Abstract
In response to inflammatory stimulation, dendritic cells (DCs) have a remarkable pattern of differentiation (maturation) that exhibits specific mechanisms to control immunity. Here, we show that in response to Lipopolysaccharides (LPS), several microRNAs (miRNAs) are regulated in human monocyte-derived dendritic cells. Among these miRNAs, miR-155 is highly up-regulated during maturation. Using LNA silencing combined to microarray technology, we have identified the Toll-like receptor/interleukin-1 (TLR/IL-1) inflammatory pathway as a general target of miR-155. We further demonstrate that miR-155 directly controls the level of TAB2, an important signal transduction molecule. Our observations suggest, therefore, that in mature human DCs, miR-155 is part of a negative feedback loop, which down-modulates inflammatory cytokine production in response to microbial stimuli.
Keywords: bic/miR-155, LPS, TAB2, TLR/IL-1 pathway
Dendritic cells (DCs) are regulators of the immune response, whose antigen processing activities are controlled in response to inflammatory stimuli (e.g., Lipopolysaccharides, LPS) (1, 2). Among antigen presenting cells, DCs are the most efficient at initiating antigen-specific responses, inducing differentiation of naive T cells. Upon stimulation, DCs begin a maturation process characterized by dramatic functional changes, such as cytokine production or up-regulation of antigen presentation capacity (3). LPS sensing by TLR-4 activates 2 major pathways resulting in DC maturation. The first results in IKK, JNK and p38 MAPK stimulation through the MyD88-IRAK-TRAF6 pathway. The second involves the Toll-IL-1 receptor domain-containing adapter-inducing IFN-β (TRIF) and IRF3, which leads to type I IFN expression and costimulatory molecules up-regulation (4). Both pathways are required for optimal NF-κΒ activation and the production of cytokines such as IL-12 or IL-1β (5). DC activation results, therefore, in the enhanced ability to stimulate and polarize T cells in vitro and in vivo (6).
Recently, microRNAs (miRNAs) have emerged as a major class of gene expression regulators linked to most biological functions. miRNAs posttranscriptionally regulate gene expression by forming imperfect base pairing with sequences in the 3′ untranslated region (3′ UTR) of target mRNAs to prevent protein accumulation by repressing translation or by inducing mRNA degradation (7). More than 800 miRNAs have been identified in mammals (miRBase v.12.0), although their functions are only now being elucidated. The enzyme responsible for regulatory RNA biogenesis, Dicer, is required for lymphocytes function, which suggests regulatory roles for miRNAs in the immune system (8). However, the relationship between inflammation, innate immunity, and miRNA expression is just beginning to be explored (9–11). In the present study, we use a microarray-based screen to identify miRNAs induced during primary human monocyte-derived DC (moDC) maturation, and the immunologically relevant pathways controlled by these miRNAs.
We confirm that in activated primary moDCs, as in mouse bone marrow derived macrophages (12), the transcription of several miRNAs is regulated, among which the most prominently induced is miR-155. miR-155 is contained within the noncoding B cell integration cluster (Bic) gene (13) and has been linked to B and T cell immunity (9, 14, 15). Importantly, we identify TAB2, an adaptor in the TLR/IL-1 signaling cascade, as a direct target of miR-155 and we further show that miR-155 is part of the negative feed-back loop controlling IL-1β and other inflammatory cytokines produced during LPS-mediated DC activation. Thus, in addition of its role in hematopoietic lineage commitment, miR-155 plays an essential role in controlling the intensity of the inflammatory response to microbes in human DCs.
Results
Identification of miRNAs Regulated During moDC Maturation.
