into the variable region (V) exons of IgH and Ig light chain genes (3, 4). Although SHM can occur throughout the V genes, the tetranucleotide sequence RGYW (R = purine, Y = pyrimidine, W = A or T nucleotide) serves as a SHM hotspot.
The activation-induced cytidine deaminase (AID) is required for the initiation of both CSR and SHM (5, 6). AID is a single-stranded (ss) DNA-specific DNA cytidine deaminase (7–9) that initiates CSR and SHM by converting cytidine (C) to uridine (U) in targeted S regions or V exons, respectively (10). Subsequently, the introduced U/G mismatches are processed by components of the base-excision repair (BER) machinery or mismatch repair (MMR) pathways to ultimately generate DNA DSBs that serve as obligatory CSR intermediates or point mutations in the context of SHM (10). Ligation of DSBs between 2 different S regions completes CSR. Transcription appears to play a major role in allowing access of the ssDNA-specific AID to duplex DNA. S regions are G/C rich with a characteristic G-rich nontemplate strand (1, 2). Transcription through S regions generates RNA/DNA hybrid structures, termed R-loops, in which the nontemplate strand is displaced as ssDNA (11–13). R loops have been shown to provide AID access to duplex S region DNA in vitro, thus providing a mechanism of AID activity on duplex DNA (7). However, V exons do not form R loops when transcribed, indicating other types of AID access mechanisms during SHM (14). Likewise, the Xenopus Sμ region, which does not form an R-loop but is very rich in the AGCT sequence, a canonical subset of the RGYW hotspot motif, can serve as an efficient CSR substrate when inserted in place of a mouse S region (15). Thus, these findings predicted that additional mechanisms exist to allow AID to access DNA that does not form R-loops on transcription.
Attempts to elucidate such additional mechanisms by which AID could deaminate duplex DNA led to the finding that AID is phosphorylated on serine residue 38 (S38) by protein kinase A (PKA) (16–18). Biochemical studies demonstrated that AID phosphorylated on S38 interacts with replication protein A (RPA), and thereby binds to and deaminates V exons and Xenopus S regions when they are transcribed by T7 RNA polymerase in vitro (14–16), an activity that may be enhanced by the presence of SHM motifs (14). Based on this body of work, it was proposed that RPA, in a complex with AID, stabilizes ssDNA bubbles, primarily at RGYW sequences, within transcribed V genes to generate deamination substrates for AID during SHM (14). In addition, because RGYW motifs, particularly the AGCT sequence, are highly enriched in S regions, the RPA-dependent mechanism was suggested to be important for CSR beyond simple access of AID to S regions via R-loops (15). Because of the intrinsic ability of RPA to coordinate with BER and MMR pathways, which process AID-mediated deaminated residues during SHM and CSR, it has been suggested that AID-RPA interaction might have additional functions downstream of deamination (14). Overall, these studies led to the notion that AID phosphorylation and RPA interaction might play a significant role in both CSR and SHM. Recent studies of zebrafish AID, which lacks an S38A equivalent site but constitutively interacts with RPA, have strongly supported the significance of the findings on mouse AID-RPA interaction (19).
In accord with a physiological role for AID phosphorylation at S38, several studies showed that AID phosphorylated on S38 substituted with alanine (AIDS38A) was only 15−30% as active as WT AID (AIDWT) in its ability to mediate CSR in retrovirally transduced AID-deficient splenic B cells, despite retaining full ssDNA deamination activity in vitro (16, 18, 19). In addition, AIDS38A showed a reduced ability to mutate transcribed reporter genes following ectopic expression in fibroblasts (18), consistent with the suggestion that AID phosphorylation by PKA might augment AID activity in SHM. In contrast, Honjo and coworkers (20) reported that the CSR activity of AIDS38A mutant approached that of AIDWT in the retroviral complementation assay, leading them to question whether mutation at this site had any significant effect on AID activity and whether AID phosphorylation at this site had any physiological role. However, we have shown that AIDS38A is a hypomorph, which retains full ssDNA deamination activity in biochemical assays (16). Therefore, because AIDWT activity is generally saturated in the retroviral complementation assays, we have suggested that the nearly normal AIDS38A CSR activity reported by Honjo and coworkers (20) might reflect expression of the protein at levels greater than normal physiological levels, leading to an overestimation of its actual physiological activity (21). To address these possibilities unequivocally, we expressed the AIDS38A mutant protein from the endogenous genomic locus and assayed effects on CSR and SHM in activated B cells.
