Abstract
Autophagy is a highly conserved process that maintains homeostasis by clearing damaged organelles and long-lived proteins. The consequences of deficiency in autophagy manifest in a variety of pathological states including neurodegenerative diseases, inflammatory disorders, and cancer. Here, we studied the role of autophagy in the homeostatic regulation of innate antiviral defense. Single-stranded RNA viruses are recognized by the members of the RIG-I-like receptors (RLRs) in the cytosol. RLRs signal through IPS-1, resulting in the production of the key antiviral cytokines, type I IFNs. Autophagy-defective Atg5−/− cells exhibited enhanced RLR signaling, increased IFN secretion, and resistance to infection by vesicular stomatitis virus. In the absence of autophagy, cells accumulated dysfunctional mitochondria, as well as mitochondria-associated IPS-1. Reactive oxygen species (ROS) associated with the dysfunctional mitochondria were largely responsible for the enhanced RLR signaling in Atg5−/− cells, as antioxidant treatment blocked the excess RLR signaling. In addition, autophagy-independent increase in mitochondrial ROS by treatment of cells with rotenone was sufficient to amplify RLR signaling in WT cells. These data indicate that autophagy contributes to homeostatic regulation of innate antiviral defense through the clearance of dysfunctional mitochondria, and revealed that ROS associated with mitochondria play a key role in potentiating RLR signaling.
Keywords: innate immunity, interferon, reactive oxygen species, virus infection, mitochondria
Autophagy is an ancient evolutionarily conserved pathway designed to maintain cellular homeostasis by degrading long-lived proteins and organelles in the cytosol. It has also been studied extensively as a critical survival mechanism during starvation conditions (1, 2). Recent studies demonstrated that autophagy is used by the cells of the innate and adaptive immune systems to combat viral infections (3, 4).
Current paradigm suggests that autophagy or the molecules required for autophagy could regulate these innate viral recognition pathways in distinct ways. RNA viruses are recognized by two distinct innate sensors (5). In plasmacytoid dendritic cells (pDC), recognition of ssRNA viruses occurs in the endosomes via Toll-like receptors 7 and 8 (6–8). Autophagy plays a key role in the recognition of certain ssRNA viruses by delivering viral replication intermediate from the cytosol to the endosome, where it engages Toll-like receptor 7 activation in pDCs (9). In contrast to pDCs, most other cell types of the body use cytosolic sensors of viral replication via retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA-5) belonging to the RLR family (10–13). A recent study reported by Jounai et al. demonstrated that innate recognition of vesicular stomatitis virus (VSV) in mouse embryonic fibroblasts (MEFs) via the RIG-I pathway is negatively regulated by the Atg5-Atg12 conjugate (14). This study showed that type I interferons and cytokine production were enhanced in Atg5−/− MEFs in response to infection with VSV, which is recognized through RIG-I (15), or to transfection with Poly I:C, which triggers MDA-5-dependent signaling (12, 16). Consequently, Atg5−/− MEFs were more resistant to VSV infection. The authors further demonstrated that the Atg5-Atg12 conjugate directly associates with RIG-I, MDA-5 and the adaptor protein IFN-β promoter stimulator 1 (IPS-1) (17–20)—also known as mitochondrial antiviral signaling (19), Cardif (21), or virus-induced signaling adaptor (20)—through the caspase recruitment domain (14). These results revealed a non-canonical role of Atg family members as suppressors of RLR signaling (22). Thus, in contrast to pDCs, fibroblasts that rely on cytosolic sensors of viral replication appear to use molecules involved in autophagy to repress RLR signaling. However, the role of the process of autophagy in the regulation of RLR signaling has not been examined.