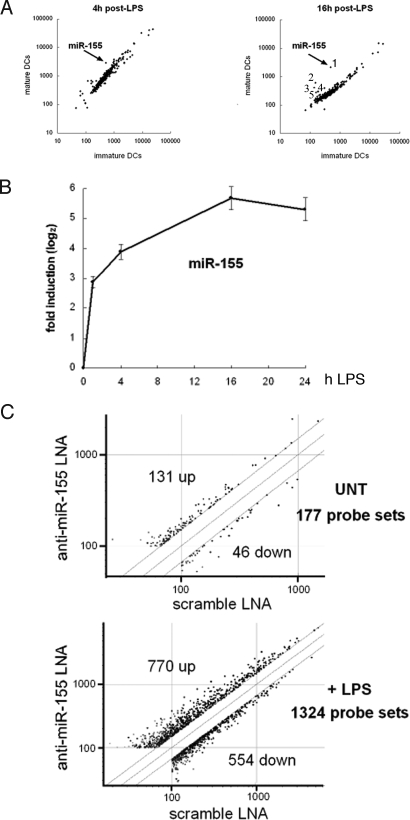
We generated a microarray containing 381 nonredundant human and rodent miRNA sequences, which was used to determine the expression levels of mature miRNAs purified from immature and 4 h or 16 h LPS-activated moDCs. Among the screened miRNAs, miR-155 showed the highest substantial induction over time (Fig. 1A). A quantitative PCR (qPCR) kinetics performed on mature miRNA indicated that miR-155 is rapidly induced, reaching a 50-fold induction 16 h after LPS treatment (Fig. 1B). Independently of our screen, 5 miRNAs (miR-142, miR-144, miR-150, miR-155 and miR-223) were recently found to be highly specific for hematopoietic cells (16). It has been reported that, besides miR-155, the miRNAs miR-146 and miR-125b are regulated in response to LPS-stimulation (12, 17–19). We performed qPCR to monitor the expression of these immunological-relevant miRNAs in LPS-activated moDCs (Fig. S1A). From this analysis, we could confirm the up-regulation of miR-155 and of 2 other miRNAs, miR-223 and miR-146a, albeit to a lesser extent than the first. In contrast, miR-125b, miR-144 and miR-142–5p were found to be down-regulated, whereas no regulation was found for miR-150. In most cases the trend of transcription regulation initiated after 4 h of LPS treatment was amplified after 16 h, suggesting that these miRNAs are involved in maintaining the directionality of the maturation process. Our results parallel data obtained in different human cell-lines, showing that miR-146a is induced by LPS, playing an inhibitory role on the inflammatory response (17).
Fig. 1.
miR-155 expression analysis in LPS-stimulated moDCs and gene expression analysis of moDCs after miR-155 knockdown. (A) Microarray analysis of miRNA expression in moDCs after stimulation with LPS. The scatter plot shows averaged (n = 16, quadruplicate hybridizations of 2 independent technical replicates, performed on 2 different DC preparations) background-subtracted raw intensities for each probe on both channels for Cy3-labeled control (immature DCs) and Cy5-labeled 4 h or 16 h LPS-treated cells (mature DCs) and their respective dye-swaps. Each dot represents one miRNA probe [(1) hsa_miR-155, (2) ambi_miR-7084, (3) hsa_miR-340, (4) hsa_miR-368, (5) mmu_miR-350]. (B) miR-155 expression was determined by relative qRT-PCR and normalized on U6 RNA levels. The data indicate the mean (± SD) of a triplicate qPCR, representing at least 3 independent experiments, each derived from a different DC preparation. (C) Total RNA was used for gene expression analysis using Affymetrix microarrays. We tested 12,190 probe sets that passed the control criteria. The scatter plot shows the anti-miR-155 LNA expression values normalized to scramble LNA values, applying a cut-off of 1.5. Each dot represents one probe set. A list of the 1324 probe sets identified in presence of LPS is available in Table S1. UNT, untreated cells.
TLR/IL-1 Signaling Pathway Is Modulated by miR-155 in Activated moDCs.
miR-155 is a key regulator of lymphoid development and immunity (14, 15), but its specific function in activated human DCs remains to be elucidated. We devised a strategy to functionally inhibit the mature form of miR-155 with the aim of identifying the signaling pathways controlled by miR-155 during DC maturation (Fig. S2). A LNA-modified oligonucleotide (20) specifically designed for miR-155 knockdown (anti-miR-155 LNA) was introduced in moDCs by nucleofection prior LPS stimulation. Twenty-four hours after transfection, a reduction in miR-155 levels of 8- and 32-fold was observed by qPCR in immature and mature moDCs respectively (Fig. S1B). We could also show that DC maturation remained normal under these conditions (Fig. S3). A comparative microarray analysis (Affymetrix U133 2.0 chip) was performed among miR-155 silenced (anti-miR-155-LNA) and control transfected (scramble-LNA) moDCs, exposed or not to LPS. As expected from the limited expression of miR-155 in nonactivated DCs, little variation in mRNA expression (177 probe sets differentially expressed employing a cut-off of 1.5) was detected upon miR-155 silencing. Conversely, in LPS-activated cells many mRNAs (1324 probe sets, 770 up-regulated and 554 down-regulated, complete list in Table S1) were affected by miR-155 inhibition (Fig. 1C). miRNAs inhibit mRNA translation or reduce mRNA stability after imperfect binding to miRNA-recognition elements (MREs) within the 3′-untranslated region (3′-UTR) of target genes. The specificity of this response is mediated by a “seed” region localized at residues 2–8 of the 5′-end of the miRNA (7). We validated our experimental approach using the Sylamer software (21) and demonstrated that miR-155 inhibition in activated DCs, clearly leads to the favored up-regulation of mRNAs containing miR-155 seeds (6 and 7 nt) in their 3′UTR, but not the seeds of anti-miR155-LNA or of the scramble-LNA oligonucleotides used as control (Fig. S4 A and B).
