Abstract
Mammalian cells employ numerous innate cellular mechanisms to inhibit viral replication and spread. Tetherin, also known as Bst-2 or CD317, is a recently identified, IFN-induced, cellular response factor that blocks release of HIV-1 and other retroviruses from infected cells. The means by which tetherin retains retroviruses on the cell surface, as well as the mechanism used by the HIV-1 accessory protein Vpu to antagonize tetherin function and promote HIV-1 release, are unknown. Here, we document that tetherin functions as a broadly acting antiviral factor by demonstrating that both human and murine tetherin potently inhibit the release of the filovirus, Ebola, from the surface of cells. Expression of the Ebola glycoprotein (GP) antagonized the antiviral effect of human and murine tetherin and facilitated budding of Ebola particles, as did the HIV-1 Vpu protein. Conversely, Ebola GP could substitute for Vpu to promote HIV-1 virion release from tetherin-expressing cells, demonstrating a common cellular target for these divergent viral proteins. Ebola GP efficiently coimmunoprecipitated with tetherin, suggesting that the viral glycoprotein directly interferes with this host antiviral factor. These results demonstrate that tetherin is a cellular antiviral factor that restricts budding of structurally diverse enveloped viruses. Additionally, Ebola has evolved a highly effective strategy to combat this antiviral response elicited in the host during infection.
Keywords: antiviral, CD317, viral budding, HIV-1 vpu, Bst-2
The innate immune system is the first line of defense against invading viral pathogens. Virus detection by the host triggers a protein signaling cascade that leads to the production of type I interferons (IFNs), such as IFN-α and IFN-β. IFNs induce a systemic antiviral cellular response aimed at limiting virus replication and spread. However, viruses have evolved several highly effective strategies to combat IFN synthesis, signaling, and the activity of IFN-induced antiviral proteins. Tetherin regulation by the HIV-1 accessory protein Vpu provides a potent example of a strategy used by a virus to escape the innate immune response (1, 2).
Tetherin, also known as Bst-2 or CD317, is type II transmembrane glycoprotein that accumulates in perinuclear compartments and at the plasma membrane (3). Expression of tetherin is IFN-α-induced in cell lines, such as 293T and HT1080, and results in retention of mature retroviral particles at the surface of infected cells (1, 2, 4). Accordingly, cell lines that express endogenous tetherin exhibit markedly reduced retroviral release compared with tetherin-negative cells (1, 2, 4). Tethered retroviral particles can be liberated from the cell surface by proteolysis, suggesting that the nascent viral envelope is no longer contiguous with the plasma membrane and that protein interactions are required to repress virus release (1, 4, 5). Following cell surface retention, virions are internalized into endosomal compartments, thereby limiting the extent of virus spread (5). Not surprisingly, retroviruses, such as HIV-1, have evolved a strategy, in the form of Vpu, to overcome the restriction imposed by tetherin. Indeed, tetherin only inhibits HIV-1 budding in the absence of Vpu (1, 2).
Ebola Zaire is a highly pathogenic negative-sense RNA virus that causes severe hemorrhagic disease. Ebola belongs to the family Filoviridae, whose members, the Ebola and Marburg viruses, produce filamentous virions. Expression of the Ebola matrix protein, VP40, alone in mammalian cells produces filamentous virus-like particles (VLPs) that bud from the cell surface and structurally resemble infectious Ebola virions (6, 7). The viral glycoprotein (GP) forms the surface spikes seen in the viral envelope membrane. Coexpression of Ebola GP with VP40 produces VLPs that incorporate GP, resemble authentic Ebola by electron microscopy, and can fuse with target cells to deliver the virion contents (7, 8).
Recently, budding of Ebola VLPs was shown to be inhibited in IFN-α-induced human cells and certain human cell lines, such as HeLa, known to express endogenous tetherin (4). These findings suggest that tetherin may block Ebola virus release; however, this link has yet to be substantiated. Additionally, if tetherin is a broad-spectrum antiviral factor that inhibits budding of diverse viral families, such as filoviruses, it is likely that these viruses have evolved strategies to promote budding in the presence of tetherin. Similarly, if this system represents an intrinsic antiviral response, then tetherins from other mammalian cells might be expected to display antiviral activity. In this study, we show that human and murine tetherin retain Ebola particles at the cell surface. Additionally, we demonstrate that the Ebola GP, likely through physical association with tetherin, is sufficient to overcome this intrinsic antiviral response and promote virus release.
