Abstract
Innate immunity is an ancient defense system used by both vertebrates and invertebrates. Previously characterized innate immune responses in plants and animals are triggered by detection of pathogens using specific receptors, which typically use a leucine-rich repeat (LRR) domain to bind molecular patterns associated with infection. The nematode Caenorhabditis elegans uses defense pathways conserved with vertebrates; however, the mechanism by which C. elegans detects pathogens is unknown. We screened all LRR-containing transmembrane receptors in C. elegans and identified the G protein-coupled receptor FSHR-1 as an important component of the C. elegans immune response to Gram-negative and Gram-positive bacterial pathogens. FSHR-1 acts in the C. elegans intestine, the primary site of exposure to ingested pathogens. FSHR-1 signals in parallel to the known p38 MAPK pathway but converges to regulate the transcriptional induction of an overlapping but nonidentical set of antimicrobial effectors. FSHR-1 may act generally to boost the nematode immune response, or it may function as a pathogen receptor.
Keywords: host-pathogen interactions, leucine-rich repeat
The evolutionarily ancient innate immune system is the critical first line of defense against microbial infection of vertebrate and invertebrate animals (1). In addition to providing an initial rapid response to pathogen attack, the innate immune system activates the adaptive immune system in jawed vertebrates (2). Innate immunity includes both constitutive and inducible defenses. In general, an inducible innate immune response can be broken down into 3 stages (3). First, the invading pathogen is recognized by host germline-encoded pattern recognition receptors (PRRs). Next, this information is processed and transmitted via the activation of signaling pathways. Finally, these signaling cascades lead to upregulation of antimicrobial effectors.
The soil nematode Caenorhabditis elegans is a tractable model for studying innate immunity. Numerous powerful molecular genetic tools are available that facilitate the identification and characterization of components of the innate immune system. Infection of C. elegans with several bacterial and fungal pathogens has shown that both host response pathways and pathogenic virulence factors are conserved, indicating the relevance of this host-pathogen model to other metazoans (4, 5).
Forward and reverse genetic analyses have identified several signaling pathways required for the C. elegans response to diverse pathogens. Of central importance is a p38 MAPK pathway, which consists of a core cassette of NSY-1/MAPKKK, SEK-1/MAPKK, and PMK-1/MAPK regulated by the upstream Toll/IL-1 adaptor TIR-1 (6–8). The p38 pathway also plays a key role in the defense against pathogens in both insects and mammals and is thought to represent an ancient component of metazoan innate immunity (9). A second conserved pathway that has a dramatic effect on the innate immune system in worms is the insulin/insulin-like growth factor (IGF) pathway (10). The DAF-2 insulin/IGF receptor activates AGE-1/PI3K, leading to the phosphorylation and cytoplasmic sequestration of the FOXO transcription factor DAF-16 and the activation of a hyperimmune state (11, 12).
Recent microarray experiments have identified a large number of genes that are transcriptionally induced by C. elegans in response to pathogen exposure (13–16). Many of these pathogen-response genes encode proteins that have in vitro antimicrobial activity or are homologous to known antimicrobial factors in other species. A subset of these putative antimicrobial effectors is regulated by the p38 MAPK signal transduction cascade, demonstrating a direct link between a specific immune signaling pathway and the induced response to pathogen infection (15).
Despite significant advances in understanding the signal transduction and subsequent antimicrobial effector response to pathogen attack, the PRRs responsible for detecting this attack and triggering a response in C. elegans have not yet been identified. Toll-like receptors (TLRs) are the canonical transmembrane PRRs in mammals, and the C. elegans genome does contain a single TLR homolog, TOL-1 (2, 5). Initial studies with TOL-1 indicated that it was not required for resistance against several pathogens tested (17). However, subsequent studies have suggested that it is important for resistance against the Gram-negative bacterium Salmonella enterica, preventing invasion of this pathogen into the pharynx (18). However, the tissue in which TOL-1 is acting to confer resistance against Salmonella and its mechanism of action are unclear. TOL-1 may or may not be a PRR, but it cannot be the only PRR in C. elegans, given that tol-1 mutants are not sensitive to infection by most pathogens that have been tested. Plants and some metazoans, including mammals, also have cytosolic PRRs referred to as nucleotide oligomerization domain (NOD) proteins (2). However, no NOD-like proteins are encoded in the C. elegans genome. Thus, we sought to identify alternate candidate C. elegans PRRs.
One distinguishing feature of the majority of PRRs known in other species, including the mammalian TLRs and NOD/CLRs, is that the ligand-binding domain of these receptors comprises multiple leucine-rich repeats (LRRs) (19). We hypothesized that C. elegans may also use LRR domains in pathogen pattern recognition. To test this, we undertook a screen of candidate LRR receptors. Here we report the identification and characterization of the LRR-containing G protein-coupled receptor FSHR-1 as a putative C. elegans immune receptor. We found that FSHR-1 is a critical component of the C. elegans innate immune response that acts in parallel to both the insulin and p38 MAPK pathways. FSHR-1 regulates transcription of a set of putative antimicrobial effectors that are induced by pathogen infection. Tissue-specific knockdown and tissue-specific misexpression experiments indicate that FSHR-1 functions in the C. elegans intestine, the major site of pathogen exposure.