Using the Ingenuity Pathway analysis software (IPA version 6.3), we were then able to identify several “canonical pathways” affected in miR-155 silenced LPS-activated moDCs (Table S2). Interestingly, the top ranked pathway was the mitogen-activated protein kinase p38 (p38 MAPK) signaling pathway, whose genes were mostly up-regulated upon miR-155 inhibition (Fig. S5). The p38 MAPK pathway mediates cellular responses to injurious stress and immune signaling, serving cell type-specific inflammatory functions that can result in cytokine and chemokine production (22). Remarkably, 6 of the 9 top ranked pathways contained the proinflammatory cytokine IL-1 and its signal transduction components, suggesting a possible role of miR-155 in modulating the TLR/IL-1 signaling pathway, which is known to cross-talk with the p38 MAPK pathway (Fig. S5).
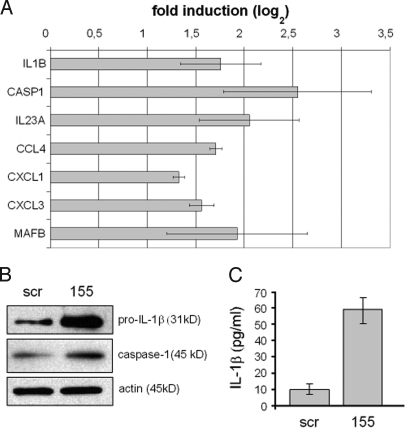
We next focused on the 770 probe sets up-regulated in miR-155 silenced LPS-activated moDCs (listed in Table S1) and looked for genes related to the inflammatory response. Among the up-regulated genes were IL-1α, IL-1β and other proinflammatory cytokines including IL-6, TNF-α and IL-23, and the chemokines MIP-1 α/β, MIP-2 α/β and MIP-3 α. Expression of the IL-1β-converting enzyme caspase 1 was also increased in response to miR-155 silencing (Table 1). Thus, an inflammation-related gene signature can be identified in miR-155 silenced moDCs. Because gene expression of proinflammatory cytokines is transient and is limited to the early phase of DC maturation (23), transcription levels of the proinflammatory genes identified upon miR-155 knockdown were monitored in activated moDCs with functional miR-155. We confirmed that the identified inflammation-related genes were rapidly and strongly up-regulated in the early phase of DC maturation (from 0 h to 4 h after LPS), followed by a relatively important down-regulation in the late phase (from 4 h to 16 h after LPS) (Table 1, fold change in miR-155 (+)). Because the expression of miR-155 occurs concomitantly with the down-regulation of these inflammatory genes mostly in this late phase of DC maturation, miR-155 could exert a major inhibitory role on the inflammatory response. In addition of inflammatory genes, the transcription factor MafB, involved in the differentiation of macrophages and DCs, was also found to be up-regulated upon miR-155 silencing. Interestingly MafB was also found to be a potential target of miR-223, which suggests the existence of regulation networks involving several miRs able to repress the same target mRNAs (Table S3). Whether miR-155 and miR-223 regulates the DC differentiation pathways or this finding only reflects an enhanced response to inflammatory cytokines will have to be clarified.
Table 1.
Regulation of selected inflammation-related genes in LPS-activated moDCs, in absence or presence of miR-155
| Affymetrix ID | Gene symbol | Description | Fold change miR-155 (−) moDCs | Fold change miR-155 (+) moDCs |
||
|---|---|---|---|---|---|---|
| 4 h vs 0 h | 16 h vs 4 h | P | ||||
| Cytokines | ||||||
| 205067_at | IL1B | interleukin 1, beta | 2.2* | 174.0 | −3.4 | 3.25 × 10−06 |
| 39402_at | 2.2* | 150.0 | −3.2 | 5.19 × 10−06 | ||
| 210118_s_at | IL1A | interleukin 1, alpha | 1.7 | 287.1 | −30.8 | 1.62 × 10−04 |
| 205207_at | IL6 | interleukin 6 | 1.6 | 239.1 | −7.3 | 5.63 × 10−06 |
| 220054_at | IL23A | interleukin 23, alpha subunit p19 | 2.3* | 20.0 | −8.6 | 7.78 × 10−04 |
| 207113_s_at | TNF | tumor necrosis factor alpha | 1.7 | 53.8 | −14.1 | 5.54 × 10−05x |
| Chemokines | ||||||
| 205114_s_at | CCL3 (MIP1A) | chemokine (C-C motif) ligand 3 | 1.8 | 37.4 | −3.6 | 1.27 × 10−05 |
| 204103_at | CCL4 (MIP1B) | chemokine (C-C motif) ligand 4 | 2.2* | 35.1 | −3.2 | 1.06 × 10−05 |
| 205476_at | CCL20 (MIP3A) | chemokine (C-C motif) ligand 20 | 2.0 | 386.0 | −5.0 | 1.94 × 10−05 |
