Abstract
Although both heterodimeric subunits of core binding factors (AML1/RUNX1 and CBFβ) essential for normal hematopoiesis are frequently mutated to form different chimeric fusion proteins in acute leukemia, the underlying molecular mechanisms and structural domains required for cellular transformation remain largely unknown. Despite the critical role of CBFβ for wild-type AML1 function and its direct involvement in chromosomal translocation, we demonstrate that both the expression and interaction with CBFβ are superfluous for AML1-ETO (AE)-mediated transformation of primary hematopoietic cells. Similarly, the hetero-oligomeric interaction with transcriptional repressor ETO family proteins and the highly conserved NHR1 domain in AE fusion are also dispensable for transforming activity. In contrast, AE-mediated transformation is critically dependent on the DNA binding and homo-oligomeric properties of the fusion. Abolishment of homo-oligomerization by a small-molecule inhibitor could specifically suppress AML1 fusion-mediated transformation of primary hematopoietic cells. Together, these results not only identify the essential molecular components but also potential avenues for therapeutic targeting of AE-mediated leukemogenesis.
Keywords: RUNX1, transformation, oligomerization, leukemia, NHR2
AML1/RUNX1 and CBFβ are 2 critical transcription factors essential for generation of hematopoietic stem cells (HSCs) (1, 2). Mice deficient in AML1 or CBFβ have almost identical phenotypes; they completely lack definitive hematopoiesis and die at approximately embryonic day 12.5. In acute myeloid leukemia in which leukemic stem cells have been functionally identified, AML1 and CBFβ represent the most commonly mutated targets (1, 2). t(8;21) resulting in AML1-ETO (AE) fusion can be found in up to 40% of AML-M2; and inv(16) leading to CBFβ-SHMMC fusion constitutes approximately 30% of AML-M4. AML1 is also fused to TEL as a result of t(12;21) in approximately 25% of childhood leukemia, the most common form of childhood cancers. Although animal modeling clearly indicates that AML1 fusions per se are not sufficient for induction of full-blown leukemia, they function to enhance self-renewal and expand targeted HSCs and early progenitors for cooperative secondary mutations to take place (3–5).
While enhanced self-renewal has emerged as a critical feature associated with various oncogenic transcription factors involved in acute leukemia (6–8), much less is known about the underlying molecular mechanisms. It is clear that most, if not all, of the transcription factors do not work as monomers but need to complex with various proteins for full activity and specificity, although the molecular composition of the associated transcriptional complexes required for self-renewal remains largely unknown. At the molecular level, AE encodes the DNA-binding runt homology domain (RHD) fused in-frame with almost the entire transcriptional repressor protein ETO containing 4 different Nervy homology regions (NHRs) (Fig. 1A), suggesting a gain of transcriptional repressor function by the oncogenic fusion (9). A well-recognized and important function of the RHD is to recruit CBFβ that enhances the stability and DNA binding ability of the wild-type AML1 (1, 2). Although its role in cellular transformation has not been demonstrated, CBFβ has been regarded as a critical partner for AE function, and specific inhibitors targeting this interaction have been recently developed for potential therapeutic intervention (10). Similarly, the functional contribution of the ETO protein in the AE-mediated transformation is not well defined. NHR1 sharing significant sequence homology with TATA-box binding protein-associated factors can interact with activation domain (AD1) of E proteins, leading to transcriptional repression through displacement of p300/CBP coactivators, but again its functional significance for transformation is still largely unknown (11, 12). NHR2, capable of interacting with SIN3/HDACs complexes, encodes a hydrophobic amino acid heptad repeat, which mediates both homo-oligomerization and hetero-oligomerization (9, 13) with ETO family members that play important roles in transcriptional repression and normal development. NHR1 and NHR2 are invariably retained in both the wild-type and the recently identified oncogenic spliced isoform (AE9a) of AE (14), suggesting their potential functions in oncogenic transformation. NHR3 has a coiled-coil structure, which can help to recruit transcriptional cofactors. NHR4 contains a myeloid-Nervy-DEAF1 homology domain with 2 zinc finger motifs that have been shown to recruit NCoR/SMRT/HDAC1/2 transcriptional repressor complexes (9). Although attempts had been made to identify the critical domains required for AE-mediated transformation, conflicting results were presented from most of these studies using well-established cell lines, which suffer from the pitfalls of carrying multiple irrelevant genetic mutations that may not reflect the normal biology of the disease [for a summary see Hug et al. (5)]. For example, NHR4 was required to inhibit differentiation of U937 cells (15) but was dispensable for transformation of NIH 3T3 cells (16). The only available structure/function data on primary cells are limited to NHR2 of the ETO portion of the fusion but cannot distinguish the functional contribution between homo-oligomerization and hetero-oligomerization (13). The lack of comprehensive structure/function data not only has significantly impeded the progress of understanding the biology of the disease but also hinders the development of specific cancer therapeutics. To this end, we performed an extensive functional analysis to dissect AE-associated protein complexes. We found dispensable functions of both CBFβ and ETO interaction for AE-mediated transformation of primary hematopoietic cells, which is, however, critically dependent on the homo-oligomerization property of the fusion. A small-molecule inhibitor that specifically dissociates homo-oligomerization could reverse AML1 fusion-mediated transformation. Together these results not only reveal critical domains and potential therapeutic targets for AE-mediated transformation of primary hematopoietic cells but also strongly endorse a prevalent homo-oligomerization-dependent mechanism for leukemia-associated transcription factors.