Results
The FSHR-1 Receptor Is Required for C. elegans Immunity.
To determine whether C. elegans uses LRR-containing cell-surface receptors in pathogen recognition, we tested all C. elegans genes that are predicted to contain both an extracellular LRR domain and at least 1 transmembrane domain for a role in the innate immune response. We removed the function of each candidate gene via RNAi and exposed the worms to the bacterial pathogen Pseudomonas aeruginosa strain PA14.
Of the 14 candidate LRR receptors in the C. elegans genome, only 1, encoded by the gene C50H2.1, located on chromosome V, was found to affect the response to PA14 when its function was inhibited by RNAi. Animals subjected to C50H2.1 RNAi are significantly more sensitive (P < 0.01) to killing by PA14 (Fig. 1A). C50H2.1 encodes FSHR-1, a glycopeptide hormone receptor homologue. FSHR-1 is the sole C. elegans member of the LRR-containing class of G protein-coupled receptors (LGR), represented in mammals by the follicle-stimulating hormone, thyroid-stimulating hormone, and luteinizing hormone receptors. The extracellular domain of FSHR-1 contains 9 LRRs, which are ≈40% similar to the corresponding region on mammalian LGRs. FSHR-1 has a 7-pass transmembrane domain and unusually large C-terminal cytoplasmic tail. C. elegans FSHR-1 has known roles in germline development and in the function of cholinergic synapses in the nervous system (20, 21).
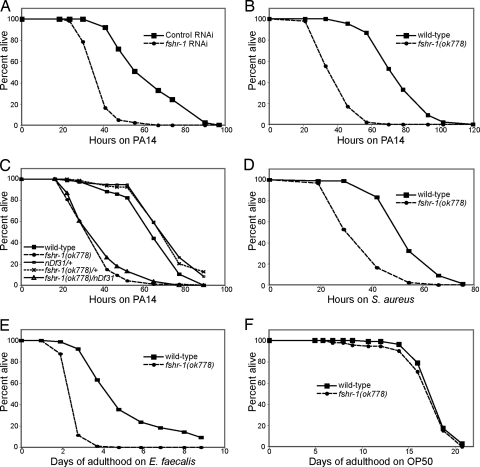
Fig. 1.
FSHR-1 is required for innate immunity. (A and B) fshr-1 (RNAi) or fshr-1(ok778) mutant worms are sensitive to killing by pathogenic P. aeruginosa PA14 relative to wild-type control worms. (C) fshr-1(ok778)/nDf31 heterozygotes are just as sensitive to PA14 as fshr-1(ok778) homozygotes. Parent nDf31/+ worms (actual genotype: nDf31/nT1[qIs51]) and sibling fshr-1(ok778)/+ worms (actual genotype: fshr-1(ok778)/nT1[qIs51]) are not sensitive to PA14. (D–F) fshr-1(ok778) mutants are sensitive to killing by pathogenic S. aureus and E. faecalis but not to E. coli OP50.
The existing deletion allele fshr-1(ok778) recapitulates the RNAi phenotype: ok778 mutant homozygotes are sensitive to pathogenic PA14 (Fig. 1B). The ok778 deletion removes most of the LRR domain and all 7 transmembrane domains. The fshr-1(ok778) allele is a genetic null: transheterozygotes of ok778 and nDf31, a deletion spanning the entire fshr-1 genomic region, have the same pathogen sensitivity phenotype as ok778 homozygotes (Fig. 1C).
In addition to their defect in a response to the Gram-negative pathogen P. aeruginosa PA14, fshr-1(ok778) mutants are also more sensitive than wild-type worms to killing by the Gram-positive bacteria Staphylococcus aureus and Enterococcus faecalis (Fig. 1 D and E). Importantly, fshr-1(ok778) mutant worms do not have a reduced lifespan on Escherichia coli strain OP50, which is relatively nonpathogenic under the conditions tested (Fig. 1F). Thus, the sensitivity phenotype of fshr-1(ok778) is not caused by nonspecific sickness. Rather, FSHR-1 is specifically required for the C. elegans innate immune response to Gram-negative and Gram-positive pathogens.
FSHR-1 Acts in Parallel to p38 MAPK and Insulin Pathways.