| 204470_at | CXCL1 (GROA) | chemokine (C-X-C motif) ligand 1 | 2.3* | 237.7 | −5.4 | 2.43 × 10−05 |
| 209774_x_at | CXCL2 (MIP2A) | chemokine (C-X-C motif) ligand 2 | 3.2 | 103.9 | −5.2 | 2.10 × 10−04 |
| 207850_at | CXCL3 (MIP2B) | chemokine (C-X-C motif) ligand 3 | 2.9* | 108.7 | −5.1 | 2.73 × 10−04 |
| Apoptosis | ||||||
| 211368_s_at | CASP1 | caspase 1 (IL-1 beta-converting enzyme) | 2.3* | 3.8 | −2.5 | 2.58 × 10−02 |
| 211366_x_at | 2.2* | 3.3 | −2.6 | 2.51 × 10−02 | ||
| 209970_x_at | 1.8* | 3.1 | −2.3 | 2.89 × 10−02 | ||
| Ematopoiesis | ||||||
| 222670_s_at | MAFB | v-maf musculoaponeurotic fibrosarcoma oncogene homolog B | 2.6* | −2.1 | −2.0 | 3.51 × 10−02 |
The fold regulation of selected inflammation-related genes affected by miR-155 knockdown has been derived from Table S1 (miR-155 (-) moDCs) and compared with the fold regulation obtained in nontransfected LPS-activated moDCs (miR-155 (+) moDCs) in their early (from 0 h to 4 h after LPS) and late (from 4 h to 16 h after LPS) phase of maturation. All the indicated fold changes are derived from Affymetrix microarray experiments and, if presents, multiple probe sets are indicated. The fold changes of miR-155 (-) moDCs derive from 1 single microarray experiment, and the most relevant genes have been confirmed by qPCR (see Figure 3). The fold changes of miR-155 (+) moDCs are a mean of gene expression values derived from 4 independent microarray experiments, each experiment including a different blood donors. For each experiment, samples have been collected from untreated moDCs (0 h) and at 2 different time points after LPS-stimulation (4 h and 16 h). The expression values were first normalized per gene. The 4 h values were then normalized to 0 h (4 h vs 0 h) and the 16 h values to 4 h (16 h vs 4 h). The p-values have been calculated with the software GeneSpring GX 9.0, applying a repeated measures ANOVA test combined to a asymptotic p-value computation, without any correction. IL-1A and IL-1B are indicated in bold.
*Fold change confirmed by qPCR.
The induction of inflammation-related mRNAs identified by microarrays analysis was also established by qPCR, confirming the impact of miR-155 knockdown on the IL-1 signaling pathway (Fig. 2A). IL-1β and caspase 1 levels were quantified by immunoblot (see SI Materials and Methods). Both IL-1β and caspase 1 proteins were considerably increased in miR-155-silenced moDCs after LPS stimulation (Fig. 2B). As already observed in LPS-activated moDCs (24), IL-1β was mostly detected in the immature pro-IL-1β form, whereas active IL-1β was much less abundant (data not shown). Levels of secreted IL-1β were also considerably increased in miR-155-silenced moDCs compared with scramble-treated cells (Fig. 2C).
Fig. 2.
Induction of IL-1β and caspase-1 in moDCs after miR-155 knockdown. Immature human moDCs were transfected either with anti-miR-155 or with scramble LNA oligonucleotides and stimulated or not with LPS for 24 h. Cells and supernatants were collected and after harvesting both total RNA and cells extracts were enriched. (A) Total RNA was used to quantify expression of the indicated genes by relative qRT-PCR, normalizing on GAPDH RNA levels. The graph shows a log2-scale fold induction calculated by normalizing the anti-miR-155 LNA expression values on the scramble LNA values. Data indicate the mean (± SD) of a triplicate qPCR. (B) Cells extracts were used to perform immunoblots to assay the immature form of IL-1β (pro-IL-1β) and caspase-1. An actin immunoblot is shown for equal loading control. (C) Supernatants were used to perform quantitative sandwich enzyme immunoassays to measure secreted IL-1β. In all panels, data are representative of at least 3 independent experiments, each derived from a different DC preparation. Symbols: scr, scramble LNA; 155, anti-miR-155 LNA.
Identification of miR-155 Direct mRNA Targets in Mature moDCs.