Here, we addressed the importance of the autophagy-dependent pathways in regulating RLR signaling in MEFs and primary macrophages. Autophagy is one of the major pathways by which damaged mitochondria are cleared from the cells through a process known as mitophagy (23). We hypothesized that autophagy could regulate the level of RLR signaling in the following two non-mutually exclusive ways. First, cells deficient in autophagy may produce more IFNs and cytokines upon RLR stimulation as a result of an increase in the number of mitochondria per cell, leading to IPS-1 accumulation on a per-cell basis. A previous study has shown that varying the amount of IPS-1 by transfection and siRNA knockdown resulted in enhanced or reduced IFN and cytokine activation, respectively (17). Second, enhanced RLR signaling in the absence of autophagy may result from the accumulation of dysfunctional mitochondria that harbor elevated levels of reactive oxygen species (ROS). Cells defective in autophagy have been shown to accumulate ROS (24). In this study, we provide evidence for both of these possibilities. We demonstrate that dysfunctional mitochondria accumulated in the absence of Atg5. These cells exhibited a corresponding increase in the levels of IPS-1 protein and an increase in RLR signaling. More importantly, we show that Atg5−/− cells accumulated ROS localized to the mitochondria. Depletion of ROS by antioxidant treatment significantly diminished the amplified RLR signaling phenotype in Atg5−/− MEFs. Further, we show that artificially increasing mitochondria-associated ROS in WT MEFs was sufficient to enhance RLR signaling. Such ROS-dependent activation of RLR, together with the increase in IPS-1 levels, account for the functional augmentation of RLR and viral clearance in the absence of autophagy. Thus, these results provide insights into the role of constitutive autophagy in maintaining cellular homeostasis by clearing dysfunctional mitochondria, the absence of which leads to accumulation of ROS and dysregulation of RLR pathways.
Results
Atg5 Deficiency in MEFs Leads to Enhanced Type I IFN Production and Suppression of Viral Infection.
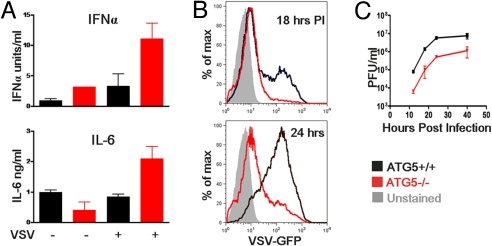
A recent report indicated that innate recognition of VSV in MEFs via the RIG-I pathway is regulated by the Atg5-Atg12 conjugate (14). Atg5−/− MEFs produced enhanced type I interferons and cytokines in response to VSV and were more resistant to VSV replication. Upon examination of Atg5+/+ or Atg5−/− MEFs in response to VSV infection (RIG-I stimulation) or Poly I:C transfection (MDA-5 stimulation), we indeed observed an increase in IFN-α, IFN-β, and IL-6 production at both the mRNA [supporting information (SI) Fig. S1 A and B] and protein (Fig. 1A) levels. Consequently, VSV replication was suppressed in Atg5−/− MEFs as measured by VSVG-GFP expression by FACS (Fig. 1B) and by plaque assays of released virus from MEF supernatant (Fig. 1C). In contrast, enhanced RLR signaling was not detected in MEFs lacking a non-autophagosome-related gene, MyD88 (Fig. S1A), indicating the specificity of the phenotype to Atg5-deficient MEFs. Thus, these results reproduced the previous data published by Jounai et al. (14), and the system provided us with a means to address the importance of autophagy in regulating RLR signaling. These data indicated that Atg5 is responsible for regulating the levels of RLR stimulation in MEFs.
Fig. 1.
Atg5-deficient MEFs show increased cytokine production to Poly I:C and VSV stimulation and are resistant to infection. WT, and Atg5−/− MEFs were incubated with VSV-GFP (at a multiplicity of infection of 4) or transfected with 1 μg/mL Poly I:C. Twelve hours later, IFNα and IL-6 production was assessed by ELISA (A). WT and Atg5−/− MEFs were infected with VSV-GFP (at a multiplicity of infection of 1) and the levels of infection were determined by measuring GFP expression by FACS at 18 and 24 h after infection (B) and by measuring viral titers in the supernatants by plaque assay at the indicated time points (C). Results are representative of 6 separate experiments.
To avoid the possible effects of secondary mutations that often take place in high passage MEFs, we used multiple lines of primary MEFs (below 10th passage) and confirmed all of our results throughout the course of our study. Further, to examine whether the phenotype of Atg5−/− MEFs is caused by overexpression of compensatory mechanisms, we complemented Atg5−/− MEFs with lentiviral transduction of Atg5 (Fig. S2). The complementation with Atg5 in the knockout MEFs resulted in significant reduction of RLR signaling. However, it did not completely revert the phenotype to that of WT MEFs, suggesting that the phenotype in Atg5−/− MEFs is caused by cumulative effects of the loss of Atg5 over time.
Atg5 Deficiency in MEFs Leads to the Accumulation of Healthy and Dysfunctional Mitochondria.