Results
Tetherin Restricts Ebola VLP Budding.
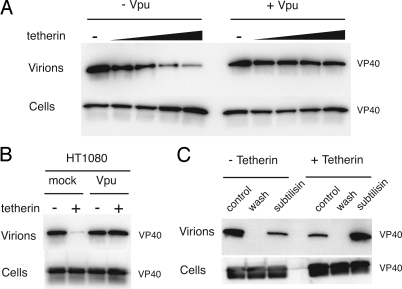
The ability of HIV-1 Vpu expression to promote Ebola VLP release suggests that tetherin may be an intrinsic immunity factor with broad activity against a variety of enveloped viruses (4). To directly address whether tetherin is capable of inhibiting Ebola budding, a transient Ebola VLP release assay was used. 293T cells, which express little endogenous tetherin (1), were cotransfected with plasmids encoding the Ebola matrix protein VP40 and tetherin. Ebola VLP release into the supernatant was significantly inhibited in the presence of tetherin (Fig. 1A). HIV Vpu effectively antagonized the activity of tetherin and fully restored Ebola budding (Fig. 1A). Tetherin-mediated inhibition of Ebola VLP release and subsequent recovery of budding upon Vpu expression also was observed in HT1080 cells (Fig. 1B). To determine whether the effect of tetherin on Ebola budding is due to accumulation of viral particles at the cell surface, as was observed for tetherin restriction of HIV-1 (5), we treated VP40- and tetherin-expressing 293T cells with the protease subtilisin. As shown in Fig. 1C, protease treatment effectively promoted release of tetherin-retained Ebola VLPs into the supernatant. These data suggest that Ebola VLPs retained at the cell surface by tetherin are anchored by proteinaceous interactions in a manner mechanistically similar to that described for HIV-1 particles.
Fig. 1.
Tetherin inhibits budding of Ebola VLPs. (A) Human 293T cells were cotransfected with Ebola FLAG-VP40, with or without HIV vpu, and 0 to 100 ng of tetherin DNA. Cell lysates and supernatants were harvested 48 h after transfection. Virions in clarified supernatants were pelleted through sucrose. FLAG-VP40 in cell lysates and pelleted virion samples was analyzed by Western blot analysis. (B) FLAG-VP40 in cell lysates and pelleted virions was analyzed in HT1080 cells transfected with 25 ng of tetherin with or without vpu. (C) Constitutive virion release was measured from cells transfected in the presence or absence tetherin (control). Supernatants were removed, and cells were incubated with PBS (wash) or subtilisin. In all experiments, the total amount of DNA per transfection was normalized by using empty pCAGGS vector.
Ebola GP Antagonizes Tetherin Activity.
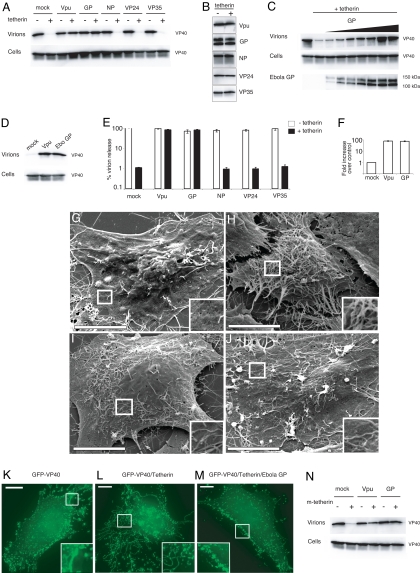
Because tetherin-restricted budding of Ebola VLPs was efficiently rescued by HIV Vpu, we next addressed whether Ebola virus encodes a protein that possesses a Vpu-like function that could overcome tetherin activity. Ebola encodes 7 genes, 4 of which, GP, nucleoprotein (NP), VP24, and VP35, have been shown previously to influence Ebola virus budding (9, 10). Plasmids encoding these 4 Ebola genes were transfected with VP40 into 293T cells in the presence or absence of tetherin to test their effects upon budding. As demonstrated in Fig. 2A, Ebola GP was capable of promoting Ebola VLP budding in tetherin-expressing cells, with the level of rescue similar to that observed when HIV-1 Vpu is used instead of GP. Ebola NP, VP24, and VP35 had no observable effect on VLP release, despite detectable expression of the Ebola proteins in cell lysates (Fig. 2B). Titration of Ebola GP in the presence of tetherin showed a dose-dependent response, with complete restoration of VLP release seen at the higher levels of GP expression (Fig. 2C). Ebola GP was similarly capable of antagonizing tetherin in another human cell line, HT1080, which normally expresses low levels of tetherin [supporting information (SI) Fig. S1]. In addition, HeLa cells, which express endogenous tetherin, exhibited limited VLP release when transfected with VP40 alone (Fig. 2D). Coexpression of either Ebola GP or HIV-1 Vpu in HeLa cells enhanced VLP release (Fig. 2D) and diminished the accumulation of VP40-positive structures at the cell surface (Fig. S2), reflecting the ability of these proteins to interfere with the activity of tetherin.