The p38 MAPK pathway and the insulin/IGF pathway are important signal transduction pathways involved in the defense of C. elegans against P. aeruginosa PA14 (6, 7, 10). To test whether the receptor FSHR-1 is a member of either of these known pathways, we examined the phenotype of double mutants with fshr-1(ok778) and members of each pathway.
daf-2 encodes the insulin/IGF receptor. In contrast to the enhanced sensitivity of fshr-1 mutants, daf-2 mutants have enhanced resistance to killing by PA14. We found that daf-2(e1368ts); fshr-1(ok778) double mutants have an immunity phenotype that is intermediate between either of the single mutants (Fig. 2A). The degree of resistance/sensitivity of the double mutants depends on the length of time the worms are shifted to the daf-2 restrictive temperature before exposure to pathogens (e.g., worms that are preshifted to the restrictive temperature for 4 h are less resistant than worms preshifted for 8 h). The double mutants are neither as sensitive to killing by PA14 as fshr-1 single mutants nor as resistant to killing as daf-2(e1368) single mutants (P < 0.001). This additive phenotype suggests that FSHR-1 and DAF-2 act in parallel pathways. daf-2 mutations also extend lifespan and constitutively activate dauer formation. However, daf-2(e1368); fshr-1(ok778) double-mutant worms have the same dauer formation and longevity phenotypes as daf-2(e1368) single mutants at the restrictive temperature [Fig. S1 and data not shown], suggesting that the effect of an fshr-1 mutation on daf-2 mutants is specific to immunity.
Fig. 2.
FSHR-1 acts in parallel to DAF-2 and the p38 MAPK pathway. (A) daf-2 and fshr-1 single and double mutants were raised at 15 °C and then shifted to the restrictive temperature of 25 °C 4 h before exposure to PA14. (B and C) Loss of components of the p38 MAPK pathway, either by genetic mutation (pmk-1) or RNAi (tir-1 and nsy-1), enhances the pathogen sensitivity of fshr-1(ok778) null mutants. Experiments with pmk-1 and fshr-1 single and double mutants were repeated 5 times, and in all cases the double mutants were significantly more sensitive than either single mutant (P < 0.01 4 out of 5 times; P < 0.05 1 out of 5 times).
The p38 MAPK PMK-1 is required for the C. elegans response to PA14, and pmk-1(km25) mutant worms are sensitive to killing by PA14, similar to fshr-1(ok778) mutants. pmk-1(km25); fshr-1(ok778) double mutants are even more sensitive than either single mutant (Fig. 2B). Because a null pmk-1(km25) mutation modestly but significantly enhances (P < 0.01) the null phenotype of fshr-1(ok778), these 2 genes likely act in parallel. Loss of function of tir-1 or nsy-1, which act upstream of pmk-1 in the p38 MAPK pathway, also significantly enhances (P < 0.005) the fshr-1(ok778) null phenotype (Fig. 2C). These data suggest that FSHR-1 cannot signal in a simple linear pathway with the p38 MAPK pathway. The simplest explanation is that the entire p38 MAPK pathway signals in parallel to the FSHR-1 receptor. However, it is also possible that FSHR-1 does act in the p38 MAPK pathway. If so, there must be at least 1 additional receptor that regulates p38 MAPK and at least 1 additional signaling pathway acting downstream of FSHR-1 to account for the additive phenotype in the fshr-1; pmk-1 double mutants.
FSHR-1 Regulates a Set of PA14-Response Genes.
Microarray analysis has defined a set of genes whose transcription is upregulated by C. elegans upon infection with PA14, many of which encode putative secreted antimicrobial factors (15). A subset of these PA14-response genes depends on the PMK-1 p38 MAPK pathway for their induction in the presence of pathogens. Of these, some also depend on PMK-1 for their basal expression in the absence of pathogens and are designated class A genes by Troemel et al. (15). Those genes that depend on PMK-1 for their induction but not their basal expression on nonpathogenic OP50 are designated class C. The remaining PA14-response genes, whose transcriptional induction in the presence of pathogens is independent of the p38 MAPK pathway, are presumably regulated via at least 1 as-yet-unknown signaling pathway. Of these, class B genes are defined as those whose basal expression on nonpathogenic OP50 depends on PMK-1, despite the fact that their induction on pathogenic PA14 does not. Finally, class D genes do not depend on PMK-1 for either their induction or basal expression.
To determine whether, like PMK-1, FSHR-1 regulates PA14-response genes, we used quantitative (q)RT-PCR to measure the transcriptional induction of 10 of these genes representing the 4 classes described above in fshr-1(ok778) mutants infected with PA14. Five of the 10 PA14-response genes tested (F56D6.2, C17H12.8, K08D8.5, F49F1.6, and F55G11.2) depend on PMK-1 for their full induction upon infection (class A or class C). Three of these 5 PMK-1-dependent genes (F56D6.2, C17H12.8, and F49F1.6) are induced by PA14 at least 10-fold less in fshr-1(ok778) mutant worms than in wild-type worms (P < 0.0001), indicating that they require FSHR-1 for their full induction in response to infection (Fig. 3A). The remaining 2 PMK-1-dependent genes tested also partially depend on FSHR-1 for their induction, but the effects of a fshr-1(ok778) mutation are more modest (P < 0.005). It is interesting to note that the effect of an fshr-1(ok778) mutation on the induction of each of these 5 PMK-1-dependent genes is not as great as the effect of a pmk-1(km25) mutation, consistent with the observation that pmk-1(km25) mutant worms are more sensitive to killing by PA14 than are fshr-1(ok778) mutant worms (Fig. 2B). Additionally, for genes such as F49F6.1 and F55G11.2, whose induction is partially reduced but not completely eliminated in both pmk-1(km25) and fshr-1(ok778) single-mutant worms, double-mutant pmk-1(km25); fshr-1(ok778) worms have even less induction than either single mutant. This result is consistent with the additive pathogen sensitivity phenotype between these 2 mutations that is described above (Fig. 2B) and supports the conclusion that FSHR-1 and PMK-1 function in parallel signaling pathways.