Results
Generation of AIDS38A Knock-In Mutant Mice.
We used gene-targeted mutation with a LoxP-flanked (Floxed) pgkNeo cassette-based targeting vector to introduce the S38A point mutation into exon 2 of the AID gene in 129 strain (TC1) mouse embryonic stem (ES) cells [Fig. S1A]. We verified homologous recombination of the targeting construct into the endogenous AID locus and the presence of the mutation via Southern blotting and PCR approaches (Fig. S1 B and C). ES cells targeted for the mutation on 1 allele were then subjected to high G418 selection to obtain ES cells homozygous for the AIDS38A mutation. The floxed pgkNeo cassette was subsequently removed from ES cells heterozygous or homozygous for the targeted mutant AID allele to yield, respectively, AIDS38A/+ and AIDS38A/S38A mutant ES cells. The latter were used to generate mice in which all lymphocytes were of the AIDS38A/S38A genotype via RAG2-deficient blastocyst complementation (RDBC) (22). The AIDS38A/+ ES cells were used to introduce the AIDS38A mutation into the germline of 129 strain mice. The majority of analyses presented here were performed with AIDS38A/S38A B cells generated via RDBC, but major findings were confirmed by analyses of B cells from AIDS38A/S38A germline mutant mice (Table S1). We have also analyzed B cells from germline mutant mice that were haploinsufficient for the AIDS38A mutant (AIDS38A/−); these mice were generated by breeding mice carrying the AIDS38A mutation (AIDS38A/+) with mice carrying an AID null mutation (AID−/−).
Development of AIDS38A/S38A or AIDS38A/− B lymphocytes appeared unperturbed with normal numbers of splenic B and T cells (Fig. S1D and data not shown). In addition, when splenic B cells were activated in culture with anti-CD40 plus IL-4 or with bacterial LPS, levels of AID in AIDS38A/S38A and AID+/+ B cells appeared roughly similar in most experiments with respect to both levels and induction kinetics (Fig. 1 A and B and Fig. S2). Likewise, the levels of AID and kinetics of induction appeared similar for AIDS38A/− and AID+/− B cells, although overall AID levels were reduced compared with those of AIDS38A/S38A and AID+/+ B cells, consistent with the reported haploinsufficiency effect on AID expression (23). Finally, the levels of AID induction generally appeared to be somewhat less in B cells activated with LPS plus anti-δ-dextran versus those activated with anti-CD40 plus IL-4 (Fig. S2). Although there is some variation in AIDS38A or AIDWT levels between experiments (Fig. 1 A and B and Fig. S2), we conclude that, on average, AID expression levels from corresponding AIDWT and AIDS38A alleles are similar under a given activation condition and note that others have recently come to the same conclusion based on independent analyses of the AIDS38A knock-in mutation (24).
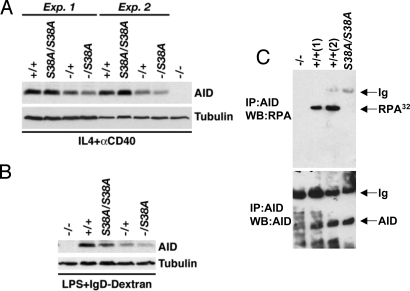
Fig. 1.
AID expression and RPA interaction in AIDS38A mutant mice. (A) AID protein levels in anti-CD40 plus IL-4–stimulated splenic B cells. Sixty micrograms of whole cell extracts from purified and activated splenic B cells from mice of the indicated genotypes harvested at day 2.5 of stimulation were separated by SDS-PAGE and immunoblotted with antibodies to AID or tubulin. Results from 2 independent experiments (Exp.) are shown. (B) Protein levels in LPS + anti-δ-dextran–treated splenic B cells. Whole cell extracts (60 μg) were analyzed as in A in response to LPS-IgD-dextran stimulation. (C) Interaction of AIDWT and AIDS38A mutant proteins with RPA. Nuclear extracts from AID-deficient (−/−), WT (+/+), and AIDS38/S38A mice were immunoprecipitated (IP) with AID, and the immunoprecipitate was analyzed by Western blotting (WB) for the presence of the 32-kDa subunit of RPA (Top) or AID (Bottom). Ig, Ig chains.
AIDS38A Fails to Interact with RPA in Vivo.