Fig. 2.
The Ebola glycoprotein is sufficient to overcome tetherin-restricted VLP and HIV budding. (A) 293T cells were transfected with FLAG-VP40, with or without tetherin, and empty vector (mock) or plasmid-encoded viral genes. VP40 in cell lysates and virions was analyzed as in Fig. 1. Viral gene expression in cell lysates was analyzed in B. No bands were detected in Western blot analysis of mock-transfected sample. (C) Cells were transfected with tetherin and increasing amounts of Ebola GP DNA (0 to 700 ng). Expression of GP was determined by Western blot analysis of cell lysates using GP antisera. (D) VLP budding was analyzed in HeLa cells. (E and F) HIV release was quantified by p24 ELISA of virions from 293T (E) or HeLa cells (F). HIV p24 levels in cell lysates were equivalent as determined by Western blot analysis. Error bars represent SEM of samples transfected in triplicate. (G–J) Scanning electron microscopy of HeLa cells transfected with pCAGGS alone (G) or with GFP-VP40 and pCAGGS (H), Ebola GP (I), or Vpu (J). (Scale bar: 10 μM.) (K–M) Immunofluorescence of GFP is shown in HT1080 cells transfected with GFP-VP40 alone (K) or in combination with tetherin (L and M) and Ebola GP (M). (Scale bar: 10.6 μM.) (N) VLP budding was analyzed in 293T cells transfected with VP40, murine tetherin, and the indicated plasmids.
To address whether the effect of GP on tetherin was specific for Ebola VLPs, we analyzed the effect of Ebola GP, NP, VP24, and VP35 on HIV-1 release. Substantial levels of HIV virions were detected in supernatants of 293Ts transfected with a plasmid encoding HIV Gag-Pol. As anticipated, coexpression of tetherin and Gag restricted HIV-1 virion release, showing a 100-fold reduction as quantified by p24 ELISA (Fig. 2E) and undetectable virion-associated Gag (p24) in cell supernatants by Western blot analysis (Fig. S3). As was seen with Ebola VLP budding, HIV-1 virion release from tetherin-expressing 293T cells was completely recovered in the presence of Ebola GP. Similar levels of rescue were seen with HIV-1 Vpu. In contrast, budding was not restored by the Ebola proteins NP, VP24, or VP35 (Fig. 2E). Additionally, HIV virion release in HeLa cells that constitutively express tetherin was enhanced 100-fold upon expression of Ebola GP (Fig. 2F).
Tetherin Retains Ebola Particles on the Cell Surface.
Protease treatment releases Ebola particles in cells expressing tetherin, suggesting that virions are retained at the cell surface. In contrast, the ability of Ebola GP to restore virion budding from tetherin-expressing cells suggests that GP inhibits this accumulation. To address these observations, microscopic analysis of cells producing Ebola VLPs was performed. Scanning electron microscopy of HeLa cells that constitutively express tetherin revealed a dramatic difference between cells expressing VP40 alone or in combination with Ebola GP. VP40 expression resulted in significant accumulation of filamentous virus particles on the cell surface (Fig. 2H), whereas the presence of Ebola GP or Vpu diminished the appearance of surface VLPs (Fig. 2 I and J). Similarly, expression of a GFP-VP40 fusion protein in HT1080 cells that express low levels of tetherin revealed numerous punctate structures that likely represent assembling virions (Fig. 2K). Cells expressing tetherin together with GFP-VP40 displayed numerous fluorescent filamentous structures on the cell surface (Fig. 2L), whereas coexpression of Ebola GP abrogated the effect of tetherin (Fig. 2M). Tetherin partially colocalized with GFP-VP40-positive filamentous and vesicular structures (Fig. S1C). In cells also expressing Ebola GP, there was substantial overlap of all 3 signals near the cell surface (Fig. S1D). These results support the role of Ebola GP in promoting virion release by antagonizing tetherin-mediated retention of virions at the cell surface.