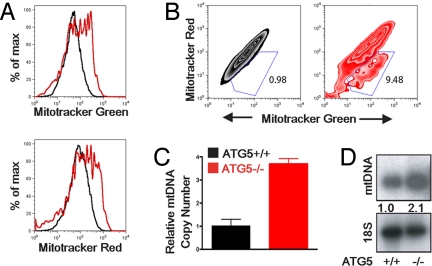
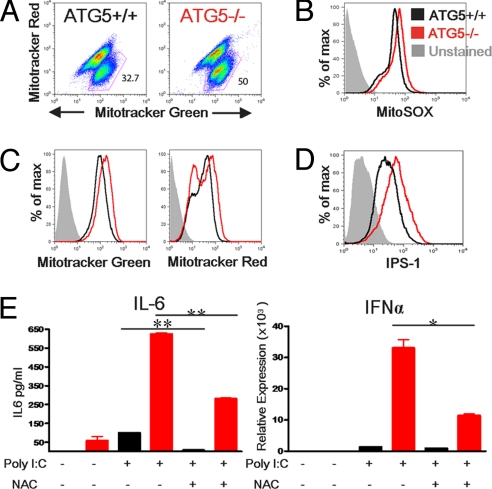
Based on the finding that an Atg5-dependent process specifically controls the RLR pathway, we focused on the importance of the interface between autophagy and mitochondria in RLR signaling. Autophagy is one of the major pathways by which damaged mitochondria are cleared from the cells through a process known as mitophagy (23). We hypothesized that autophagy may regulate the level of RLR signaling in the following manners: cells deficient in autophagy may produce more IFNs and cytokines as a result of (i) an increase in the number of mitochondria per cell, such that more IPS-1 accumulates on a per-cell basis; and (ii) an increase in intracellular ROS resulting from the accumulation of damaged mitochondria. To test these possibilities, we first examined the number of both intact and dysfunctional mitochondria in WT and Atg5−/− MEFs. Our results indicated that Atg5−/− MEFs contain roughly 2-fold more mitochondria on a per-cell basis compared with WT MEFs (Fig. 2A and Fig. S3D). We then used two types of mitochondria-specific label to distinguish respiring (MitoTracker Red) versus total (MitoTracker Green) mitochondria. The increase in mitochondria was accounted for by an increase in both the intact (MitoTracker Green-positive, MitoTracker Red-positive) and dysfunctional non-respiring (MitoTracker Green-positive, MitoTracker Red-negative) mitochondria (Fig. 2B and Fig. S3 A and B). To provide an independent confirmation of the increase in the number of mitochondria, we quantified the amount of mitochondrial DNA present in the cells in relation to genomic DNA by quantitative PCR (Fig. 2C) and by Southern blotting for mitochondrial DNA (Fig. 2D). These analyses also revealed a 2 to 3-fold increase in the total amount of mitochondrial DNA present in Atg5−/− MEFs compared with WT MEFs.
Fig. 2.
Dysfunctional mitochondria accumulate in Atg5-deficient MEFs. (A and B) Atg5+/+ and Atg5−/− MEFs were stained with 100 nM MitoTracker Green (which stains the lipid membrane of the mitochondria) and 100 nM MitoTracker Red (which fluoresces upon oxidation in respiring mitochondria). Histograms (A) and contour plots (B) of FACS analysis are depicted. (C) Mitochondrial DNA copy number was measured by quantitative PCR and normalized to nuclear DNA levels in a ratio of mtDNA COI over 18S rDNA. Relative mitochondrial DNA copy numbers are depicted. (D) Mitochondrial DNA levels were assessed by Southern blotting using a mitochondrial DNA-specific probe. Numbers indicate relative intensity of mtDNA normalized to cellular DNA. Data are representative of 3 similar experiments.
IPS-1 Overexpression in Atg5−/− MEFs.
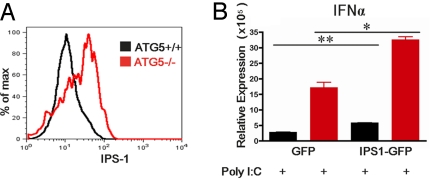
Next, as IPS-1 is associated with the outer membrane of mitochondria, we reasoned that the increase in the number of mitochondria might be accompanied by an increase in IPS-1 levels in a cell. Upon FACS analysis of Atg5+/+ and Atg5−/− MEFs for the levels of IPS-1 by intracellular staining, it became apparent that Atg5−/− MEFs indeed showed a 2-fold increase (Fig. S3D) in IPS-1 by mean fluorescence intensity compared with the WT MEFs (Fig. 3A). This increase in IPS-1 was confirmed by Western blot (Fig. S4). These data indicated that the deficiency in autophagy leads to an approximately 2-fold increase in the total number of mitochondria, many of which are dysfunctional, and a parallel increase in the levels of mitochondria-associated IPS-1 in Atg5−/− MEFs.