Fig. 1.
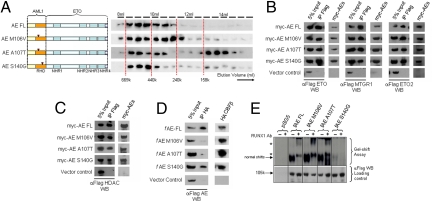
Identification and biochemical characterization of AE point mutants defective in CBFβ interaction. (A) Left: Gel filtration analysis on AE point mutants. Right: Western blot using α-Flag antibody on protein fractions collected from indicated elution volumes; dotted lines indicate the positions of the corresponding size standards. (B–D) Coimmunoprecipitation assay between (B) myc-tagged AE constructs and Flag-tagged ETO family members, (C) myc-tagged AE constructs and Flag-tagged HDAC1, or (D) Flag-tagged AE constructs and HA-tagged CBFβ. Antibodies used for immunoprecipitation (IP) and Western blot (WB) are indicated. (E) Top, Gel shift assay using lysates from 293 cells transfected with indicated constructs together with P-end-labeled CD11a probe in the absence or presence (for supershift) of AML1 antibody. Asterisks indicate supershifted bands. Bottom, Western blot of cell lysates used for gel shift experiment using α-Flag antibody.
Results
Given the heterodimeric nature and the crucial functions of CBFβ interaction with the wild-type AML1, we first sought to dissect the functional requirement of CBFβ interaction in AE-mediated transformation. On the basis of previous structural, biochemical, and mutagenesis data, we designed 3 different single-point mutants on the RHD (M106V, A107T, and S140G) that have been shown in the context of AML1 to selectively disrupt their ability to interact with CBFβ and/or DNA binding (supporting information Fig. S1A) (17–19). Specifically, M106V and A107T AML1 mutants would have compromised CBFβ binding ability, whereas S140G AML1 mutant has additional defective DNA binding. To further assess the properties of these point mutations in the context of AE fusion, we generated equivalent AE point mutants and subjected them to various biochemical assays. Analogous to the wild-type AE, all of the mutants were capable of forming high-molecular-weight complexes, although A107T mutants might contain a slightly higher proportion of homodimers (Fig. 1A). In addition, these mutants exhibited a similar ability as the wild-type fusion protein to interact with components of transcriptional complexes, including different members of ETO family proteins ETO1, ETO2, and MTGR1 (Fig. 1B), as well as histone deacetylase 1 (HDAC1) (Fig. 1C), indicating that most of the biochemical properties of these mutants indeed have not been altered. However, when assessed for their abilities to recruit CBFβ and bind DNA, both AE M106V and AE A107T mutants had almost completely lost their capacity for interacting with CBFβ, whereas AE S140G mutant could still efficiently coprecipitate with CBFβ (Fig. 1D). In contrast, AE S140G had a severely compromised DNA binding property, whereas both AE M106V and AE A107T mutants could still bind to DNA, although the AE A107T mutant might have a weaker DNA binding property toward certain targets (Fig. 1E and data not shown).