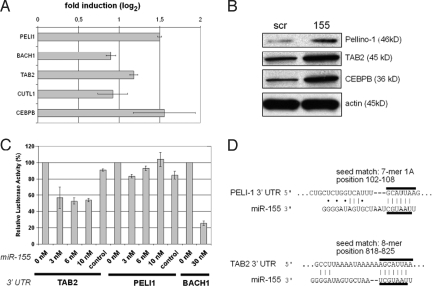
Although we could show that miR-155 controls the IL-1β pathway and maybe other inflammatory cytokine signaling cascades, its mode of action and the nature of its direct target(s) remained to be elucidated. Several miR-155 targets have been characterized exclusively in the murine hematopoietic system, including PU.1 (25) and AID (26, 27) in B cells, c-Maf in T cells (15), Bach1, Sla, Cutl1, Csf1r, Jarid2, Cebpβ, PU.1, Arntl, Hif1α and Picalm in RAW 264.7 myeloid cells (28), but none of them has been validated in human DCs and no relationship to the IL-1β signaling pathway revealed so far. We used the miRNA target prediction software TargetScan (version 4.2, www.targetscan.org) to scan for “seed matches” the 770 probe sets up-regulated in miR-155 silenced moDCs (Table S4). Importantly, among the 23 identified genes, we found the previously validated miR-155 targets BACH1, CUTL1 and CEBPB (28), suggesting that some of the targets are conserved between the murine and human immune systems. However, IL-1β and caspase 1 were not identified as candidate targets. Interestingly, 2 important adaptor proteins of the TLR/IL-1 signal transduction cascade, Pellino-1 and TAB2, displayed potential seed matches for miR-155 and were further investigated.
Both, Pellino-1 and TAB2 are part of a molecular complex containing the TNF receptor-associated factor 6 (TRAF6) and allow activation of the inflammatory response upon TLR4 or IL-1 receptor triggering (29, 30). Up-regulation of Pellino-1, TAB2, BACH1, CUTL1 and CEBPB mRNAs was confirmed by qPCR in miR-155-silenced moDCs. The fold-increase of Pellino-1 and TAB2 mRNAs was relatively modest, although still in the higher range of what was observed for the other known miR-155 targets (Fig. 3A). Immunoblot demonstrated that both molecules were induced at the protein level upon miR-155 inhibition in a similar manner to the IL-6 positive regulator CEBPB, used here as a positive control (Fig. 4B).
Fig. 3.
TAB2 is a direct target of miR-155 in moDCs. In A and B moDCs were treated, harvested and analyzed as described in Fig. 3. (A) qRT-PCR analysis for the indicated genes, normalizing on GAPDH RNA levels. (B) Immunoblots to assay Pellino-1, TAB2 and CEBPB. An actin immunoblot is shown for equal loading control. Symbols: scr, scramble LNA; 155, anti-miR-155 LNA. (C) miR-155 directly repress TAB2 mRNAs through 3′ UTR interactions. The full-length 3′ UTRs of the human genes BACH1 (positive control), Pellino-1, TAB-2 were cloned into a reporter vector, downstream of luciferase. These vectors were then cotransfected with the indicated amount of miR-155 or miR control precursor in 293T cells, and luciferase activity was quantified. The graph shows the percentage of remaining luciferase activity calculated by normalizing the miR-155 expression values on the miR-control values. Data are representative of at least 3 independent experiments. (D) Predicted interaction between the miR-155 seed and the seed matches on human Pellino-1 and TAB2 3′UTR mRNAs, determined with the software TargetScan. The type of seed match and the relative position on the 3′UTR mRNA are indicated. A-T and G-C base pairs are indicated with a line, whereas G:U bounds are indicated with a dot.
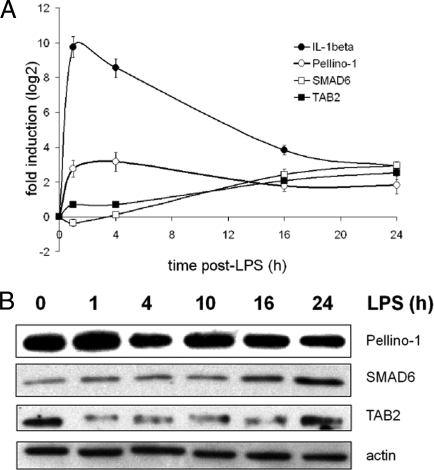
Fig. 4.
Regulation of IL-1β, TAB2, Pellino-1 and SMAD6 in LPS-activated moDCs. Immature human moDCs were stimulated with LPS and harvested at the indicated time points. (A) qRT-PCR kinetics analysis for the indicated genes, normalizing on GAPDH RNA levels. Data indicate the mean (± SD) of a triplicate qPCR, representing at least 3 independent experiments, each derived from a different DC preparation. (B) Immunoblot kinetics analysis for Pellino-1, SMAD6 and TAB2. An actin immunoblot is shown for equal loading control. Data are representative of at least 3 independent experiments.