Fig. 3.
IPS-1 overexpression in Atg5−/− MEFs. (A) Intracellular staining of IPS-1 protein analyzed by FACS in Atg5+/+ and Atg5−/− MEFs. (B) Atg5+/+ and Atg5−/− MEFs were transduced with lentivirus expressing IPS-1-GFP fusion protein or GFP (control). Cells expressing only high levels of GFP were sorted by FACS and stimulated with 1 μg/mL Poly I:C complexed to Lipofectamine. Twelve hours later, IFN-α and IFN-β mRNA levels were assessed by RT quantitative PCR. *P < 0.05, **P < 0.005. Similar results were obtained from 2 separate experiments.
We next asked if the increase in IPS-1 levels in the Atg5−/− MEFs can solely account for the increase in IFN synthesis following Poly I:C transfection. To this end, we transduced Atg5+/+ and Atg5−/− MEFs with lentivirus expressing IPS-1 fused with GFP. Cells expressing high levels (MFI levels between 6 × 102 and 6 × 104) of GFP were sorted by FACS and stimulated with Poly I:C. If the differential expression levels of IPS-1 were the sole reason for the enhanced RLR signaling in Atg5-deficient MEFs, we would expect to see similar levels of IFN synthesis by normalizing the levels of IPS-1 in WT and Atg5−/− MEFs. IPS-1-expression by lentiviral transduction led to an increase in IFN synthesis (Fig. 3B). However, despite the similar levels of IPS-1 expression in both groups, Atg5−/− MEFs still produced higher levels of IFN in response to Poly I:C transfection (Fig. 3B). Therefore, these data indicated that the increase in IPS-1 levels does not fully account for the increase in RLR signaling in the absence of Atg5.
Absence of Autophagy Leads to ROS Accumulation in Mitochondria.
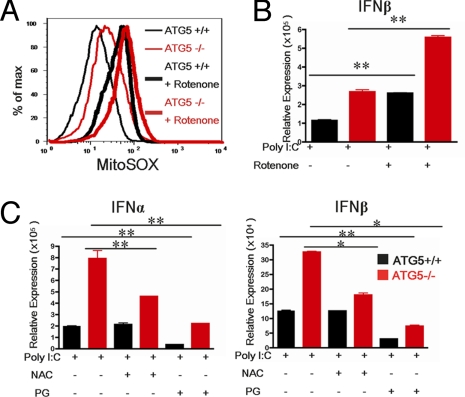
The results thus far indicated that the lack of autophagy, in addition to the increase in IPS-1 levels, results in hyperstimulation of RLR in Atg5-deficient MEFs. Given that the absence of autophagy leads to the accumulation of dysfunctional mitochondria (Fig. 2), we reasoned that the presence of the dysfunctional mitochondria is somehow potentiating RLR activation in concert with the elevated levels of IPS-1. To test this hypothesis, we examined whether the phenotype of the Atg5−/− MEFs might reflect an increase in ROS production in the dysfunctional mitochondria. In non-phagocytic cells such as the fibroblasts, mitochondria are the major sites for generation of ROS, including superoxide (O2·−), hydroxyl radical (HO·), and hydrogen peroxide (H2O2). Previous reports have shown that cells defective in autophagy accumulate ROS (24). To examine ROS levels in the mitochondria, we used the mitochondrial-specific ROS indicator MitoSOX to highly selectively detect superoxide in the mitochondria of live cells. MitoSOX is targeted to the mitochondria and its oxidation by superoxide leads to the generation of red fluorescence. Our data indicated that MitoSOX fluorescence was enhanced in Atg5−/− MEFs compared with WT (Fig. 4A and Fig. S3D). Combined with the fact that there are more mitochondria with lower MitoTracker Red signal (i.e., loss of membrane potential; Fig. 2), and higher ROS accumulation (i.e., gain of oxidative lesions; Fig. 4), these data suggest that cells require the Atg5 gene to clear damaged mitochondria. Therefore, constitutive autophagy fine-tunes the clearance rate of the dysfunctional mitochondria.
Fig. 4.