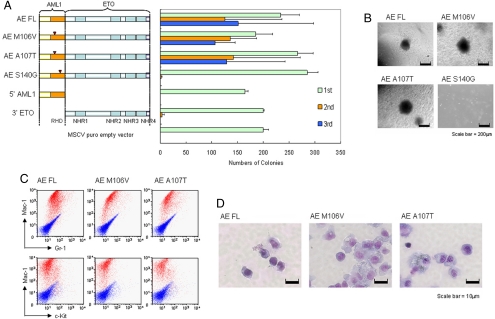
We then used a retroviral transduction/transformation assay (RTTA), which has been successfully used as a surrogate in vitro assay for assessing self-renewal/transformation properties of various leukemia-associated transcription factors (6–8, 20), to study the biological effect of AE on primary hematopoietic cells. As expected, primary murine hematopoietic cells transduced with vector control, truncated 5′ AML1, or 3′ ETO quickly exhausted their proliferative capacity in the second round of plating and failed to give third-round colonies (Fig. 2A). In contrast, full-length AE could enhance replating ability and transform primary hematopoietic cells to give rise to the third and subsequent rounds of colonies that expressed c-kitlo, Mac-1, and Gr-1 markers (Fig. 2 A–C and data not shown), confirming essential functional contributions from both AML1 and ETO portions of the protein for myeloid transformation. Analogous to the wild-type AE, both AE M106V and AE A107T mutants with significantly compromised CBFβ binding ability could still competently transform primary hematopoietic cells and gave rise to third-round colonies (Fig. 2A) with similar morphology and immunophenotypes (Fig. 2 B–D). Moreover, with AE M106V and AE A107T, like the wild-type AE, transduced cells could also be replated to form colonies in subsequent fourth and fifth replatings (Fig. S1B). In contrast, cells transduced with AE S140G mutants that could still interact with CBFβ but were defective in DNA binding quickly exhausted their proliferative ability and failed to give third-round colonies (Fig. 2A), although all of the mutants were expressed at the expected size and a comparable level (Fig. S1C). Similar results were obtained using a different AE R174A DNA binding-defective mutant that also failed to transform primary hematopoietic cells (data not shown). Together, these results suggest that CBFβ interaction is neither necessary nor sufficient for AE-mediated transformation of primary hematopoietic cells, which may depend on the DNA binding property of the fusion (21).
Fig. 2.
Enhanced self-renewal of primary hematopoietic cells by CBFβ interaction-deficient AE point mutants. (A) Left, Schematic diagram indicating the retroviral constructs of AE and the point mutants used in the RTTA. Right, Bar chart represents the corresponding number of colonies in each round of platings. Error bars indicate SD of 3 independent experiments. (B) Typical third-round colony morphology of the indicated retroviral-transduced primary bone marrow cells. (C) Phenotypic analysis of cells transformed by the indicated constructs. Red profiles represent stainings obtained with antibodies specific for the indicated surface markers. Blue profiles show unstained controls. (D) Typical morphology of primary bone marrow cells transduced with indicated constructs after the third plating.
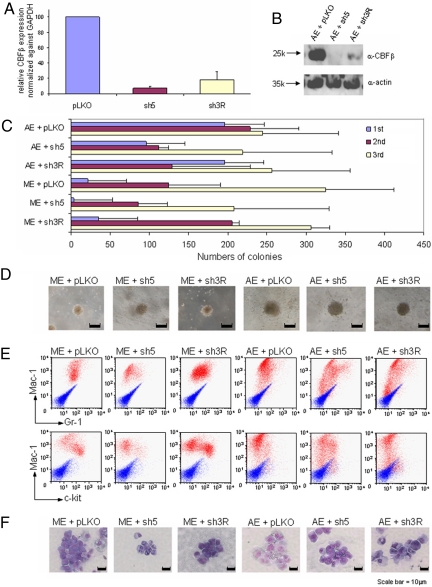
To further confirm our findings and investigate the effect of global reduction of CBFβ in AE-mediated transformation of primary hematopoietic cells, 2 independent shRNA constructs were developed and validated for their ability to knock down the expression of endogenous CBFβ. As compared with the vector control, sh3R and sh5 could potently knock down the expression of CBFβ transcript in primary hematopoietic cells by an average of 80% and 95%, respectively (Fig. 3A). At the protein level, both sh3R and sh5 could knock down the level of endogenous CBFβ in primary transduced cells by more than 95%, as demonstrated by Western blot analysis (Fig. 3B). When these shRNAs were individually cotransduced with either MLL-ENL (control) or AE in the RTTA, MLL-ENL cells transduced with CBFβ shRNAs were still capable of forming third and subsequent rounds of colonies, suggesting that CBFβ knockdown did not have a major impact on MLL-mediated transformation (Fig. 3 C and D and data not shown). Similar to MLL fusion, AE-transduced cells were also capable of giving rise to almost the same number of third and subsequent rounds of transformed colonies in the presence of CBFβ shRNAs, as compared with the vector control (Fig. 3 C and D and data not shown). To examine whether CBFβ knockdown would have an effect on differentiation, analyses on the fifth-round colonies transduced with vector control or CBFβ shRNAs revealed very similar morphology and immunophenotypes, although a slight reduction of c-kit-positive cells was occasionally observed in MLL-ENL cells cotransduced with CBFβ sh5 (Fig. 3 E and F), indicating that global knockdown of CBFβ expression has, if any, very limited effect on AE-transduced primary cells. Together with the point mutant data, these results demonstrate that despite its crucial role in wild-type AML1 functions, CBFβ is largely dispensable for DNA binding-dependent AE-mediated transformation.