We tested whether miR-155 could directly repress the identified mRNA targets through 3′ UTR interactions. Thus, the full-length 3′ UTRs of the human genes BACH1, Pellino-1, TAB-2 were cloned into a reporter vector, downstream of firefly luciferase cDNA. These vectors were then used to assess whether miR-155 could repress luciferase activity in 293T cells. BACH1 3′UTR, used here as a positive control, showed a reduction to 30% of total luciferase reporter activity, in presence of miR-155. Although TAB2 3′UTR displayed a reduced light emission down to 50%, Pellino-1 3′UTR did not display any significant reduction of luciferase levels, if compared with the control (Fig. 3C). Interestingly, when the 3′UTRs of their murine homologues were tested in the same experimental setting, mouse Pellino-1 (mPellino-1) 3′UTR displayed significantly higher repression levels (20%) than its human counterpart (Fig. S6). Computational prediction indicates that Pellino-1 mRNA 3′ UTR comprizes a 7-mer 1A seed match specific to miR-155, whereas TAB2 contains a 8-mer seed match (Fig. 3D). In the case of Pellino-1 mRNA, the doubt subsists on the reality of its direct targeting by miR-155, because only a modest inhibitory effect could be observed in the murine system. However, TAB2 3′UTR is clearly a newly identified direct miR-155 target, which probably exerts a negative feedback control on the TLR/IL-1 signaling pathway in the late phase of DC activation, thus potentially impairing IL-1β production and its autocrine function.
TAB2 Is Posttranscriptionally Regulated in Activated DCs.
Decreased levels of TAB2 could lead to a reduced recruitment of TRAF6, thus down-modulating the activation of TAK1 and of the NF-κB pathway (31). Moreover, upon TGF-β stimulation, the signaling protein Mothers against decapentaplegic homolog 6 (Smad6) forms a complex with Pellino-1. Smad6-Pellino-1 interaction abrogates signaling mediated by the complex of IRAK1, Pellino-1 and TRAF6 that forms after stimulation with IL-1β. Blockade of IRAK1-Pellino-1-TRAF6 signaling prevents the degradation of the inhibitor IκBα and subsequent nuclear translocation of NF-κB, and the expression of pro-inflammatory genes (32). Therefore, differences in the levels of TAB2, Pellino-1 and Smad-6 proteins could have potent anti-inflammatory activity, especially in a context in which Smad6 could be over-expressed and TAB2 down-modulated.
mRNAs levels for Pellino-1, Smad6 and TAB2 were measured by qPCR during LPS-induced moDC maturation (Fig. 4A). TAB2 mRNA levels increased 5-fold over 24 h of DC activation, whereas Pellino-1 mRNA expression was first induced 9-fold at 4 h after LPS, followed by a steadily down-regulation. In comparison, the levels of Smad6 mRNA continuously increased reaching 8-fold induction, 24 h after LPS-stimulation. As expected from miR-155 expression kinetics, TAB2 protein levels were quickly and strongly down-regulated upon maturation, despite a continuous increase in the transcription of the TAB2 mRNA. In addition, Pellino-1 protein levels did not overly decrease, whereas Smad6 levels were strongly induced (Fig. 4B). Thus, upon DC activation, the molecular ratio between Pellino-1 and Smad6 is modified in favor of Smad6, whereas TAB2 protein is lost presumably because of miR-155-mediated inhibition, potentially contributing to the negative control of IL-1R signaling.
Discussion
Interaction of resting immature DCs with TLR ligands leads to a cascade of proinflammatory cytokines (33). Although the inflammatory response is critical for the control of pathogenic infections, excessive production of proinflammatory cytokines is harmful to the host and can lead to autoimmunity. In addition, several cytokines are able to trigger DC-activation via autocrine or paracrine pathways (34). In this context, IL-1 forms an important part of the inflammatory response, increasing the expression of adhesion factors on endothelial cells to enable transmigration of leukocytes to sites of infection (35). Importantly, IL-1 is an activator of DCs (36, 37) and is involved in the pathogenesis of numerous diseases with an inflammation component (38). Thus, inflammation may cause more damage than healing, if its magnitude and duration are not strictly controlled by negative regulators that act at the molecular level.
Here, we demonstrate that miRNAs play a key role in modulating the IL-1 pathway in human DCs, by directly targeting adaptor molecules of the TLR/IL-1 signaling cascade. Similarly to what was observed in human monocytes (17), we have found that both miR-155 and miR146a are up-regulated upon LPS-stimulation of human primary DCs. Interestingly, miR146 has been proposed to target the 3′ UTRs of the TRAF6 and IRAK-1 genes, and to control Toll-like receptor and cytokine signaling through a negative feedback loop, thus reducing inflammatory cytokines production.