Murine Tetherin Functions as an Antiviral Factor.
The tetherin proteins from rodents are divergent from the primate orthologs such that the mouse and human proteins display only 36% sequence identity. To address whether these distinct proteins have antiviral activity, we expressed the mouse tetherin cDNA in 293T cells. As seen in Fig. 2N, murine tetherin inhibited Ebola VLP release at levels comparable to those seen with human tetherin. Similar to the studies with human tetherin, expression of Ebola GP efficiently restored Ebola VLP budding in the presence of murine tetherin (Fig. 2N). The broad-spectrum activity of murine tetherin was addressed by analyzing HIV-1 virion release. HIV-1 budding was effectively suppressed by murine tetherin (Fig. S4). Interestingly, HIV-1 Vpu appears to be only partially able to restore HIV-1 or Ebola VLP budding in cells expressing murine tetherin (Fig. 2N and Fig. S4). Thus, in contrast to HIV-1 Vpu, the ability of Ebola GP to efficiently restore virion release suggests that GP may have evolved a mechanism to counteract tetherin in a wide variety of mammalian hosts.
Features of Ebola GP Important to Counteract Tetherin.
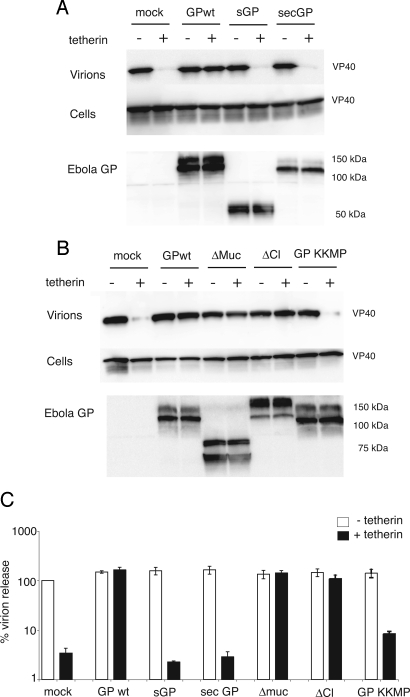
Ebola infection produces several forms of the viral GP, including nonstructural GP (sGP), which shares the N-terminal 295 residues with full-length GP and is the primary product of the GP gene (11, 12). Infection also yields soluble GP, which is shed from the cell surface by cleavage at the extracellular base of GP by the tumor necrosis factor α-converting enzyme (TACE) protease (13). We tested whether sGP and a ΔTM form of GP (secGP), representing the TACE-cleaved protein, could restore Ebola VLP release in the presence of tetherin. Fig. 3A shows that neither sGP nor secGP was able to recover tetherin-restricted Ebola VLP budding, suggesting that full-length GP is the only natural product of the GP gene that can counteract tetherin.
Fig. 3.
Effect of Ebola GP mutants on tethered virions. (A and B) Cell lysates and purified virions from 293T cells transfected with FLAG-VP40, tetherin, and Ebola GP were analyzed by Western blot analysis. Expression of the various GP mutants was examined by Western blot analysis of cell lysates by using sGP antisera. (A) The effect of sGP and secGP on VLP budding was determined. (B) The GP mutants, Δmuc, ΔCl (deletion of the furin cleavage site), and GP-KKMP, were tested. (C) HIV budding in GP- and tetherin-expressing 293Ts was determined by p24 ELISA. Error bars represent SEM of samples transfected in triplicate.
We next sought to determine the contribution of the highly glycosylated mucin domain to the antitetherin activity of Ebola GP. The mucin domain within GP is known to contribute to cellular pathogenesis by affecting surface levels of a variety of cellular proteins (14, 15). It appears that this region of GP, however, is not required to counteract tetherin because deletion of the mucin domain had no effect upon the ability of GP to promote virion release from tetherin-expressing cells (Fig. 3B). Additionally, a GP mutant possessing a deletion of the furin cleavage site, which prevents processing of GP0 into GP1/GP2 monomers (16, 17), also rescued Ebola budding (Fig. 3B). The effect of these mutants also was substantiated by using the HIV release assay (Fig. 3C).