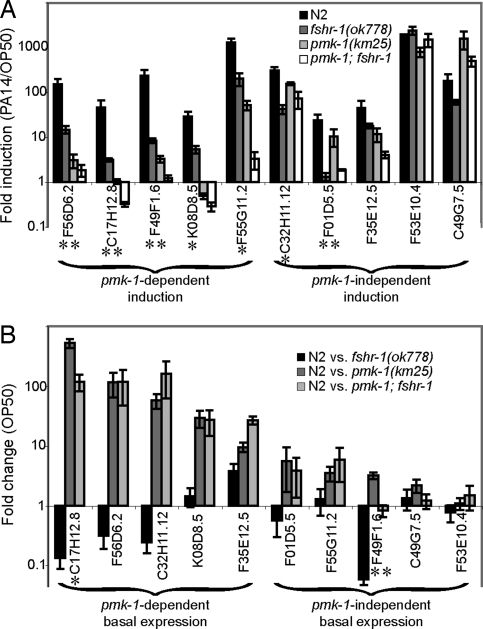
Fig. 3.
FSHR-1 regulates PA14-response genes. (A and B) qRT-PCR was used to analyze the relative transcription of PA14-response genes in wild-type and mutant worms fed OP50 or PA14. Error bars represent SEM for 3 independent biologic replicates. (A) Fold induction was calculated as the ratio of normalized expression on PA14 divided by expression on OP50. *Genes with greater than 5-fold reduction in their induction in fshr-1(ok778) mutant worms relative to wild-type worms with P < 0.01. **Genes with greater than 10-fold reduction in their induction in fshr-1(ok778) mutants relative to wild-type with P < 0.01. (B) Fold change in basal expression was calculated as the ratio of wild-type expression to mutant expression in worms fed OP50. Troemel et al. (15) reported that F01D5.5 was regulated basally by PMK-1, but in our experiments the difference did not reach statistical significance. *Genes with greater than 5-fold lower basal expression in wild-type worms relative to fshr-1(ok778) mutants. **Genes with greater than 10-fold lower basal expression in wild-type worms relative to fshr-1(ok778) mutants.
The remaining 5 PA14-response genes tested (C32H11.12, F35E12.5, F01D5.5, F53E10.4, and C49G7.5) do not depend on PMK-1 for their induction upon infection (class B or class D). Three of these 5 PMK-1-independent PA14-response genes (F35E12.5, F53E10.4, and C49G7.5) are also independent of FSHR-1 because their induction upon exposure to PA14 is not significantly affected in fshr-1(ok778) mutants (Fig. 3A). The induction of F01D5.5 expression upon PA14 exposure, however, is similar in wild-type and pmk-1(km25) mutants but is substantially reduced in fshr-1(ok778) mutants (P < 0.001), indicating that the induction of this gene is independent of PMK-1 but dependent on FSHR-1. Likewise, C32H11.12 induction is independent of PMK-1 but modestly reduced in fshr-1(ok778) mutants (P < 0.005); thus, C32H11.12 partially depends on FSHR-1 for its induction in the presence of PA14. These data suggest that FSHR-1 and PMK-1 regulate the induction of largely overlapping but nonidentical sets of PA14-response target genes.
By definition, the PA14-response genes described above are expressed at lower basal levels in worms that have not been exposed to pathogen but instead have been fed exclusively nonpathogenic OP50. PMK-1 also controls the basal expression in the absence of pathogen exposure of a subset of these genes (class B or class D) (15). The set of genes whose basal expression is regulated by PMK-1 partially overlaps the set of genes whose pathogen induction is regulated by PMK-1 (15).
To determine whether PMK-1 and FSHR-1 regulate the basal expression of similar sets of genes, we used qRT-PCR to measure the relative expression of each of the 10 selected PA14-response genes in wild-type and mutant worms grown on OP50 in the absence of infection. Five of the 10 genes tested (C17H12.8, F56D6.2, C32H11.12, K08D8.5, and F35E12.5) require PMK-1 p38 MAPK for their basal expression (class A or class B): wild-type worms fed nonpathogenic OP50 express each of these genes at a dramatically higher level than pmk-1(km25) mutant worms fed OP50 (Fig. 3B). The remaining 5 genes tested (F01D5.5, F55G11.2, F49F1.6, C49G7.5, and F53E10.4) do not require PMK-1 for their basal expression because there is not a substantial difference between transcript levels in wild-type and mutant worms fed OP50 (class C or class D). In contrast, FSHR-1 is not required for activating the basal expression of any of the 10 genes tested. Basal transcription in 8 of the 10 genes is not significantly different between wild-type and fshr-1(ok778) mutant worms. F49F1.6 and C17H12.8 may be repressed by FSHR-1 in the absence of pathogen because each gene was expressed at significantly lower levels in wild-type worms than in fshr-1(ok778) mutants (P < 0.01). Therefore, FSHR-1 and PMK-1 have distinct effects on the basal transcription of PA14-response genes.