The mammalian p38 MAPK was originally discovered as an evolutionary conserved protein kinase, whose activity is induced by LPS and IL-1 (39, 40), and p38 has been recently shown to have distinct cell type-specific functions in inflammation, coordinating pro- and anti-inflammatory gene expression (22). Intracellular signaling pathways that are concurrently activated by the same stimulus often interact with one another through cross-regulatory feedback mechanisms. TAB2 acts as a multifunctional signaling molecule, facilitating both IL-1-dependent TRAF6 ubiquitination, assembly of the IL-1 signaling complex and activation of JNK, p38, and NF-kappa-B. We have demonstrated that TAB2 mRNA is directly targeted by miR-155. Therefore, we propose a model in which low miR-155 expression in the early phase of DC maturation enables the activation of the p38 MAPK pathway, favoring IL-1 β expression, which in turn would induce, in an autocrine fashion, the genes directly under the control of the IL-1-receptor and its signaling cascade. In contrast, high miR-155 and miR-146 expression in the late phase of DC maturation would inhibit p38 MAPK activation, silencing gene expression of IL-1 and other inflammatory cytokines. Thus, by targeting different molecular actors of the TLR/IL-1 signaling transduction pathway, miR-155 and miR-146 could exert together a regulation activity, which would limit the over-production of inflammatory cytokines during the late phase of LPS-mediated DC-activation. This loop could also be completed by the Pellino-1/Smad6 inhibitory complex, although we were not able to confirm that Pellino-1 is a direct target of miR-155 (Fig. S7).
In murine macrophages, miR-155 over-expression has also been postulated to increase TNF-α translation and to render mice hypersensitive to LPS/d-galactosamine-induced septic shock (18). Our results clearly contrast with these observations, and suggest that miR-155 is critical for the fine-tuning of the inflammatory response in human DCs. This hypothesis is supported by the recent finding that miR-155 is also induced by TGF-β, which has well characterized anti-inflammatory function (41).
We have found that the IL-6 signaling pathway is regulated by miR-155 in LPS-activated moDCs. Among the potential direct mRNA targets of miR-155 there was the transcription factor C/EBPβ, which activates IL-6 gene expression (42, 43). Thus, besides TAB2, other proteins involved in DC function and cytokine production could be the direct targets of miR-155. In fact, miR-155 has been reported to control the expression of the transcription factors c-Maf and PU.1 and to influence B cell development (15, 25). In the present work, we have identified a possible regulation of the c-Maf-related transcription factor, MafB, mediated by miR-155 and possibly miR-223. Because the ratio of PU.1 and MafB expression is a key to determine the fate of monocyte differentiation in DCs or macrophages (44), these miRs could maintain the proper equilibrium of expression among these different transcription factors, all necessary for DC lineage commitment.
Because of the potentially harmful effects of proinflammatory cytokines, it would not be surprising that >1 negative feedback loop controls inflammation, including miR-223 and miR-146. Regulatory mechanisms that operate at the transcriptional and posttranscriptional levels are complementary to the “safe guards” that negatively regulate cytokine processing at the protein level. In the case of IL-1β, besides regulation of gene expression, modulation takes place at the level of protein synthesis, processing and secretion. Thus, improving our understanding on the control of inflammatory cytokines expression, should ultimately lead to the development of drugs better suited to treat inflammatory diseases. This report highlights specific miRNAs and their targets genes, which could be attractive candidates for the development of new therapeutic interventions.
Materials and Methods
Human peripheral blood mononuclear cells (PBMC) were isolated from human leukapheresis by Ficoll-PaqueTM PLUS (Amersham Biosciences), washed 4 times with RPMI and CD14+ cells were immunomagnetically purified with AutoMACS (Miltenyi Biotech). Purified CD14+ monocytes were analyzed using a FACSCalibur (Becton Dickinson), confirming the purity of CD14+ cells to be 95%. To promote differentiation into iDC, purified CD14+ cells (0.5 × 106 cells per ml) were plated in 6-well plates (2 × 106 cells per well) and cultured in RPMI medium 1640 supplemented with 10% FCS, non essential amino acids, penicillin/streptomycin 100 ng/ml (>1,000 units/ml), recombinant hGM-CSF and 20 ng/ml (>100 units/ml) hIL-4 (PeproTech) for 5 days. At days 2 and 4, half of the volume of the medium was replaced with fresh medium. For DC maturation, 100 ng/ml LPS (Escherichia coli type 026:B6; Sigma) was added to the cells at day 5. For transfection experiments, immature DCs were harvested at day 5 of culture and resuspended in the Amaxa electroporation buffer (Human dendritic cell Nucleofector Kit, Amaxa) to a concentration of 2 × 107 cells per ml. A total of 24 μL of a 5 nmol of solution of anti-miR-155 LNA (miRCURY LNA Knockdown, hsa-miR-155, CCCCTATCACGATTAGCATTAA, Exiqon) or Scramble-LNA (Scramble-miR, GTGTAACACGTCTATACGCCCA, Exiqon) was mixed with 0.1 mL of cell suspension, transferred to a 2.0-mm electroporation cuvette, and nucleofected with an Amaxa Nucleofector apparatus. After electroporation, cells were transferred to complete medium, and cultured in 6-well plates at 37 °C. One hour after electroporation, LPS was added to the cells that were incubated for further 24 h until analysis.
Supplementary Material
Acknowledgments.