Fig. 3.
AE-mediated self-renewal is independent on CBFβ expression. (A) RNAs from primary bone marrow cells transduced with virus carrying indicated shRNA were isolated 7 days after transduction and selection, reverse transcribed, and subjected to quantitative PCR. Error bars indicate SD of 2 independent experiments carried out in duplicate. (B) Western blot on primary bone marrow cells transduced with virus carrying indicated shRNAs using α-CBF or α-actin antibodies (for loading control). (C) Bar chart represents the numbers of colonies in each plating of primary hematopoietic cells cotransduced with either AML-ETO or MLL-ENL (control) together with one of the shRNAs as indicated at left. Error bars indicate SD of 6 different experiments. (D) Typical third-round colony morphology of the indicated retroviral-transduced primary bone marrow cells. (E) Immunophenotypic analysis of cells cotransduced by MLL-ENL or AE and indicated shRNAs in the fifth round of plating. Red profiles represent stainings obtained with antibodies specific for the indicated surface markers. Blue profiles show unstained controls. (F) Typical fifth-round cell morphology of primary bone marrow cells transduced with indicated constructs.
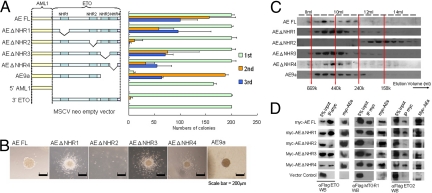
To further dissect the critical molecular components associated with the ETO portion of the fusion, AE mutants targeting individual NHRs were subjected to RTTA. Deletion of any single NHR1, NHR3, or NHR4 did not have a significant impact on transformation, indicating dispensable or redundant functions shared by these domains (Fig. 4 A and B). Consistently, the recently identified oncogenic spliced isoform AE9a (14), with deletion of both NHR3 and NHR4 could still form third and subsequent rounds of colonies (Fig. 4 A and B and data not shown). In contrast, AE ΔNHR2 mutant with a specific deletion of NHR2 had severely compromised transformation ability and failed to form third-round colonies (Fig. 4 A and B), although all of the mutants were expressed at the expected size and comparable level (Fig. S2A). Further biochemical analyses revealed that deletion of NHR2 did not adversely affect homodimerization (Fig. S2B) but completely abolished its ability to form high-molecular-weight homo-oligomeric complexes (Fig. 4C). Identification of NHR2 with oligomeric ability as a critical region for AE-mediated transformation is reminiscent of the recent finding on RARα fusion in acute promyelocytic leukemia, whereby the formation of homotetrameric instead of dimeric complexes is required for transformation of primary hematopoietic cells (22). However, in addition to homo-oligomerization, NHR2 is also responsible for hetero-oligomeric interaction with various proteins in the complexes (9, 13). Loss of NHR2 had abolished its ability to form hetero-oligomers with ETO corepressor family proteins (Fig. 4D). Together these results indicate that AE-mediated transformation absolutely depends on NHR2, which mediates both homo- and hetero-oligomerization. It is not clear, however, whether one or both of these properties are critical.
Fig. 4.