FSHR-1 Acts in the Intestine.
FSHR-1 is expressed in several somatic tissues, most strongly in the intestine and neurons (20, 21). We hypothesized that FSHR-1 carries out its role in the C. elegans innate immune response in the intestine, given that this tissue comes into direct contact with any pathogenic bacteria the worm has eaten. The C. elegans strain rde-1(ne219); kbIs7 permits RNAi-mediated knockdown of gene expression only in the intestine but not in other tissues (Materials and Methods, Fig. S2 A and B and SI Materials and Methods). RNAi of fshr-1 in worms of this genetic background thus removes FHSR-1 function specifically in the intestine. Intestinal-specific fshr-1 RNAi results in a strong pathogen sensitivity phenotype (P < 0.001), similar to systemic fshr-1 RNAi, indicating that FSHR-1 function is required in the intestine for its role in innate immunity (Fig. 4A).
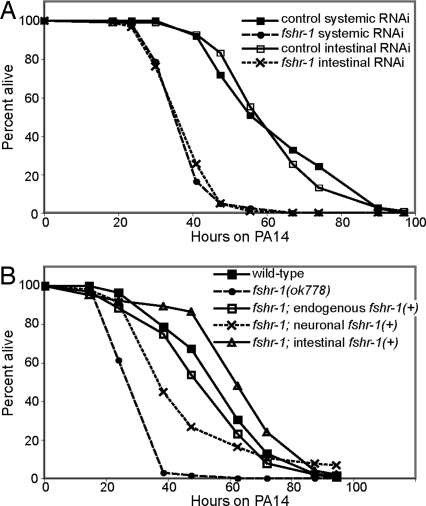
Fig. 4.
FSHR-1 expression in the intestine is necessary and sufficient for pathogen resistance. (A) Intestinal-specific RNAi of fshr-1 causes worms to be as sensitive to PA14 as systemic RNAi of fshr-1. (B) Wild-type FSHR-1 expressed from endogenous or intestinal promoters fully rescues the fshr-1(ok778) mutant phenotype. FSHR-1 expressed from a neuronal promoter only partially rescues the mutant phenotype.
To determine whether intestinal FSHR-1 is also sufficient for its immune function, we expressed wild-type fshr-1(+) from the control of endogenous (fshr-1), neuronal (ric-19), or intestinal (ges-1) promoters and tested the ability of each transgene to rescue a fshr-1(ok778) mutation. Expression of fshr-1(+) from either the endogenous or the intestinal ges-1 promoter fully rescues the pathogen sensitivity of fshr-1(ok778) null mutants (Fig. 4B). Four additional independent lines of ges-1::fshr-1 also fully rescue fshr-1(ok778) (Fig. S3A). Interestingly, expression of fshr-1(+) in neurons weakly suppresses the sensitivity of fshr-1(ok778) mutants (Fig. 4B). This weak neuronal suppression may indicate an immune role for FSHR-1 in neurons, or it may be nonspecific. For example, most of the worms expressing fshr-1(+) in neurons produce virtually no embryos when they are fed PA14; in contrast, control worms and worms expressing fshr-1(+) under the control of the fshr-1 or ges-1 promoters produce large quantities of embryos. Sterility has been shown to cause significant pathogen resistance in C. elegans (22), so it is possible that the apparent sterility caused by neuronally expressed fshr-1(+) may lead to the partial suppression of fshr-1(ok778) pathogen sensitivity, rather than a more specific role for neuronal FSHR-1 in pathogen defense. Regardless of whether FSHR-1 can function in the neurons, the strong suppression of the fshr-1(ok778) null phenotype by intestinally expressed fshr-1(+) indicates that FSHR-1 is acting in the intestine. Therefore, FSHR-1 function in the intestine is both necessary and sufficient for its role in the C. elegans innate immune response.
Discussion
We have identified a novel role for the G protein-coupled receptor FSHR-1 in the innate immune response of C. elegans. Analyses of the pathogen sensitivity of single and double mutants indicate that the FSHR-1 receptor does not act in a simple linear pathway with either the p38 MAPK or insulin signaling pathways and thus likely defines a novel pathway or branch in the C. elegans innate immune network. FSHR-1 is required for the induction of a set of putative antimicrobial effectors upon exposure to PA14, indicating that the FSHR-1 signal is responsive to the presence or absence of pathogen. Surprisingly, this set of PA14-response genes whose induction depends on FSHR-1 is very similar but not identical to the set of genes whose induction depends on the p38 MAPK PMK-1. We propose a model in which FSHR-1 and p38 MAPK signal in parallel to each other but converge on a common set of target effector genes in response to attack by pathogenic PA14 (Fig. 5). Unlike p38 MAPK, FSHR-1 does not activate the basal expression of any pathogen-response genes tested and may in fact negatively regulate some of them. Although FSHR-1 is produced in multiple tissues and has multiple roles in the worm, it performs its innate immune function in the intestine.