We thank the Centre National de la Recherche Scientifique Affymetrix facility, the Portugese Foundation for Science and Technology, and Anton Enright and Harpreet Saini at European Bioinformatics Institute Cambridge for expert data analysis with the Sylamer software. This work was supported by grants from the Ministère de la Recherche et de la Technologie, Réseau National Génopole, La Ligue Nationale Contre le Cancer and the Human Frontier of Science Program (P.P. and M.S.) and the Swiss National Science Foundation (M.C. and W.R.). P.P. and M.S. are European Molecular Biology Organization Young Investigators and are part of the DC-THERA Network of Excellence.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. GSE13296).
This article contains supporting information online at www.pnas.org/cgi/content/full/0811073106/DCSupplemental.
References
- 1.Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 3.Reis e Sousa C. Dendritic cells in a mature age. Nat Rev Immunol. 2006;6:476–483. doi: 10.1038/nri1845. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 5.Shen H, Tesar BM, Walker WE, Goldstein DR. Dual signaling of MyD88 and TRIF is critical for maximal TLR4-induced dendritic cell maturation. J Immunol. 2008;181:1849–1858. doi: 10.4049/jimmunol.181.3.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudziak D, et al. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- 7.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Otsuka M, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: New regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 10.Lindsay MA. MicroRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Hoefig KP, Heissmeyer V. MicroRNAs grow up in the immune system. Curr Opin Immunol. 2008;20:281–287. doi: 10.1016/j.coi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 12.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 14.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tili E, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 19.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 20.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Van Dongen S, et al. Detecting microRNA binding and siRNA off-target effects from expression data. Nat Meth. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim C, et al. The kinase p38 alpha serves cell type-specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat Immunol. 2008;9:1019–1027. doi: 10.1038/ni.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Q, et al. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294:870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 24.Gardella S, et al. Secretion of bioactive interleukin-1beta by dendritic cells is modulated by interaction with antigen specific T cells. Blood. 2000;95:3809–3815. [PubMed] [Google Scholar]
- 25.Vigorito E, et al. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng G, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connell RM, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schauvliege R, Janssens S, Beyaert R. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: A role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006;580:4697–4702. doi: 10.1016/j.febslet.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 30.Takaesu G, et al. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol Cell. 2000;5:649–658. doi: 10.1016/s1097-2765(00)80244-0. [DOI] [PubMed] [Google Scholar]
- 31.Lee SW, Han SI, Kim HH, Lee ZH. TAK1-dependent activation of AP-1 and c-Jun N-terminal kinase by receptor activator of NF-kappaB. J Biochem Mol Biol. 2002;35:371–376. doi: 10.5483/bmbrep.2002.35.4.371. [DOI] [PubMed] [Google Scholar]
- 32.Choi KC, et al. Smad6 negatively regulates interleukin 1-receptor-Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat Immunol. 2006;7:1057–1065. doi: 10.1038/ni1383. [DOI] [PubMed] [Google Scholar]
- 33.Blanco P, Palucka AK, Pascual V, Banchereau J. Dendritic cells and cytokines in human inflammatory and autoimmune diseases. Cytokine Growth Factor Rev. 2008;19:41–52. doi: 10.1016/j.cytogfr.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banchereau J, Pascual V, Palucka AK. Autoimmunity through cytokine-induced dendritic cell activation. Immunity. 2004;20:539–550. doi: 10.1016/s1074-7613(04)00108-6. [DOI] [PubMed] [Google Scholar]
- 35.Dinarello CA. The interleukin-1 family: 10 years of discovery. FASEB J. 1994;8:1314–1325. [PubMed] [Google Scholar]
- 36.Wesa AK, Galy A. IL-1 beta induces dendritic cells to produce IL-12. Int Immunol. 2001;13:1053–1061. doi: 10.1093/intimm/13.8.1053. [DOI] [PubMed] [Google Scholar]
- 37.Eriksson U, et al. Activation of dendritic cells through the interleukin 1 receptor 1 is critical for the induction of autoimmune myocarditis. J Exp Med. 2003;197:323–331. doi: 10.1084/jem.20021788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 39.Han J, Lee JD, Bibbs L, Ulevitch RJ. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 40.Freshney NW, et al. Interleukin-1 activates a novel protein kinase cascade that results in the phosphorylation of Hsp27. Cell. 1994;78:1039–1049. doi: 10.1016/0092-8674(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 41.Kong W, et al. MicroRNA-155 Is Regulated by TGF{beta}/Smad Pathway and Contributes to Epithelial Cell Plasticity by Targeting RhoA. Mol Cell Biol. 2008 doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spooner CJ, Guo X, Johnson PF, Schwartz RC. Differential roles of C/EBP beta regulatory domains in specifying MCP-1 and IL-6 transcription. Mol Immunol. 2007;44:1384–1392. doi: 10.1016/j.molimm.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Kuilman T, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 44.Bakri Y, et al. Balance of MafB and PU. 1 specifies alternative macrophage or dendritic cell fate. Blood. 2005;105:2707–2716. doi: 10.1182/blood-2004-04-1448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.