Autophagy-deficient MEFs have increased levels of mitochondrial ROS. Atg5+/+ and Atg5−/− MEFs treated with 1 μM of rotenone for 12 h. (A) Levels of mitochondria associated ROS in Atg5+/+ and Atg5−/− MEFs were analyzed by MitoSOX labeling. (B) After 12 h rotenone pretreatment, cells were transfected with 1 μg/mL Poly I:C. Expression levels of IFN-β were assessed by RT quantitative PCR 12 h after transfection. (C) MEFs were pretreated with 10 mM of the anti-oxidant NAC or 100 μM of PG for 15 min and transfected with Poly I:C (1 μg/mL). IFN-α and IFN-β production was assessed by RT quantitative PCR at 12 h after transfection. *P < 0.05, **P < 0.005. Data are representative of 3 separate experiments.
ROS Accumulation Is Required to Enhance RLR Signaling.
To test whether increased ROS contributes to the enhanced cytokine production in Atg5−/− MEFs, we pretreated WT and Atg5−/− MEFs with an antioxidant, N-acetyl-L-cysteine (NAC), before Poly I:C stimulation. The NAC treatment significantly reduced IFN-α and IFN-β in the Atg5−/− MEFs to almost WT levels (Fig. 4C). Further, treatment of MEFs with another antioxidant, propyl gallate (PG), significantly reduced Poly I:C-induced IFN and cytokine production in all groups (Fig. 4C). These data demonstrated that mitochondrial ROS accumulated in the absence of Atg5, and that ROS is required for the enhancement of RLR signals. In addition, data from the PG treatment experiments imply that constitutive levels of ROS are used by RLR for signaling.
Raising Mitochondrial ROS Levels Is Sufficient to Enhance RLR Signaling.
Next, we tested whether an increase in mitochondrial ROS is sufficient to enhance RLR stimulation in WT cells. To this end, we treated cells with rotenone to induce ROS accumulation localized to the mitochondria. Rotenone is a well known inhibitor of electron transfer from complex I to ubiquinone of the electron transport chain (25), resulting in ROS accumulation (26). Rotenone treatment resulted in enhanced mitochondrial ROS (Fig. 4B and Fig. S3H). Importantly, treatment of both WT and Atg5−/− MEFs with rotenone resulted in a significant increase in IFN production in response to Poly I:C transfection (Fig. 4B). Therefore, excess ROS induction in the mitochondria is sufficient to potentiate enhanced RLR stimulation even in the presence of constitutive autophagy.
Previous studies have indicated that IPS-1 is cleaved by endogenous caspases upon stress and apoptosis (27, 28). One possible explanation for our observation is that ROS-mediated stress signals cleave IPS-1 only in WT but not Atg5−/− MEFs and suppress RLR signaling. To directly test this hypothesis, WT and Atg5−/− MEFs were transfected with Poly I:C in the presence of rotenone (to induce ROS accumulation on mitochondria) or antioxidant NAC (to reduce ROS) and cleavage of IPS-1 was measured by Western blot (Fig. S4). These analyses revealed no preferential cleavage of IPS-1 in WT compared with Atg5−/− MEFs. Another possibility is that ROS might enhance RIG-I levels in cells. To this end, we measured RIG-I mRNA in WT and Atg5−/− MEFs following Poly I:C stimulation following rotenone or NAC treatment. Although Poly I:C induced RIG-I transcription in both WT and Atg5−/− MEFs, neither rotenone nor NAC treatments affected the increase in RIG-I levels (Fig. S5). Importantly, RIG-I mRNA levels were comparable in Poly I:C-stimulated WT and Atg5−/− MEFs that were treated with either rotenone or NAC. Therefore, these data indicated that, in the absence of autophagy, ROS accumulation on mitochondria potentiates RLR signaling independent of IPS-1 cleavage or RIG-I transcription.
Atg5-Deficient Primary Macrophages Exhibit Amplified RLR Signaling.
Thus far, our study used MEFs generated from Atg5+/+ or Atg5−/− animals. To examine whether the phenotype observed with MEFs translates to primary cells, we generated neonatal liver macrophages from Atg5+/+ or Atg5−/− pups. Analyses of the number and the respiratory status of mitochondria revealed that Atg5-deficeint macrophages accumulated both healthy and damaged mitochondria (Fig. 5 A and C and Fig. S3 E and G), similar to the MEFs (Fig. 2). Consistent with our observation with MEFs, Atg5−/− macrophages sustained higher levels of ROS on mitochondria (Fig. 5B) and demonstrated an accumulation of IPS-1 (Fig. 5D and Fig. S3F). Finally, when primary macrophages were stimulated with Poly I:C, much higher levels of IFN-α and IL-6 were produced from Atg5−/− macrophages compared with WT cells (Fig. 5E). These data indicated that autophagy controls the clearance of damaged mitochondria in primary cells, and in its absence, leads to amplification of RLR signaling in both MEFs and macrophages.