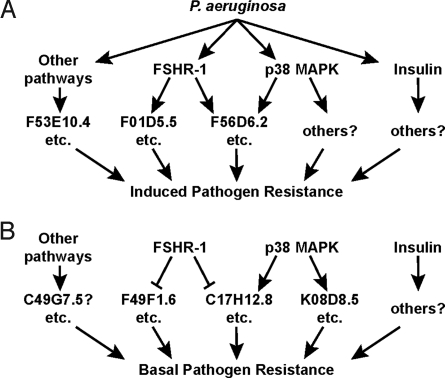
Fig. 5.
A model for a C. elegans innate immune network. (A and B) This model shows the integration of signaling from 3 known innate immune pathways. (A) Each pathway regulates the induction of a set of effectors in response to P. aeruginosa. The FSHR-1 and p38 MAPK pathways regulate partially overlapping sets of pathogen-response genes. Specific examples of target genes mentioned in this work are listed for each category. (B) These signaling pathways also regulate the basal expression in the absence of pathogen of several genes. Examples mentioned here are listed for each category.
FSHR-1 was identified by screening 14 C. elegans genes by RNAi that encode proteins predicted to contain both an extracellular LRR domain and at least 1 transmembrane domain for susceptibility to P. aeruginosa. Of these 14 genes, only fshr-1 RNAi resulted in enhanced P. aeruginosa-mediated killing. Subsequently, fshr-1 RNAi and an fshr-1 null allele were tested and found to also be susceptible to S. aureus and E. faecalis. It is possible that some of the other LRR transmembrane proteins also function as putative immune receptors, and these are now being tested with a variety of pathogenic bacteria and yeasts.
How does FSHR-1 act in the intestine to promote pathogen resistance? One possibility is that FSHR-1 acts in a regulatory role, generally enhancing the ability of the intestine to respond to pathogen attack. FSHR-1 might act as a hormone receptor in this scenario, receiving hormone signals from other tissues. The canonical ligand bound by members of the class of G protein-coupled receptor to which FSHR-1 belongs is the heterodimeric glycopeptide hormone FSHα/β (23). Worms do not have an identifiable FSHα subunit, and the gene that most closely resembles a FSHβ subunit does not, in our preliminary studies, seem to play an important role in C. elegans innate immunity. An intriguing possibility is that FSHR-1 may be activated by a noncanonical host ligand in response to an upstream signal from a PRR.
Another possibility is that FSHR-1 is a PRR and thus is responsible for the initial sensing of infection in C. elegans. FSHR-1 acts in the intestine, a tissue that is the primary site of infection and in which it has the potential for direct contact with pathogens. PRRs can sense infection by binding directly to a pathogen, by recognizing pathogen-produced virulence factors, or by recognizing the damage these virulence factors cause to host tissues (24). fshr-1 mutants are sensitive to killing by at least 1 Gram-negative pathogen and 2 Gram-positive pathogens. The surface architecture of Gram-negative and Gram-positive bacteria is quite distinct, but there are shared components. If FSHR-1 does directly recognize these pathogens or their products, it must either bind a factor conserved among diverse pathogens or have a very low specificity that enables it to recognize multiple ligands. There is precedent for promiscuous pathogen receptors: the scavenger-like receptor Eater, which is required for the Drosophila innate immune response, directly binds Gram-positive and Gram-negative bacteria as well as yeast (25). Alternatively, FSHR-1 could detect pathogens indirectly by binding host-derived byproducts of the damage caused by infection. If FSHR-1 were a PRR, it would be the first example of a G protein-coupled receptor functioning as a pathogen receptor in any animal.
Localization of the FSHR-1 protein could help distinguish between these scenarios. FSHR-1 is expressed in the neurons and intestine, but its subcellular localization has not been dissected (20, 21). If FSHR-1 acts as a regulatory receptor that detects a systemic signal and triggers or enhances the intestine's response, we would predict that FSHR-1 would localize more basolaterally within the intestinal cells. In contrast, if FSHR-1 is a PRR that is an initial sensor of intestinal infection, we would predict that FSHR-1 would localize apically, where it would be more directly exposed to pathogens. Further studies will be necessary to distinguish between these possibilities.
The mammalian FSH receptor (FSHR) is expressed on ovarian granulosa cells, where it activates production of the critical steroid hormone estradiol and regulates gametogenesis (26). Tantalizing new evidence suggests that these granulosa cells may have some immune-related functions (27). No canonical immune cells are present in ovarian follicles; however, granulosa cells in the follicle respond to exposure to E. coli by suppressing estradiol production, suggesting that pathogens may somehow be recognized by this tissue. The TLR4/CD14/MD-2 receptor complex, which when present on immune cells recognizes LPS from Gram-negative bacteria such as E. coli, is also expressed by granulosa cells, although these proteins have not yet been shown to be required for the granulosa cell response to E. coli. Because FSHR regulates estradiol production in granulosa cells, it is tempting to speculate that FSHR could also play a role in the recognition and/or response to pathogens in follicular granulosa cells.