Finally, we sought to determine whether full-length GP with altered cellular localization was capable of antagonizing tetherin. We tested the effect of GP tagged with the endoplasmic reticulum (ER) retention motif, KKMP. This mutant is efficiently retained in the ER, with minimal surface expression as determined by immunofluorescence microscopy and flow cytometry (18). GP-KKMP failed to restore tetherin-restricted VLP budding, despite ample expression of the GP in cell lysates (Fig. 3B). GP-KKMP was also 10-fold less efficient than WT GP at recovering budding of HIV in the presence of tetherin (Fig. 3C). These results indicate that cellular localization of full-length GP is critical in overcoming the inhibitory effects of tetherin.
Ebola GP Interacts with Tetherin.
To begin addressing the mechanism by which Ebola GP antagonizes tetherin, we performed coimmunoprecipitation analysis in 293T cells expressing GP and AU1 epitope-tagged tetherin. Lysates from transfected cells were incubated with anti-AU1 antibodies, precipitated, and washed, and the complexed proteins were analyzed by Western blot analysis. Ebola GP was immunoprecipitated only in the presence of tetherin (Fig. 4A). Only the lower-molecular mass Ebola GP band, an endoglycosidase H-sensitive form of GP, was effectively precipitated by tetherin. Comparison with the input lanes suggests that at least 10% of the Ebola GP is in complexes with tetherin. As a control for nonspecific precipitation, the exogenous membrane protein Tva was coexpressed with tetherin. In this case, no Tva was detected in the tetherin precipitates (data not shown). Performing the analysis by using a polyclonal antibody to Ebola GP for immunoprecipitation showed that tetherin was specifically and effectively precipitated in the presence of GP (Fig. 4B). Together, these results demonstrate a specific interaction between GP and tetherin.
Fig. 4.
Ebola GP interacts with tetherin. (A and B) 293T cells were transfected with the indicated plasmids, and RIPA lysates were harvested at 24 h. Proteins were immunoprecipitated with anti-AU1 (A) or GP antisera (B) and Western blotted as indicated. The input represents one tenth of the immunoprecipitated sample. Tetherin appears as 3 bands representing the addition of 2 N-linked glycans. (C) Constitutively released VLPs from 293T cells were harvested (control) or treated with PBS (wash), subtilisin, or DTT. Pelleted VLPs and cell lysates were analyzed as described previously.
DTT Is Not Sufficient to Release Tethered VLPs.
A current model to describe how tetherin retains virions at the cell surface implicates intermolecular disulfide formation between tetherin monomers in apposing cellular and viral membranes (1, 19). To examine the importance of disulfide bonding in virion tethering, we incubated 293T cells expressing VP40 and tetherin with 50 to 500 mM DTT for 10 min at 37 °C. In other tests, 150 mM DTT was sufficient to reduce cell surface Ebola GP, releasing free GP1 into the supernatant (data not shown). Although proteolysis by subtilisin was capable of liberating tethered virions under these conditions, DTT alone had no detectable effect on VLP (Fig. 4C) or HIV budding (Fig. S5). These data suggest that disulfide bonding alone does not mediate virion tethering. However, a role for disulfide bonding in conjunction with other proteinaceous interactions cannot be ruled out.
Discussion
The ability of Ebola virus to evade the innate immune response, especially IFN induction, is likely a fundamental step in pathogenesis. In fact, Ebola possesses several strategies to antagonize IFN production and signaling. Ebola VP35 prevents IFN production by blocking activation of IFN regulatory factor 3, and VP24 inhibits IFN responsiveness by blocking the nuclear accumulation of tyrosine-phosphorylated STAT1 (20, 21). These proteins likely suppress de novo IFN-induced synthesis of viral restriction factors, but constitutive expression in various cell types remains a restrictive barrier to viral infection. In this study we demonstrate that the Ebola GP possesses the ability to counteract tetherin, a newly identified member of the innate response that is both IFN-induced in many primary cells and constitutively expressed on the primary targets of Ebola virus infection, particularly monocytes and dendritic cells (22).