A portion of AIDWT binds to RPA in B cells (14). Likewise, PKA-mediated phosphorylation of unphosphorylated AIDWT on S38 in vitro confers ability to bind to RPA (16). In contrast, the AIDS38A mutant protein fails to substantially bind RPA even after exposure to PKA (16). To investigate whether endogenous AID-RPA interaction is dependent on the integrity of the AID S38 residue, we first generated nuclear extracts from AIDWT/WT and AIDS38A/S38A mutant B cells following activation in culture for 3 days with anti-CD40 plus IL-4. Subsequently, we immunoprecipitated endogenous WT and mutant AID proteins with anti-AID antibodies and assayed the immunoprecipitates via Western blotting with antibodies against the 32-kDa subunit of RPA (RPA32). Although the endogenous AIDWT protein showed robust interaction with endogenous RPA32 as previously described (14, 16, 19), the endogenous AIDS38A protein showed no detectable interaction with RPA (Fig. 1C, Top), even though levels of AIDWT and AIDS38A proteins in the respective immunoprecipitates appeared similar (Fig. 1C, Bottom). These results demonstrate that the interaction between endogenous AID and RPA is dependent on the integrity of the AID S38 residue.
Decreased CSR of AIDS38A/S38A Mutant Cells to IgG1 and IgG3.
To examine the effects of the endogenous AIDS38A mutation on CSR, we purified splenic AIDS38A/S38A B cells from the RDBC chimeras or homozygous germline mutant mice and compared their ability to undergo class switching to IgG1 or IgG3 with that of WT B cells following culture in the presence of anti-CD40 plus IL-4 or LPS plus anti-δ-dextran, respectively. Switching to IgG1 and IgG3 was assayed at days 3.0, 3.5, and 4.5 by staining B cells with anti-IgG1– or anti-IgG3–specific antibodies followed by flow cytometric analyses. On average, about 40% of AIDWT/WT B cells stimulated with anti-CD40 plus IL-4 switched to IgG1 at day 3 poststimulation and about 50–60% switched to IgG1 at days 3.5 and 4.5 poststimulation (Fig. 2, Fig. S3A, and Table S1). In contrast, only about 2% of AIDS38A/S38A B cells activated in parallel switched to IgG1 at day 3 (about 5% of WT levels), about 10% switched to IgG1 at day 3.5 (about 18% of WT levels), and about 18% switched to IgG1 at day 4.5 (about 40% of WT levels) (Fig. 2, Fig. S3A, and Table S1). The increased relative levels of AIDS38A/S38A B cells that switched to IgG1 compared with WT B cells at later time points may reflect WT switching reaching a maximum by day 3.5, although AIDS38A/S38A continues to rise (Fig. 2). In this case, the values at day 3 may better estimate the relative ability of AIDS38A to function in CSR relative to AIDWT. Following stimulation with LPS plus anti-δ-dextran, on average, ≈7.5% of WT B cells underwent CSR to IgG3 at day 3, with levels rising to 12.5% at day 3.5 and to about 18% at day 4.5 (Fig. 3, Fig. S3B, and Table S1). In contrast, switching to IgG3 in AIDS38A/S38A B cells was barely above AID-deficient background (less than 1%) at all 3 time points (Fig. 3, Fig. S3B, and Table S1). Overall, our finding show that mouse B cells homozygous for the AIDS38A mutation showed severely impaired CSR to IgG1 and barely detectable CSR to IgG3.
Fig. 2.
CSR activity in AIDS38A/S38A and AIDS38A/− mice for IgG1. B cells were isolated from the spleen of mice of the indicated genotypes and stimulated for CSR to IgG1 for 3, 3.5, and 4.5 days using anti-CD40 antibodies and IL-4. Levels of CSR in these B cells were analyzed using FACS as described in Materials and Methods. The percentage of IgG1+B220+ cells for each genotype was plotted against the days in culture. Each bar graph represents the mean of multiple experiments (Table S1), with the error bars representing SD from the mean. AID-deficient splenic B cells activated with anti-CD40 plus IL-4 were used as controls, and less than 0.1% of the cells stained for IgG1 at all 3 time points (Table S1).
Fig. 3.