NHR2 that mediates both homo- and hetero-oligomerization is necessary for enhanced self-renewal. (A) Left, Schematic diagram indicating the retroviral constructs used in the RTTA. Right, Bar chart represents the corresponding number of third-round colonies in each plating. Error bars indicate SD of 3 independent experiments. (B) Typical third-round colony morphology of the indicated retroviral-transduced primary bone marrow cells. (C) Gel filtration analysis. The panel shows the Western blot using anti-Flag antibody on protein fractions collected from indicated elution volumes, with dotted lines indicating corresponding size standards. (D) Coimmunoprecipitation assay between myc-tagged AE constructs and Flag-tagged ETO family members. Antibodies used for immunoprecipitation (IP) and Western blot (WB) are indicated.
To distinguish the functional contribution of homo- vs. hetero-oligomeric properties of NHR2 in AE-mediated transformation, we made use of a mutant FKBP (FKBPF36M) module, which will specifically form homo-oligomers with itself (but not endogenous FKBP) in a regulated and reversible manner (23) and has been successfully used to replace and demonstrate the role of homo-oligmerization in RARα fusion-mediated transformation (20). By inserting mutant FKBP oligomeric modules into the transformation-incompetent AE ΔNHR2 construct, we generated an AML1-NHR1-FKBP-NHR3/4 construct that will allow homo-oligomerization independent of NHR2 (Fig. 5A). Analogous to the wild-type AE, AML1-NHR1-FKBP-NHR3/4 still maintained its ability to interact with HDAC1 (Fig. S3A) and CBFβ (Fig. S3B). Further biochemical analyses showed that the synthetic FKBP oligomeric modules could restore the ability of AE ΔNHR2 to form high-molecular-weight homo-oligomeric complexes (Fig. 5A) but importantly not hetero-oligomerization with ETO family proteins (Fig. 5B). When tested in the RTTA, AML1-NHR1-FKBP-NHR3/4, in contrast to the AE ΔNHR2 mutant, could enhance replating ability and transform primary hematopoietic cells (Fig. 5C) with very similar morphology and immunophenotypes as the wild-type AE-transformed cells (Fig. 5 D and E). Analogous to the wild-type AE, AML1-NHR1-FKBP-NHR3/4-transduced cells could also form colonies in the subsequent fourth and fifth rounds of replatings (Fig. S3C). Thus, the FKBP homo-oligomeric module could resurrect the transformation property to the otherwise incompetent AE ΔNHR2 construct, indicating that homo-oligomerization represents the major function of NHR2 in AE-mediated transformation, although we could not exclude possible recruitments of other proteins by mutant FKBP.
Fig. 5.
Rescue of transformation-defective AEΔNHR2 mutant by synthetic homo-oligomerization FKBP modules. (A) Left, Schematic diagram of the constructs used in the gel filtration analysis. Right, Western blot using α-Flag antibody on protein fractions collected from indicated elution volumes, with dotted lines indicating corresponding size standards. Coimmunoprecipitation assay between myc-tagged AE constructs and (B) Flag-tagged ETO family proteins. IP, immunoprecipitation; WB, Western blot. (C) Left, Schematic diagram of AE and the synthetic AML-NHR1-FKBP-NHR3/4 constructs used in RTTA. Right, Bar chart represents the corresponding numbers of colonies in the each round of plating. Error bars indicate SD of 3 independent experiments. (D) Typical third-round colony morphology of the indicated retroviral-transduced primary bone marrow cells. (E) Immunophenotypic analysis of cells transduced by AE or AML1-NHR1-FKBP-NHR3/4. Dot plots represent stainings obtained with antibodies specific for the indicated surface markers. Contour plots indicate unstained controls. (F) Bar chart represents the corresponding numbers of third-round colonies transduced with constructs indicated in the left with or without treatment of AP21998.
Finally, to further assess the role of homo-oligomerization in maintenance of the AML1 fusion-mediated transformation, we took advantage of a highly selective small-molecule inhibitor, AP21998, which can specifically interrupt the homo-oligomerization interface within the mutant FKBP modules (20). As confirmed by gel filtration analyses, AP21998 could effectively dissociate AML1-NHR1-FKBP-NHR3/4 homo-oligomeric complexes (Fig. 5A). When AP21998 was incubated with the wild-type AE-transduced cells in RTTA, it did not have any effect on colony formation, suggesting that the drug is not generally toxic to cells (Fig. 5F). In contrast, AP21998 could specifically abolish the colony-forming ability of AML1-NHR1-FKBP-NHR3/4-transduced cells, thus strongly indicating that the formation of higher-order homo-oligomeric complexes is essential for AML1 fusion-mediated transformation of primary hematopoietic cells (Fig. 5F).