The mechanism by which GP counteracts tetherin to restore virus budding remains unclear. We have shown that GP colocalizes and coimmunoprecipitates with tetherin in transfected cells. Yet, despite this interaction, steady-state levels of tetherin appear to be unchanged (Fig. 4B). This finding suggests that Ebola GP does not divert tetherin toward a degradative pathway. One explanation for the antagonistic effect of Ebola GP is that the GP interaction physically disrupts the ability of tetherin to retain virions. Alternatively, the GP–tetherin interaction may alter the localization of tetherin intracellularly or at the cell surface, which would be consistent with observations that Vpu downmodulates cell surface levels of tetherin (2). Coimmunoprecipitation of tetherin exclusively with a lower-molecular mass, putative immaturely glycosylated form of GP suggests that the interaction occurs during GP maturation. Interestingly, only the higher-molecular mass Ebola GP species, which does not coprecipitate with tetherin, is incorporated into virions (data not shown). It remains possible that other unidentified cellular factors are involved in either the observed GP–tetherin interaction or the ability of Ebola GP to promote virus budding.
Murine tetherin shares ≈36% protein sequence homology with the human ortholog; however, several structural features are conserved. Both proteins possess an N-terminal transmembrane domain, a C-terminal GPI anchor, and 3 cysteines in the extracellular loop. Despite this limited sequence conservation, expression of murine tetherin efficiently restricted Ebola and HIV-1 budding (Fig. 2K and Fig. S4). Because mice are not natural targets for infection by either of these viruses, the function of the murine ortholog suggests that tetherin is a nonspecific innate antiviral inhibitor of enveloped virus budding. We also found that Ebola GP effectively blocks the antiviral function of both murine and human tetherin, suggesting that sequences or features within these 2 proteins required for recognition by GP are conserved. Moreover, the ability of GP to affect murine and human tetherin likely reflects an adaptation of Ebola virus to efficiently infect a broad spectrum of mammalian hosts that express restrictive yet diverse tetherin proteins. An analysis of Ebola GP's effect upon tetherin proteins from various species, including bats that appear to be the host reservoir for Ebola, should directly address this question.
The Ebola glycoprotein is a class I viral fusion protein with an overall architecture similar to that of other class I proteins, including HIV Env and influenza HA. Comparison of the glycoprotein sequences for the 4 subtypes of Ebola indicates that the amino-terminal ≈200 residues within GP1 and all of GP2 are highly conserved. An extremely divergent mucin-like region is localized to the C-terminal region of GP1. Several groups have previously shown that the GP mucin domain causes downmodulation of cell surface proteins (14, 15). Therefore, we initially hypothesized that the mucin domain within Ebola GP would prove critical in interfering with tetherin activity. However, we found that the mucin domain was dispensable for the antitetherin activity of Ebola GP. Studies are underway to determine how other GP domains, such as the receptor-binding domain and the GP2 fusion machinery, contribute to promote VLP budding in the presence of tetherin. A recently published structure of the trimeric Ebola glycoprotein (23) should aid in elucidating the molecular requirements within Ebola GP for interaction with tetherin.
Ebola is likely not the only enveloped virus that has evolved a mechanism to evade tetherin in the absence of Vpu. Indeed, most strains of HIV-2, with the exception of some isolates, such as SIVcpz, lack a Vpu-like protein (24). Instead, the HIV-2 Env protein appears to enhance retroviral budding in HeLa and IFN-induced 293T cells (25, 26), suggesting that HIV-2 Env antagonizes tetherin. Studies have linked the activity of HIV-2 Env to a GYxxΘ motif in the cytoplasmic tail and to unmapped regions in the ectodomain of the protein (27). Unlike HIV-2 Env, Ebola GP possesses a cytoplasmic tail consisting of only a few amino acids, and no GYxxΘ motif is present in the membrane proximal region of GP2 or throughout the protein. Despite the absence of the GYxxΘ motif, our data demonstrate that a membrane anchor is vital for GP activity because secGP, lacking only the transmembrane domain and cytoplasmic tail, could not promote VLP budding in the presence of tetherin. This could be due to loss of a domain that specifically modulates tetherin activity. Alternatively, loss of secGP activity could be due to altered GP localization or changes in local GP concentrations that are seen with secreted proteins. It is clear that GP localization does appear important, because ER-retained GP was ineffective at restoring virus budding. Additional studies will be needed to address how GP structure and localization facilitate budding of virions in tetherin-expressing cells.