Fig. 5.
Atg5-deficient primary macrophages exhibit amplified RLR signaling. Neonatal liver macrophages were generated from Atg5+/+ or Atg5−/− pups. Cells were stained with 100 nM MitoTracker Green and 100 nM MitoTracker Red as in Fig. 2, and both dot plots (A) and histograms (C) of FACS analysis are depicted. Additionally, cells were labeled with MitoSOX as in Fig. 4 (B) or with antibody to IPS-1 as in Fig. 3. (D) Primary macrophages were transfected with 10 μg/mL Poly I:C in the presence or absence of NAC, and 12 h later, the levels of IL-6 were assessed by ELISA and the levels of IFNα were assessed by RT quantitative PCR (E). *P < 0.05, **P < 0.005. Results are representative of 2 similar experiments.
Discussion
In this study, we examined the role of autophagy in RLR signaling. We demonstrated that dysfunctional and healthy mitochondria accumulate in the absence of Atg5. This resulted in a parallel accumulation of IPS-1 protein on a per-cell basis, leading to enhanced IFN and cytokine production upon RLR engagement. However, overexpression of IPS-1 in WT cells did not confer the capacity to elevate RLR signaling to the levels found in Atg5−/− MEFs. This led us to look for additional mechanisms by which lack of autophagic clearance of dysfunctional mitochondria might affect RLR signaling. We found that ROS accumulate in the mitochondria of Atg5−/− MEFs. ROS associated with damaged mitochondria was found to be both necessary and sufficient to support elevated RLR-induced gene transcription. Further, we demonstrated that all of these processes are also essential in regulating RLR signaling in primary macrophages. Collectively, our data demonstrated the critical importance of autophagy in maintaining cellular homeostasis—both by clearing damaged and excess mitochondria and regulating the levels of IPS-1. More importantly, we discovered that mitochondrial-associated ROS plays a key role in the positive regulation of RLR signaling.
Our data revealed that IPS-1 levels increased in correlation with the number of mitochondria in Atg5−/− cells. IPS-1 is expressed on the outer membrane of the mitochondria because of the presence of a mitochondrial membrane targeting sequence (19). We did not observe any increase in IPS-1 mRNA levels in Atg5−/− cells compared with WT MEFs (data not shown), indicating that the increase in IPS-1 protein levels is caused by post-transcriptional regulation. We ruled out the possibility that such post-transcriptional regulation is caused by ROS-mediated protection of IPS-1 from cleavage by an endogenous caspase. Thus, the corresponding increase in the IPS-1 protein levels with mitochondria number suggests that mitochondrial location of IPS-1 might provide protection from degradation, and/or that the level of IPS-1 is titrated by the number of available mitochondrial membrane sites. Although IPS-1 increase might account for a part of the enhanced RLR stimulation in Atg5−/− cells, based on the results of antioxidant treatment, ROS accumulation is largely responsible for the RLR phenotype of Atg5−/− cells. ROS can occur in different locales in a cell, including mitochondria, endoplasmic reticulum, peroxisomes, plasma membrane, and within the cytosol. ROS are also generated by phagocytes for the elimination of pathogens. In addition, ROS induction occurs through a variety of ligand-receptor interactions as well. Of these, the ROS generated in the mitochondria as a byproduct of the electron transport chain account for the majority of cellular ROS. To protect against the damage from ROS, the cell possesses a formidable array of antioxidant enzymes capable of attenuating ROS. Oxidative stress results from an imbalance between oxidant production and the antioxidant capacity of the cell to prevent oxidative injury. Autophagy is a key mechanism by which damaged mitochondria are removed from the cells. In its absence, damaged mitochondria harboring elevated levels of ROS accumulate (24). The toxic effects of oxidative stress have been implicated in a large number of human diseases and aging. However, although ROS are better known for the DNA, protein, and lipid damage caused in their excess, recent studies have unveiled their duality as important signal transduction mediators via reversible oxidization of reactive cysteine residues within a protein, which can modify protein function. In particular, ROS are a well known activator of NFκB, AP-1, MAPK, and PI3K pathways (29). A role for ROS in antiviral IFN production has not previously been reported, and it is important to identify the molecular mechanism by which ROS augment IFN. Our results indicated that mitochondrial ROS positively regulate RLR signaling. These results suggest that a putative target of ROS is either a negative regulator of RLR that is inactivated by oxidation, or a positive regulator of RLR that is activated by oxidation. It is tempting to speculate that such target of oxidation is also associated with the mitochondria. It will be important to determine the molecular target(s) of ROS in RLR signaling pathway.