Discussion
Functional dissection of protein complexes associated with oncogenic transcription factors, which often represent the initiating events in acute leukemia (24), is critical for understanding the transformation mechanisms and identifying potential therapeutic targets for intervention of self-renewal pathways corrupted in cancer stem cells (20, 22, 25–27). CBFβ, which plays an important role for wild-type AML1 functions, in part by increasing its DNA binding ability (28) and stability (29), has been thought to be a critical component and potential target for AE-mediated transformation (1, 10). However, the discovery of (i) transformation-competent AE point mutants with defective CBFβ binding and (ii) efficient AE-mediated transformation in CBFβ-deficient primary cells indicate that CBFβ interaction is not essential for the oncogenic AML1 fusion complexes, which may have acquired additional properties that can override the CBFβ dependence (as discussed later). These results also argue against a dominant negative mechanism of CBFβ functions but favor a gain of function by AE. Notably, DNA binding-independent pathways have been suggested for AML1/RUNX1 DNA-binding mutants identified in familial platelet disorder with predisposition to AML patients, in which the AML1 mutants can also function independent of CBFβ interaction (30). Although our data evidently reveal a dispensable function of CBFβ in AE-mediated transformation of primary hematopoietic cells, it is still possible that small-molecule inhibitors targeting the CBFβ/AML1 interaction may have a more profound impact on CBFβ-SMMHC-transformed cells or cells highly dependent on the wild-type interaction, such as in patients with AML1 amplification. Consistently, the reported CBFβ/AML1 allosteric inhibitor suppresses non-CBF leukemia cell lines (e.g., HL60 and U937) that express a high level of AML1 (10), suggesting that its cytotoxic effect can be, at least in part, due to the negative impact on the wild-type CBFβ/AML1 interaction. This merits further investigation.
On the other hand, although NHR1 is always retained in both the wild-type and oncogenic spliced form of AE and acts as a docking site for positive and negative transcriptional regulators (31), it is not essential or may have redundant functions with other AE domains, such as NHR2–4, for transformation of primary hematopoietic cells. Conversely, NHR2 is the only functional domain indispensable for AE-mediated transformation. Although the tetrameric property of NHR2 that mediates both homo-oligomerization and hetero-oligomerization with other proteins such as ETO family repressors has been proposed as a critical property for various AE activity (9, 13, 32), the lack of an AE mutant that selectively loses the hetero-oligomerization property has made it impossible to clearly define the functional contributions of this domain (13). By replacing NHR2 with synthetic FKBP mutant homo-oligomeric modules, we have identified homo-oligomerization as the major and indispensable component for AE-mediated transformation of primary hematopoietic cells. Forced homo-oligomerization has recently been proposed as an essential element for RARα fusion-mediated transformation (20, 33), which enhances recruitment of transcriptional corepressor complexes such as SIN3/SMRT/HDACs to downstream targets. Given the critical function of self-association in AE-mediated transformation, it is tempting to speculate that the reduced dependence of the fusion on a CBFβ and ETO interaction may be due to its acquired homo-oligomeric properties, which may enhance the overall transcriptional activity and inhibit proteasome-mediated degradation (refs. 32, 34 and data not shown). Consistently, we also demonstrated that AE-mediated transformation can be abolished by a small-molecule inhibitor that specifically reverses homo-oligomerization. Although the relatively large hydrophobic interface in NHR2 may pose a technical challenge for inhibitor development (13), expression of a small fusion peptide consisting of the entire NHR2 that grossly interfered with both oligomerization functions of NHR2 could revert the differentiation block and induced death of AE-transformed cells (35). We also suggest that the NHR2 homo-oligomeric interface can potentially be targeted by macrodrugs, such as single antibody domains, which have been successfully used for inhibiting specific protein-protein interactions to suppress cellular transformation mediated by oncogenic RAS (36). Thus, identification of homo-oligomerization as the essential property for AE-mediated transformation not only defines a prevalent mechanism shared by the most common leukemia-associated transcription factors (20, 33, 37, 38) but also reveals a challenging but promising avenue for therapeutic intervention for these otherwise non-druggable targets in human cancers.
Materials and Methods
Constructs and Antibodies.