Materials and Methods
Cell Lines and Plasmids.
The 293T, HT1080, and HeLa cells were maintained in DMEM supplemented with 10% FBS using standard procedures. N-terminally FLAG-tagged VP40 was engineered into pCAGGS. Vpu was derived from the HIV-1 molecular clone NL4-3 and inserted into pCAGGS. The cDNAs encoding human and mouse tetherin were obtained from Open Biosystems. An AU1 tag was added to the N terminus of human tetherin. We obtained plasmids encoding Ebola NP, VP24, and VP35 from R. Harty (University of Pennsylvania, Philadelphia, PA). pCAGGS-NP was C-terminally appended with a FLAG epitope tag. The HIVgag-pol-encoding plasmid, psPAX (Addgene), and the Ebola GP variants sGP, secGP, Δmuc, ΔCl, and GP-KKMP have been described elsewhere (12–14, 16, 17).
VLP Release Assay.
Cells (1.5 × 105 per well) were plated in 24-well plates before transfection with pCAGGS-FLAG-VP40 and plasmids encoding tetherin, HIV Vpu, or various Ebola genes using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. For HIV release assays, psPAX was transfected in place of FLAG-VP40. Cell lysates and supernatants were harvested 48 h after transfection. Supernatants were clarified by low-speed centrifugation, and virions were subsequently pelleted through a 20% sucrose cushion at 40,000 × g for 20 min. Virions were resuspended in PBS for 1 h at 4 °C, and viral protein content was assessed by immunoblot or HIV p24 ELISA. Resuspended virions and cell lysates were analyzed by SDS/PAGE, and immunoblots were probed for Ebola VP40 by using anti-FLAG HRP (Sigma). HIV protein expression was detected by using the monoclonal anti-p24 antibody, no. 24-3, or anti-Vpu, no. 969 (National Institutes of Health AIDS Research and Reference Reagent Program). Expression of Ebola proteins in cell lysates was confirmed by using rabbit antisera raised against Ebola sGP (for detection of all Ebola GP constructs) or anti-FLAG-HRP (for Ebola NP detection; Sigma). Mouse anti-VP24 and rabbit anti-VP35 were gifts from R. Harty. HIV p24 titers were determined by using an HIV p24 ELISA kit (Cell BioLabs) according to the manufacturer's protocol.
Protease Stripping and DTT Treatment of Virions.
Constitutively released virions were harvested from the culture supernatant as described above 48 h after transfection of 293T cells with FLAG-VP40 and tetherin plasmids. For the remaining samples, the media was replaced with 50, 100, or 500 mM DTT, 1 mg/mL subtilisin, or PBS alone for 10 min at 37 °C. Reactions were quenched with 5 mM PMSF. Cell lysates were harvested, and virions in supernatants were pelleted through sucrose as described previously.
Coimmunoprecipitation.
Transfected 293T cells were lysed in RIPA buffer [50 mM Tris·HCl, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, protease inhibitors (Roche), pH 7.4] and sonicated. Lysates were incubated with polyclonal goat anti-AU1 (Bethyl Laboratories) or polyclonal anti-sGP antibodies for 1 h at 4 C. Sepharose Protein-G beads (Invitrogen) were added, and samples were incubated for an additional 4 h at 4 °C with shaking. Samples were washed 4 times in RIPA buffer and were resuspended in 2× SDS/PAGE sample buffer. Proteins were analyzed by immunoblotting with either anti-AU1 or anti-sGP antibodies.
Scanning Electron Microscopy.
Scanning electron microscopy experiments were carried out at the CDB/CVI Microscopy Core (School of Medicine, University of Pennsylvania, Philadelphia, PA). Samples were prepared for scanning electron microscopy as described previously (28). Samples were mounted on stubs and sputter coated with gold palladium. Specimens were observed and photographed by using a Philips XL20 scanning electron microscope (FEI) at 10-kV accelerating voltage. For details about immunofluorescence microscopy, see SI Methods.
Supplementary Material
Acknowledgments.