In addition to its well-characterized role in the mammalian gonad, recent studies have revealed a role for FSHR in the mammalian skeletal system (28). FSHR is expressed in precursor and differentiated osteoclasts, multinucleate cells of the monocyte-macrophage lineage. FSHR stimulates the formation and function of osteoclasts, which are responsible for decalcifying and degrading bone matrix. A close connection between the vertebrate skeletal and immune systems has long been known (29). Not only are hematopoietic immune cells generated in the bone marrow, but also many regulatory molecules and mechanisms are shared between these 2 systems. It would be interesting to test whether mammalian FSHR in osteoclasts provides yet another link between skeletal regulation and immunity.
We report here that FSHR-1 acts in the intestine as part of the C. elegans innate immune response. It is not known whether FSHR is expressed in the mammalian intestine. Just as an unexpected role for FSHR in the skeletal system has recently been discovered, it will be interesting to determine whether FSHR could act in the mammalian intestine in addition to its best-characterized role in the gonads.
Materials and Methods
C. elegans Strains.
C. elegans strains were maintained at 20 °C, unless otherwise noted, as previously described (30). The fshr-1 deletion allele ok778 was generated by the Oklahoma Medical Research Foundation Knockout Consortium and outcrossed to wild-type N2 worms 4 times. To determine whether ok778 is a null, heterozygous nDf31/fshr-1(ok778) worms were generated by crossing AU102 nDf31/nT1[qIs51(myo-2::gfp)] hermaphrodites with fshr-1(ok778) males. Non-Green progeny must therefore be nDf31/ok778 transheterozygotes and were tested for their sensitivity to PA14. Parent nDf31/nT1 and green sibling fshr-1(ok778)/nT1 worms were used as controls. AU131 pmk-1(km25); fshr-1(ok778) and AU132 daf-2(e1368); fshr-1(ok778) strains were generated via standard genetic techniques. To test the intestinal sufficiency of FSHR-1, the transgenic strains AU175 fshr-1(ok778); agEx43[fshr-1(+)], AU209 fshr-1(ok778); agEx52[ric-19::fshr-1(+)], and AU218 fshr-1(ok778); agEx58[ges-1::fshr-1(+)] were generated. agEx43 comprises a genomic fshr-1 PCR fragment containing 4 kb of sequence upstream of the start site, the entire ORF, and 1 kb of sequence downstream of the stop codon. agEx52 comprises 1.2 kb of upstream ric-19 regulatory sequence fused via PCR to the fshr-1 ORF and 1 kb downstream sequence. agEx58 comprises 3 kb of upstream ges-1 regulatory sequence fused via PCR to the fshr-1 ORF and 1 kb downstream sequence. Each PCR product was injected at a concentration of ≈10 ng/μL into fshr-1(ok778) worms with a myo-2::Mcherry coinjection marker. For technical reasons, the fshr-1 ORF used in these arrays contains endogenous introns. It is possible that regulatory elements are contained in these introns and may drive expression in unintended tissues; however, the introns alone are not sufficient for expression of functional FSHR-1 (Fig. S3B).
RNA Interference.
RNAi bacterial clones were obtained from the Ahringer and Vidal RNAi libraries (31–33). Bacteria were grown to saturation in LB + 50 μg/mL carbenicillin, spread on 6-cm RNAi plates [nematode growth medium (NGM) + 25 μg/mL carbenicillin + 5 mM isopropyl β-D-thiogalactopyranoside], and incubated at 25 °C for 48 h (34). Synchronized L1 larvae were placed on each plate and incubated at 20 °C for 40 h. L4 larvae or young adult worms were used in immunity assays. The strain VP303 rde-1(ne219); kbIs7[nhx-2::rde-1, rol-6] was graciously provided by K. Strange for use in intestine-specific RNAi (35). RDE-1 is required for RNAi, so worms mutant for rde-1 are refractory to RNAi. kbIs7 is an integrated transgene containing rde-1(+) expressed from an intestinal-specific promoter, thereby rescuing the rde-1(ne219) mutant phenotype exclusively in the intestines.
Immunity and Longevity Assays.
P. aeruginosa strain PA14 killing assays were performed at 23 °C, unless otherwise noted, as previously described (36). Five-fluoro-2′-deoxyuridine (FUDR; 75 μg/mL) was added to the assay plates to reduce the growth of progeny. To test the immunity or longevity phenotypes of any strains containing daf-2(e1368ts), all worms in the experiment were raised to the L4 stage at the permissive temperature of 15 °C, shifted to the restrictive temperature of 25 °C for 4 h, and transferred to PA14 killing plates or OP50 lifespan plates at 25 °C. Killing assays with S. aureus strain NCTC8325 and E. faecalis strain mmH594 were performed at 23 °C and 25 °C, respectively (36). Lifespan assays were performed at 23 °C, unless otherwise noted, on NGM plates containing 75 μg/mL FUDR seeded with E. coli strain OP50. Data from immunity and longevity assays were statistically analyzed as previously described (36). Briefly, a log-rank analysis was used to calculate mean survival for each population of worms. The mean survival values were compared via a 2-tailed Student's t-test.