AML1-ETO, AML1-ETO deletion mutants, CBFβ, and ETO family constructs have been previously reported (39). All of the AML1-ETO single-point mutant constructs were cloned using megaprimer site-directed mutagenesis and verified by DNA sequencing. Inducible homo-oligomerization AML1-NHR1-FKBP-NHR3/4 synthetic construct was made by replacing NHR2 in AML1-ETO with 4 tandem repeats of self-oligomerizing FKBP mutant (FKBPF36M) carrying Phe to Met mutation (Ariad Pharmaceuticals). The shRNA target sequences are sh3R 5′-GAGGGACTGCAGTTGGTTT-3′ in pSuper (Oligoengine) and sh5 5′-GCTCGAAGAAGAACTCGAGAA-3′ in pLKO (Open Biosystems). Detailed cloning strategies are available upon request. The following antibodies were used: murine c-kit (2B8 clone), Mac-1 (M1/70 clone), Gr-1 (RB6 8C5 clone) (all from eBiosciences), Runx1 (N-20), Myc (A14), HA (Y-11) (all from Santa Cruz). CBFb (141,4,1) (Abcam), and Flag (M2) (Sigma-Aldrich).
RTTA.
RTTA was performed as previously described (20, 22, 25). In vitro transformation was defined as enhanced self-renewal of primary hematopoietic cells beyond 3 rounds of plating in the serial replating assay. For cotransduction experiments, spinoculation was performed twice on 2 consecutive days. For the inhibitor study, AP21998 was added to a final concentration of 1 mM in half of the wells for indicated rounds of replating.
Immunophenotype analysis, coimmunoprecipitation assay, gel filtration assay, gel shift binding assay, and quantitative RT-PCR were carried out as described (20, 22, 39). Additional information about Gel Filtration Assay and Quantitative RT-PCR can be found in SI Methods.
Supplementary Material
Acknowledgments.
We thank Mel Greaves, Arthur Zelent, and Jon Wilson for constructive advice on the manuscript, Dong-Er Zhang for AE-9a cDNA, and Amanda Wilson for technical assistance. This work was supported by the Leukaemia Research Fund and the Association for International Cancer Research (AICR). C.W.E.S. is an AICR fellow and a European Molecular Biology Organization young investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810558106/DCSupplemental.
References
- 1.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 2.Ito Y. RUNX genes in development and cancer: Regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- 3.Mulloy JC, et al. Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood. 2003;102:4369–4376. doi: 10.1182/blood-2003-05-1762. [DOI] [PubMed] [Google Scholar]
- 4.McCormack E, Bruserud O, Gjertsen BT. Review: Genetic models of acute myeloid leukaemia. Oncogene. 2008;27:3765–3779. doi: 10.1038/onc.2008.16. [DOI] [PubMed] [Google Scholar]
- 5.Hug BA, Lee SY, Kinsler EL, Zhang J, Lazar MA. Cooperative function of Aml1-ETO corepressor recruitment domains in the expansion of primary bone marrow cells. Cancer Res. 2002;62:2906–2912. [PubMed] [Google Scholar]
- 6.Cozzio A, et al. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.So CW, et al. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 2003;3:161–171. doi: 10.1016/s1535-6108(03)00019-9. [DOI] [PubMed] [Google Scholar]
- 8.Huntly BJ, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Hug BA, Lazar MA. ETO interacting proteins. Oncogene. 2004;23:4270–4274. doi: 10.1038/sj.onc.1207674. [DOI] [PubMed] [Google Scholar]
- 10.Gorczynski MJ, et al. Allosteric inhibition of the protein-protein interaction between the leukemia-associated proteins Runx1 and CBFbeta. Chem Biol. 2007;14:1186–1197. doi: 10.1016/j.chembiol.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- 12.Plevin MJ, Zhang J, Guo C, Roeder RG, Ikura M. The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain. Proc Natl Acad Sci USA. 2006;103:10242–10247. doi: 10.1073/pnas.0603463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, et al. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity. Cancer Cell. 2006;9:249–260. doi: 10.1016/j.ccr.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Yan M, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 15.Gelmetti V, et al. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank RC, Sun X, Berguido FJ, Jakubowiak A, Nimer SD. The t(8;21) fusion protein, AML1/ETO, transforms NIH3T3 cells and activates AP-1. Oncogene. 1999;18:1701–1710. doi: 10.1038/sj.onc.1202459. [DOI] [PubMed] [Google Scholar]