CSR activity in AIDS38A/S38A and AIDS38A/− mice for IgG3. B cells isolated from mice of the indicated genotypes were stimulated for CSR to IgG3 for 3, 3.5, and 4.5 days using LPS and anti-δ-dextran. Levels of CSR in these B cells were analyzed using FACS. The percentage of IgG3+B220+ cells was plotted against time in culture. Each bar graph represents the mean of multiple experiments (Table S1), with the error bars representing SD from the mean. AID-deficient splenic B cells activated with LPS plus anti-δ-dextran were used as controls, and less than 0.4% of the cells stained for IgG1 at all 3 time points (Table S1).
Haploinsufficiency for the AIDS38A Mutation Leads to an Extremely Severe Defect in CSR.
Small differences in the levels of ectopically expressed AID proteins in B cells may significantly alter CSR, SHM, and translocation-generating activities (21, 23, 25). In addition, in a BALB/c background, haploinsufficiency for the AIDWT gene leads to correspondingly reduced AID levels and reduced levels of CSR (23). In this context, the more substantial impairment of CSR by AIDS38A/S38A B cells to IgG3 when stimulated with LPS plus anti-δ-dextran versus CSR to IgG1 when stimulated with anti-CD40 plus IL-4 might be explained, at least in part, by the apparently lower levels of AID induction with the former conditions (Fig. S2 A and B; also in Discussion). To explore the effects of endogenous levels on CSR in cells expressing AIDWT versus AIDS38A further, we assayed relative CSR activity of AIDWT/− and AIDS38A/− B cells in which AID expression levels were reduced to roughly 50% those of AIDWT/WT and AIDS38A/S38A mutant B cells (Fig. 1).
Following appropriate stimulations with anti-CD40 plus IL-4 or LPS plus anti-δ-dextran, respectively, AIDWT/− B cells underwent IgH class switching to IgG1 at levels that were similar to those of AIDWT/WT B cells (e.g., about 55% at day 3.5; Fig. 2) but had slightly reduced levels of CSR to IgG3 compared with AIDWT/WT B cells (e.g., about 12.6% in AIDWT/WT vs. 8.8% in AIDWT/− B cells; Fig. 3), with the latter reduction consistent with the haploinsufficiency phenotype previously reported (23). Remarkably, however, appropriately stimulated AIDS38A/− B cells showed extremely impaired IgH class switching to both IgG1 (less than 4% of WT) and IgG3 (levels were not greater than background observed with AID−/− B cells) as compared with those of AIDWT/− controls at all days of stimulation (Figs. 2 and 3, Fig. S4, and Table S1). Therefore, reduction of AID levels by about 50% (Fig. 1) has a much more severe effect on the level of CSR in B cells expressing endogenous AIDS38A than in those expressing endogenous AIDWT, consistent with our prior predictions (21).
Reduced Somatic Hypermutation in AIDS38A/S38A Peyer's Patch B Cells.
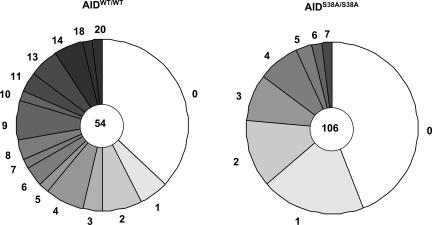
B220+PNAhi germinal center B cells isolated from Peyer's patches of 2-month-old AIDS38A/S38A RDBC chimeras and WT mice were analyzed for SHM of a 672-bp region immediately 3′ of VHJ558DJH4 rearrangements (26). In comparison to the age-matched AIDWT/WT B cells (analyses from 3 different mutant chimeras and 2 different WT mice), we observed a substantial decrease (greater than 70%) in the frequency of SHM of this downstream JH4 sequence in AIDS38A/S38A B cells (Fig. 4). From our preliminary analyses, we failed to observe any gross perturbation on the SHM spectra of AIDS38A/S38A B cells (data not shown). However, to make any firm conclusions regarding potential minor changes in the SHM spectra, greater numbers of sequences need to be analyzed. We conclude that the AIDS38A mutant protein is significantly compromised in its ability to mediate SHM.
Fig. 4.
SHM in Peyer's patch B cells. SHM within a 672-bp region of DNA downstream of JH4 was detected using AIDWT/WT and AIDS38A/S38A Peyer's patch germinal center B cells. Three 12-week-old AIDS38A/S38A mice and 2 age-matched WT control mice were used. Mutation error rate attributable to PCR using Pfu (6.76 mutations/10−5 bp) was obtained by analyzing SHM in B cells obtained from AID-deficient mice.