In conclusion, our study revealed that autophagy plays a crucial role in the homeostatic clearance of damaged mitochondria, the absence of which leads to accumulation of ROS and dysregulation of RLR signaling. It is known that many viruses have evolved to encode genes to block autophagy in infected cells (30). Combined with the role of the Atg5-Atg12 conjugate in RLR inhibition (14), an implication of our study is that, during chronic infection with a virus capable of autophagy inhibition, over-stimulation of RLR signaling might lead to excess or prolonged secretion of cytokines with immunopathologic consequences. In addition, accumulation of mitochondrial ROS as a result of decreased autophagy is expected to have effects on many other pathophysiological processes in addition to the pathway described here. In particular, our study provides insight into the link between two previously reported processes—the increased accumulation of damaged mitochondria and ROS and the decreased efficiency of autophagy with aging, both of which have been implicated in many late-onset diseases such as neurodegenerative diseases, cardiovascular disease, and cancer. Future investigation on the potential link among autophagy, ROS, and human diseases will provide a basis for guided therapeutic interventions.
Methods
Cells and Viruses.
Atg5+/+ and Atg5−/− MEF cells were generated from embryos of Atg5+/−× Atg5+/− (31) according to standard protocol and were kind gifts of Herbert W. Virgin (St. Louis, MO). MyD88−/− MEFs were a kind gift of Ruslan Medzhitov (New Haven, CT). Several independent lines of MEFs were used in each experiment. Only primary MEFs under 10th passage were used in all experiments. MEFs were propagated in high-glucose DMEM (Gibco) and supplemented with 10% heat-inactivated FBS, 1% Hepes, and 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco). VSV-G-GFP virus (32) was used in all experiments. Primary macrophages were prepared from Atg5+/+ or Atg5−/− neonatal liver according to a previously described method (ref. 33 and SI Methods). VSV-G-GFP was a kind gift of John Rose (New Haven, CT).
Stimulation of Cells.
MEFs were stimulated with delivery of Poly I:C (1 μg/mL) complexed to Lipofectamine 2000 (Invitrogen). ROS induction was accomplished by treatment of cells with rotenone (Sigma-Aldrich) at 1 μM beginning from 12 h before stimulation with Poly I:C until the completion of the experiment. Antioxidant treatment of cells consisted of either NAC (Alfa Aesar) at 10 mM or PG (MP Biomedicals) at 100 μM from 15 min before stimulation with Poly I:C until the completion of the experiment.
RT Quantitative PCR.
RNA isolation was performed using the RNeasy kit (Qiagen) according to manufacturer's instructions. Isolated RNA was used to synthesize cDNA using SuperScript II cDNA synthesis (Invitrogen) and qPCR was performed on a Stratagene MX3000P unit using SyberGreen (Qiagen) with the primers described in SI Materials and Methods.
ELISA.
Levels of IFN-α present in the supernatant was quantified by ELISA as previously described (34). Levels of IL-6 were measured using an ELISA kit (BD Biosciences) according to manufacturer's instruction.
Flow Cytometric Analyses.
Mitochondrial mass was measured by fluorescence levels upon staining with MitoTracker Green FM and MitoTracker Red CMXRos (Molecular Probes/Invitrogen) at 100 nM for 25 min at 37 °C. Mitochondria-associated ROS levels were measured by staining cells with MitoSOX (Molecular Probes/Invitrogen) at 5 μM for 40 min at 37 °C. Cells were then washed with PBS solution, trypsinized, and resuspended in PBS solution containing 1% FBS for FACS analysis. Intracellular staining to measure IPS-1 levels was performed using the BD Cytofix/Cytoperm kit according to manufacturer's instructions, with the primary anti-mitochondrial antiviral signaling antibody (Cell Signaling) and the secondary PE anti-rabbit IgG antibody (Jackson).
Mitochondrial DNA Quantification.
Relative mitochondrial DNA amount was assessed by isolating total DNA from the cells using the QIAamp DNA Mini Kit (Qiagen) and performing quantitative PCR using two independent reactions for mitochondrial and nuclear primer sets for each sample as previously described (35) using established primers (36), and is described in detail in the SI Materials and Methods. Southern blot for mitochondrial DNA was carried out using a previously described method (37) and described in detail in the SI Materials and Methods.
Construction of Lentivirus Expression Systems.