Quantitative RT-PCR.
Quantitative RT-PCR was performed as previously described (15), with primer sequences graciously provided by E. Troemel. All C. elegans strains tested were grown in parallel, and 3 independent biologic replicates were tested, in duplicate, for each primer set. Statistical analysis was performed using Prism (GraphPad). The normalized values for induction or basal expression for the 3 replicates were compared using a 1-sample t-test. Strong induction dependence was defined as at least a 10-fold difference in induction between wild-type and mutant worms with P < 0.01. Moderate induction dependence was defined as at least a 5-fold difference between wild-type and mutant worms with P < 0.01.
Supplementary Material
Acknowledgments.
We thank Anna Ausubel for technical assistance; and E. Troemel, R. Feinbaum, and J. Irazoqui for many helpful discussions and for critical reading of the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources. This work was supported by NIH grants P01 AI044220 and R01 AI064332 (to F.M.A.), a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and an NIH National Institute of Allergy and Infectious Dieases K08 award (to D.H.K.), and a Ruth L. Kirschstein National Research Service Award postdoctoral fellowship (F32 AI068376 to J.R.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813048106/DCSupplemental.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 3.Schulenburg H, Kurz CL, Ewbank JJ. Evolution of the innate immune system: The worm perspective. Immunol Rev. 2004;198:36–58. doi: 10.1111/j.0105-2896.2004.0125.x. [DOI] [PubMed] [Google Scholar]
- 4.Sifri CD, Begun J, Ausubel FM. The worm has turned—Microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Kim DH, Ausubel FM. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans. Curr Opin Immunol. 2005;17:4–10. doi: 10.1016/j.coi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Kim DH, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, et al. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc Natl Acad Sci USA. 2004;101:10990–10994. doi: 10.1073/pnas.0403546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liberati NT, et al. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci USA. 2004;101:6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuzawa A, et al. ROS-dependent activation of the TRAF6-ASK1–p38 pathway is selectively required for TLR4-mediated innate immunity. Nat Immunol. 2005;6:587–592. doi: 10.1038/ni1200. [DOI] [PubMed] [Google Scholar]
- 10.Garsin DA, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 11.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 12.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 13.Mallo GV, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 14.O'Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troemel ER, et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2(11):e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pujol N, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 18.Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 2008;9:103–109. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kedzierski L, Montgomery J, Curtis J, Handman E. Leucine-rich repeats in host-pathogen interactions. Arch Immunol Ther Exp (Warsz) 2004;52:104–112. [PubMed] [Google Scholar]
- 20.Cho S, Rogers KW, Fay DS. The C. elegans glycopeptide hormone receptor ortholog, FSHR-1, regulates germline differentiation and survival. Curr Biol. 2007;17:203–212. doi: 10.1016/j.cub.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 21.Sieburth D, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 22.Miyata S, Begun J, Troemel ER, Ausubel FM. DAF-16-dependent suppression of immunity during reproduction in Caenorhabditis elegans. Genetics. 2008;178:903–918. doi: 10.1534/genetics.107.083923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan QR, Hendrickson WA. Structure of human follicle-stimulating hormone in complex with its receptor. Nature. 2005;433:269–277. doi: 10.1038/nature03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 25.Kocks C, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 26.Simoni M, Gromoll J, Nieschlag E. The follicle-stimulating hormone receptor: Biochemistry, molecular biology, physiology, and pathophysiology. Endocr Rev. 1997;18:739–773. doi: 10.1210/edrv.18.6.0320. [DOI] [PubMed] [Google Scholar]
- 27.Herath S, et al. Ovarian follicular cells have innate immune capabilities that modulate their endocrine function. Reproduction. 2007;134:683–693. doi: 10.1530/REP-07-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun L, et al. FSH directly regulates bone mass. Cell. 2006;125:247–260. doi: 10.1016/j.cell.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 29.Takayanagi H. Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7:292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 30.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraser AG, et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 32.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 33.Rual JF, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahringer J, editor. Reverse genetics. [Accessed August 1, 2008];2006 Available at: http://www.wormbook.org/chapters/www_introreversegenetics/introreversegenetics.html.
- 35.Espelt MV, Estevez AY, Yin X, Strange K. Oscillatory Ca2+ signaling in the isolated Caenorhabditis elegans intestine: Role of the inositol-1,4,5-trisphosphate receptor and phospholipases C beta and gamma. J Gen Physiol. 2005;126:379–392. doi: 10.1085/jgp.200509355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell JR, Ausubel FM. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. In: Ewbank J, Vivier E, editors. Innate Immunity, Methods in Molecular Biology. Vol 415. Totowa, NJ: Humana Press; 2007. pp. 403–427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.