Discussion
In this report, we generate and analyze chimeric or germline mutant mice in which mature B lymphocytes express the AIDS38A protein from the endogenous AID locus. These analyses unequivocally establish the fact that mutation of the AID S38 PKA phosphorylation site to alanine markedly impairs CSR and SHM in vivo. Therefore, our findings, along with very similar findings from a contemporaneous independent study (24), lay to rest the apparent controversy surrounding whether or not the AIDS38A mutant protein has substantially reduced CSR activity (20, 21). Although it still might be argued that the defective CSR and SHM phenotype of B cells that express AIDS38A from their endogenous AID alleles somehow reflects general adverse effects of S38A mutation unrelated to AID phosphorylation (20), several lines of cumulative evidence argue against this possibility. First, the ssDNA deamination activity of AIDS38A is comparable to that of AIDWT (16), indicating that the mutation does not alter core catalytic activity of the protein. Second, unphosphorylated AIDWT, or AIDS38A that cannot be phosphorylated by PKA in vitro, loses its ability to bind RPA and mediate deamination of transcribed double-stranded (ds) DNA, despite retaining ssDNA deamination activity (14, 16). Finally, second site mutations that restore RPA binding and transcription-dependent dsDNA deamination activity to AIDS38A in the absence of PKA phosphorylation significantly rescue impaired CSR activity of the AIDS38A mutant protein (19). Together, these results provide compelling evidence that impaired CSR and SHM activity of the AIDS38A mutant protein largely results from defective AID phosphorylation and associated inability to bind RPA.
During CSR, both R-loop–dependent and R-loop–independent mechanisms have been implicated in the targeting of AID to S regions (2, 7, 13, 15). In this context, one interpretation of the major CSR defect in AIDS38A/S38A homozygous mice is that an R-loop–independent mechanism, which relies on AID phosphorylation and RPA binding, has a quite predominant in vivo role in promoting AID access to S regions. It is conceivable that the AID-RPA complex might be particularly important for AID access to the template S region strand (which remains bound to RNA in R-loops), perhaps via antisense transcription through S regions (27). Another possibility, not mutually exclusive, is that RPA recruited in the context of the AID-RPA complex has functions downstream of cytidine deamination. For example, RPA recruited to S regions via an AID-RPA complex might recruit other factors that participate in the generation of DSBs or their subsequent ligation (2). Finally, although all results to date indicate that the primary role of AID phosphorylation is to promote RPA association (19), it remains conceivable that AID S38 phosphorylation has additional roles, for example, promoting release of AID from an inactive complex that sequesters it in the cytoplasm or in an inhibitory ribonucleoprotein complex (9).
The dramatic haploinsufficiency phenotype of the AIDS38A mutation relative to AIDWT indicates a remarkable dependency of CSR activity on AIDS38A protein levels. In this context, reduction of AIDWT levels by about 50% has, at most, modest effects on CSR in B cells activated in vitro, whereas reduction of AIDS38A by a similar amount dramatically diminishes CSR, nearly abolishing it under LPS plus anti-δ-dextran activation conditions for switching to IgG3 (Figs. 1 and 3). There could be multiple nonmutually exclusive explanations for this remarkable phenotype. One intriguing possibility is that the AID protein is normally titrated by a “stoichiometric” inhibitor, such that increase in AID protein greater than the titrated level or AID modifications, such as S38 phosphorylation, result in generation of active AID protein. Whatever the mechanistic explanation for the haploinsufficiency phenotype, we note that our CSR assays were done on B cells activated in vitro under conditions that conceivably could lead to AID levels greater than those generated in at least some normal in vivo immune responses. If so, the influence of AID S38 phosphorylation in vivo might (at least in some cases) be better reflected by our findings regarding the dramatic effect of haploinsufficiency with respect to the AIDS38A mutation. Similarly, lower levels of AID induction in LPS plus anti-δ-dextran, versus anti-CD40 plus IL-4 conditions, might result in the more dramatic negative effects on the AIDS38A mutation on CSR to IgG3, although we cannot rule out the possibility that the smaller size of the Sγ3 and/or its different sequence compared with Sγ1 could also make it a poorer AID target and lead to a more dramatic effect of the S38A mutation.