The ATG5 sequence was cloned from MEF cDNA and restriction enzyme sites were inserted for 5′ BamHI and 3′ XhoI. The IRES-EGFP sequence was cut from the pIRES EGFP vector (Clontech) using 5′ XhoI and 3′ NotI. These segments were ligated and then the ATG5-IRES-GFP expression segment was ligated into the pLentivirus Vector construct at the 5′ BamHI and 3′ NotI sites. The IPS-1 sequence was excised from the expression vector pEF-BOS-Flag-IPS-1 (38) using 5′ XhoI and 3′ KpnI and ligated into the pEGFP-NI Vector (Clontech). IPS-1-GFP fusion sequence was excised using 5′ BglII and 3′ NotI and ligated into the pLentivirus Vector construct at the 5′ BglII and 3′ NotI sites. Integrity of the inserted genes was confirmed by sequencing. HEK293T cells were transfected with either ATG5-IRES-GFP or IPS1-GFP pLentivirus Vector plasmids along with a lentivirus packaging plasmid. Cell culture supernatants containing lentivirus particles were collected at 48 and 72 h after transfection, filtered, ultracentrifuged, and used to transduce MEFs.
Statistical Analyses.
Student t test was used for all statistical analyses.
Supplementary Material
Acknowledgments.
We are grateful to Dr. Noboru Mizushima for providing the Atg5+/− mice and Dr. Herbert W. Virgin for providing MEFs. We thank Anthony Rongvaux and Philip West for reagents. This work was supported by National Institutes of Health (NIH) Public Health Service grants AI054359, AI062428, and AI064705 (to A.I.). M.C.T. was supported in part by NIH Predoctoral Virology Training grant T32 AI055403. M.S. was supported by a post-doctoral fellowship from the Uehara Foundation and is a recipient of the postdoctoral award from the Mochida Memorial Foundation for Medical and Pharmaceutical Research. H.K.L. was supported by a Richard K. Gershon fellowship. A.I. holds an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807694106/DCSupplemental.
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Diebold SS, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 7.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 8.Lund JM, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HK, et al. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 10.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 11.Foy E, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci USA. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses essential role of IPS-1 in innate immune responses against RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 13.Kato H, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Jounai N, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stetson DB, Medzhitov R. Antiviral defense: interferons and beyond. J Exp Med. 2006;203:1837–1841. doi: 10.1084/jem.20061377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 18.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 19.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 22.Takeshita F, et al. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy. 2008;4:67–69. doi: 10.4161/auto.5055. [DOI] [PubMed] [Google Scholar]
- 23.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
- 25.Degli Esposti M. Inhibitors of NADH-ubiquinone reductase: an overview. Biochim Biophys Acta. 1998;1364:222–235. doi: 10.1016/s0005-2728(98)00029-2. [DOI] [PubMed] [Google Scholar]
- 26.Li N, et al. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem. 2003;278:8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- 27.Rebsamen M, Meylan E, Curran J, Tschopp J. The antiviral adaptor proteins Cardif and Trif are processed and inactivated by caspases. Cell Death Differ. 2008;15:1804–1811. doi: 10.1038/cdd.2008.119. [DOI] [PubMed] [Google Scholar]
- 28.Scott I, Norris KL. The mitochondrial antiviral signaling protein, MAVS, is cleaved during apoptosis. Biochem Biophys Res Commun. 2008;375:101–106. doi: 10.1016/j.bbrc.2008.07.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–L1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- 30.Orvedahl A, Levine B. Viral evasion of autophagy. Autophagy. 2008;4:280–285. doi: 10.4161/auto.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 32.Dalton KP, Rose JK. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology. 2001;279:414–421. doi: 10.1006/viro.2000.0736. [DOI] [PubMed] [Google Scholar]
- 33.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 34.Lund J, et al. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eaton JS, et al. Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. J Clin Invest. 2007;117:2723–2734. doi: 10.1172/JCI31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown TA, Clayton DA. Release of replication termination controls mitochondrial DNA copy number after depletion with 2′,3′-dideoxycytidine. Nucleic Acids Res. 2002;30:2004–2010. doi: 10.1093/nar/30.9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bayona-Bafaluy MP, et al. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc Natl Acad Sci USA. 2005;102:14392–14397. doi: 10.1073/pnas.0502896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasai M, Matsumoto M, Seya T. The kinase complex responsible for IRF-3-mediated IFN-beta production in myeloid dendritic cells (mDC) J Biochem. 2006;139:171–175. doi: 10.1093/jb/mvj025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.