We thank Yuri Veklich in the Microscopy Core Facility of the Department of Cell and Developmental Biology and Penn Cardiovascular Institute for assistance with scanning electron microscopy. We also thank Bob Doms for manuscript comments and Yoshihiro Kawaoka (University of Wisconsin, Madison, WI) for providing anti-GP monoclonal antibodies. This work was funded by Public Health Service Grants T32-GM07229 (to J.R.F.), T32-AI07632 (to C.A.-G.), T32-AI55400 (to R.L.K. and J.R.F.), and U54-AI57168 (to P.B.) and funds from the Philip Morris External Research Foundation (to P.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811014106/DCSupplemental.
References
- 1.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 2.Van Damme N, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kupzig S, et al. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 4.Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neil SJ, Eastman SW, Jouvenet N, Bieniasz PD. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2:e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: Implications for filovirus budding. Proc Natl Acad Sci USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noda T, et al. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol. 2002;76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons G, et al. Folate receptor alpha and caveolae are not required for Ebola virus glycoprotein-mediated viral infection. J Virol. 2003;77:13433–13438. doi: 10.1128/JVI.77.24.13433-13438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson RF, Bell P, Harty RN. Effect of Ebola virus proteins GP, NP and VP35 on VP40 VLP morphology. Virol J. 2006;3:31. doi: 10.1186/1743-422X-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Licata JM, Johnson RF, Han Z, Harty RN. Contribution of ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J Virol. 2004;78:7344–7351. doi: 10.1128/JVI.78.14.7344-7351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez A, et al. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J Virol. 1998;72:6442–6447. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volchkova VA, Feldmann H, Klenk HD, Volchkov VE. The nonstructural small glycoprotein sGP of Ebola virus is secreted as an antiparallel-orientated homodimer. Virology. 1998;250:408–414. doi: 10.1006/viro.1998.9389. [DOI] [PubMed] [Google Scholar]
- 13.Dolnik O, et al. Ectodomain shedding of the glycoprotein GP of Ebola virus. EMBO J. 2004;23:2175–2184. doi: 10.1038/sj.emboj.7600219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmons G, Wool-Lewis RJ, Baribaud F, Netter RC, Bates P. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J Virol. 2002;76:2518–2528. doi: 10.1128/jvi.76.5.2518-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang ZY, et al. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat Med. 2000;6:886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]
- 16.Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wool-Lewis RJ, Bates P. Endoproteolytic processing of the ebola virus envelope glycoprotein: Cleavage is not required for function. J Virol. 1999;73:1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francica JR, Matukonis MK, Bates P. Requirements for cell rounding and surface protein down-regulation by Ebola virus glycoprotein. Virology. 2008;383:237–247. doi: 10.1016/j.virol.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlinger HG. HIV/AIDS: Virus kept on a leash. Nature. 2008;451:406–408. doi: 10.1038/nature06364. [DOI] [PubMed] [Google Scholar]
- 20.Cardenas WB, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reid SP, et al. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vidal-Laliena M, et al. Characterization of antibodies submitted to the B cell section of the 8th Human Leukocyte Differentiation Antigens Workshop by flow cytometry and immunohistochemistry. Cell Immunol. 2005;236:6–16. doi: 10.1016/j.cellimm.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iida S, et al. Compatibility of Vpu-like activity in the four groups of primate immunodeficiency viruses. Virus Genes. 1999;18:183–187. doi: 10.1023/a:1008041323852. [DOI] [PubMed] [Google Scholar]
- 25.Bour S, Schubert U, Peden K, Strebel K. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: A Vpu-like factor? J Virol. 1996;70:820–829. doi: 10.1128/jvi.70.2.820-829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritter GD, Jr, Yamshchikov G, Cohen SJ, Mulligan MJ. Human immunodeficiency virus type 2 glycoprotein enhancement of particle budding: Role of the cytoplasmic domain. J Virol. 1996;70:2669–2673. doi: 10.1128/jvi.70.4.2669-2673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abada P, Noble B, Cannon PM. Functional domains within the human immunodeficiency virus type 2 envelope protein required to enhance virus production. J Virol. 2005;79:3627–3638. doi: 10.1128/JVI.79.6.3627-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braet F, De Zanger R, Wisse E. Drying cells for SEM, AFM and TEM by hexamethyldisilazane: A study on hepatic endothelial cells. J Microsc. 1997;186:84–87. doi: 10.1046/j.1365-2818.1997.1940755.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.