The SHM defect observed in AIDS38A mice supports the proposed model that RPA association with AID has some role in promoting AID access to transcribed V exons during SHM (14). Whether RPA has additional roles in the processing of deaminated DNA during SHM requires detailed characterization of the SHM spectrum in AIDS38A/S38A mutant B cells. We note, however, that SHM does occur, albeit at reduced levels, in AIDS38A/S38A B cells, indicating that AID must have access to V region exons in a S38-phosphorylation–independent and RPA-independent fashion. Given that transcription is clearly an important factor in AID access to V region exons (3), a possible access mechanism, related to R-loop access, would be via spliceosome-associated factors, such as CTNNBL1 (28–30). Another mechanism of AID access to V genes could be through transient changes in the DNA topology during transcription elongation by RNA polymerase II, which could potentially expose ssDNA structures (31). Conversely, others found that haploinsufficiency for the AIDS38A mutation also very severely compromises SHM, similar to its effects on CSR (24), indicating that endogenous AID activity also works at near-threshold levels for SHM. In that context, it is conceivable that AID levels also may vary in the context of particular routes of endogenous B-cell SHM, allowing phosphorylation to take on more or less important roles. Finally, in the context of both SHM and CSR, phosphorylation of AID at S38A adds to the growing list of posttranscriptional mechanisms for this potent mutator (32–35) and, correspondingly, might be important in allowing activated B cells to strike a balance between AID activity on the IgH locus versus non-Ig genes (36–38).
Materials and Methods
Mice.
AIDS38A mice, mutated at AID position 38 (serine-to-alanine substitution), were generated using the gene targeting strategy outlined in Fig. S1. The 5′ and 3′ arms of the targeting vector were, respectively, 4.8 kb and 4.5 kb long. The 3′ arm contained the AGT-to-GCT mutation corresponding to the S38-to-alanine mutation. A LoxP-flanked neomycin resistance gene was used for positive selection, and a herpes simplex virus thymidine-kinase gene was used for negative selection. The targeting construct was linearized and transfected into 129SvE ES cells. The genotype of the selected ES cells was confirmed by Southern blot analysis in addition to PCR amplification and sequencing using a primer that anneals to a location outside of the targeting construct. To generate AIDS38A/S38A homozygous mutant ES cells, we subjected the AIDS38A/+ ES cells to high G418 concentrations. The neomycin resistance cassette, flanked by loxP sites, was removed from targeted ES cells by expression of the Cre-recombinase through an adenoviral vector. Deletion of the resistance cassette was monitored by Southern blotting. The AIDS38A/S38A ES cells were injected via RDBC to generate AIDS38A/S38A chimeric mice. AIDS38A/+ ES cells were used to generate AIDS38A/+ germline mice, which were crossed to AID−/− C57BL/6 mice (5) to produce the AID+/− and AIDS38A/− mice used for analysis. The expression of AIDWT and AIDS38A mutant proteins was detected using polyclonal antibodies specific to AID protein raised in rabbits (7). All experiments with mice were done according to protocols approved by the Children's Hospital Boston Animal Care Facility.
Class Switch Recombination Assay.
B cells were isolated from 8-week-old mouse spleens of various genotypes through negative purification of B cells in magnetic-activated cell sorting columns with anti-CD43 antibody–coated magnetic beads (Miltenyi Biotech). The isolated B cells were incubated in RPMI medium containing 50 μM β-mercaptoethanol with 15% FCS. The cells were stimulated in culture with anti-CD40 antibodies (BD Pharmingen) plus IL-4 (20 ng/mL) or with LPS (25 μg/mL) and anti-δ-dextran (3 ng/mL) (a kind gift from R. Casellas, Bethesda, MD) for CSR to IgG1 or IgG3 isotype, respectively. Cells were harvested after days 3.0, 3.5, or 4.5 and assayed for surface IgG1 or IgG3 production using flow cytometry with antibodies specific for these isotypes.
Somatic Hypermutation Analysis.
Peyer's patch B220+PNAhigh cells of age-matched 8-week-old mice were obtained using the BD FACSAria cell sorter (Becton Dickinson). DNA was isolated from collected populations using the DNeasy Blood and Tissue Kit (QIAgen). PfuTurbo DNA polymerase (Stratagene) was utilized to amplify VHJ558DJH rearrangements using the oligonucleotides J558FR3Fw (GCCTGACAT CTGAGGACTCTGC) and JH4intronRv (CCT CTC CAG TTT CGG CTG AAT CC) (26). PCR products were purified using the QIAquick PCR Purification Kit (QIAgen) and cloned directly using the pGEM-T Easy Vector System (Promega) as suggested by the manufacturer. Recombinant clones were identified via blue/white color selection. Clones containing VHJ558DJH rearrangements were identified by colony hybridization using a full-length JH4-specific probe. Cloned inserts of appropriate size corresponding to VHJ558DJH4 rearrangements were sequenced, and a 672-bp region immediately downstream of JH4 was analyzed.