- 17.Akamatsu Y, et al. Redox regulation of the DNA binding activity in transcription factor PEBP2. The roles of two conserved cysteine residues. J Biol Chem. 1997;272:14497–14500. doi: 10.1074/jbc.272.23.14497. [DOI] [PubMed] [Google Scholar]
- 18.Akamatsu Y, Tsukumo S, Kagoshima H, Tsurushita N, Shigesada K. A simple screening for mutant DNA binding proteins: Application to murine transcription factor PEBP2alpha subunit, a founding member of the Runt domain protein family. Gene. 1997;185:111–117. doi: 10.1016/s0378-1119(96)00644-0. [DOI] [PubMed] [Google Scholar]
- 19.Warren AJ, Bravo J, Williams RL, Rabbitts TH. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFbeta. EMBO J. 2000;19:3004–3015. doi: 10.1093/emboj/19.12.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwok C, Zeisig BB, Dong S, So CW. Forced homo-oligomerization of RARalpha leads to transformation of primary hematopoietic cells. Cancer Cell. 2006;9:95–108. doi: 10.1016/j.ccr.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Grisolano JL, O'Neal J, Cain J, Tomasson MH. An activated receptor tyrosine kinase, TEL/PDGFbetaR, cooperates with AML1/ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci USA. 2003;100:9506–9511. doi: 10.1073/pnas.1531730100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeisig BB, et al. Recruitment of RXR by homotetrameric RARalpha fusion proteins is essential for transformation. Cancer Cell. 2007;12:36–51. doi: 10.1016/j.ccr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Rollins CT, et al. A ligand-reversible dimerization system for controlling protein-protein interactions. Proc Natl Acad Sci USA. 2000;97:7096–7101. doi: 10.1073/pnas.100101997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 25.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 26.Villa R, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J, et al. RXR is an essential component of the oncogenic PML/RARA complex in vivo. Cancer Cell. 2007;12:23–35. doi: 10.1016/j.ccr.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Gu TL, Goetz TL, Graves BJ, Speck NA. Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1) Mol Cell Biol. 2000;20:91–103. doi: 10.1128/mcb.20.1.91-103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang G, et al. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cammenga J, et al. RUNX1 DNA-binding mutants, associated with minimally differentiated acute myelogenous leukemia, disrupt myeloid differentiation. Cancer Res. 2007;67:537–545. doi: 10.1158/0008-5472.CAN-06-1903. [DOI] [PubMed] [Google Scholar]
- 31.Wei Y, et al. A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription. Nat Struct Mol Biol. 2007;14:653–661. doi: 10.1038/nsmb1258. [DOI] [PubMed] [Google Scholar]
- 32.Minucci S, et al. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol Cell. 2000;5:811–820. doi: 10.1016/s1097-2765(00)80321-4. [DOI] [PubMed] [Google Scholar]
- 33.Sternsdorf T, et al. Forced retinoic acid receptor alpha homodimers prime mice for APL-like leukemia. Cancer Cell. 2006;9:81–94. doi: 10.1016/j.ccr.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 34.Kramer OH, Muller S, Buchwald M, Reichardt S, Heinzel T. Mechanism for ubiquitylation of the leukemia fusion proteins AML1-ETO and PML-RARalpha. FASEB J. 2008;22:1369–13679. doi: 10.1096/fj.06-8050com. [DOI] [PubMed] [Google Scholar]
- 35.Wichmann C, et al. Targeting the oligomerization domain of ETO interferes with RUNX1/ETO oncogenic activity in t(8;21)-positive leukemic cells. Cancer Res. 2007;67:2280–2289. doi: 10.1158/0008-5472.CAN-06-3360. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka T, Williams RL, Rabbitts TH. Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS. EMBO J. 2007;26:3250–3259. doi: 10.1038/sj.emboj.7601744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.So CW, Lin M, Ayton PM, Chen EH, Cleary ML. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4:99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- 38.Martin ME, et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4:197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 39.Qiu J, Wong J, Tweardy DJ, Dong S. Decreased intranuclear mobility of acute myeloid leukemia 1-containing fusion proteins is accompanied by reduced mobility and compartmentalization of core binding factor beta. Oncogene. 2006;25:3982–3993. doi: 10.1038/sj.onc.1209431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.