AID Immunoprecipitation Assay.
Nuclear extracts from AID−/−, AIDWT, and AIDS38A mutant B cells were prepared as described previously (7). The extracts were precleared using protein A sepharose and thereafter treated with anti-AID antibodies (20 μg) and protein A-sepharose for 6 h at 4 °C in buffer A (20 mM Tris [pH 7.5], 1 mM DTT, 10 μM ZnCl2, 0.5 mM EDTA, 500 mM NaCl, and 10% (vol/vol) glycerol). To determine RPA interaction, the immunoprecipitate was washed in buffer A + 150 mM NaCl and then analyzed by Western blotting using anti-RPA32 antibodies (7, 16).
Supplementary Material
Acknowledgments.
We thank Tasuku Honjo for providing us with AID-deficient mice. We thank Aimee Williams, Lisa Aquaviva, and Yuko Fujiwara for technical assistance in generating and maintaining mice for experiments used in this work and Mark Curry of the Dana-Farber Cancer Research Institute for cell sorting. This work was supported by National Institutes of Health Grant AI31541 (to F.W.A.). F.W.A. is an Investigator of the Howard Hughes Medical Institute. J.C. is supported by a Scholars Grant from the Bressler Foundation and a Scholars Grant from the Damon Runyon Cancer Research Institute, U.B. is a Special Fellow of the Leukemia and Lymphoma Society of America, and B.S. is supported by an Ellison Medical Foundation/American Federation for Aging Research senior postdoctoral fellow research grant.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812304106/DCSupplemental.
References
- 1.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 3.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 4.Peled JU, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 5.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 6.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 8.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 11.Tian M, Alt FW. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- 12.Yu K, et al. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 13.Shinkura R, et al. The influence of transcriptional orientation on endogenous switch region function. Nat Immunol. 2003;4:435–441. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 15.Zarrin AA, et al. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 16.Basu U, et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 17.Pasqualucci L, Kitaura Y, Gu H, Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc Natl Acad Sci USA. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride KM, et al. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci USA. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basu U, Wang Y, Alt FW. Evolution of phosphorylation-dependent regulation of activation-induced cytidine deaminase. Mol Cell. 2008;32:285–291. doi: 10.1016/j.molcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinkura R, Okazaki IM, Muto T, Begum NA, Honjo T. Regulation of AID function in vivo. Adv Exp Med Biol. 2007;596:71–81. doi: 10.1007/0-387-46530-8_7. [DOI] [PubMed] [Google Scholar]
- 21.Basu U, Chaudhuri J, Phan RT, Datta A, Alt FW. Regulation of activation induced deaminase via phosphorylation. Adv Exp Med Biol. 2007;596:129–137. doi: 10.1007/0-387-46530-8_11. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: An assay of gene function in lymphocyte development. Proc Natl Acad Sci USA. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takizawa M, et al. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBride KM, et al. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med. 2008;205:2585–2594. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramiro AR, et al. Role of genomic instability and p53 in AID-induced c-myc-IgH translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolly CJ, Klix N, Neuberger MS. Rapid methods for the analysis of immunoglobulin gene hypermutation: Application to transgenic and gene targeted mice. Nucleic Acids Res. 1997;25:1913–1919. doi: 10.1093/nar/25.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perlot T, Li G, Alt FW. Antisense transcripts from immunoglobulin heavy-chain locus V(D)J and switch regions. Proc Natl Acad Sci USA. 2008;105:3843–3848. doi: 10.1073/pnas.0712291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conticello SG, et al. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol Cell. 2008;31:474–484. doi: 10.1016/j.molcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Gomez-Gonzalez B, Aguilera A. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci USA. 2007;104:8409–8414. doi: 10.1073/pnas.0702836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- 31.Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci USA. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-IgH translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng G, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoufouchi S, et al. Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med. 2008;205:1357–1368. doi: 10.1084/jem.20070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramiro AR, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 38.Pasqualucci L, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.