Abstract
We report the cloning and characterization of TOX3, a high mobility group box protein involved in mediating calcium-dependent transcription. TOX3 was identified as a calcium-dependent transactivator using the Transactivator Trap screen. We find that TOX3 interacts with both cAMP response element (CRE)-binding protein (CREB) and CREB-binding protein (CBP), and knockdown of the endogenous TOX3 by RNAi leads to significant reduction of calcium-induced c-fos expression and complete inhibition of calcium activation of the c-fos promoter. The effects of TOX3 on calcium-dependent transcription require the CRE elements. These observations identify TOX3 as an important regulator of calcium-dependent transcription and suggest that TOX3 exerts its effect on CRE-mediated transcription via its association with the CREB–CBP complex.
Keywords: CBP, CREB, c-fos, activity
Neuronal activity plays an important role in the development of the nervous system and regulates cell survival, axonal and dendritic growth, and synaptic plasticity. Many of these effects are mediated by calcium-dependent transcription (1). To identify factors that mediate calcium-dependent transcription in neurons, our laboratory developed a screening strategy called Transactivator Trap (2). We have previously described the cloning of CREST (2), NeuroD2 (3), and LMO4 (4) as calcium-dependent transcription using the Transactivator Trap strategy. Here we report the cloning and characterization of TOX3, a cAMP response element (CRE)-binding protein (CREB) and CREB-binding protein (CBP) interacting protein that plays a critical role in regulating calcium-dependent transcription.
TOX3 is a high mobility group (HMG) box protein related to TOX, a protein that has been implicated in the regulation of thymocyte selection (5). HMG box proteins bind to the minor groove of DNA and are nonchromosomal nuclear proteins that help to remodel the nucleosome (6). Our investigation of TOX3 was driven by its calcium-activation properties and its association with CREB. CREB has long been known to be the major mediator of stimulus-induced transcription activation in neurons. CREB was first discovered as a protein that binds to the cAMP-responsive element and mediates cAMP-dependent transcription (7, 8). CREB-dependent transcription was subsequently found to be inducible by calcium and growth factors (9, 10). Calcium regulation of CREB-mediated transcription requires phosphorylation of CREB at Ser-133. Phosphorylation of Ser-133 allows CREB to bind to CREB-binding protein (CBP) (11). CBP itself can be phosphorylated by CaMKIV (12–16), and it is a critical mediator of CREB-dependent transcription. We find that TOX3 interacts with both CREB and CBP and plays a critical role in mediating calcium-dependent transcription in neurons.
Results
Identification of TOX3 as a Calcium-Dependent Transactivator.
We used the Transactivator Trap screen (2) to search for novel mediators of calcium-dependent transcription. Briefly, the screen uses the modular nature of transcription factors that enables the transactivation domain to function without the DNA-binding domain. In addition, the screen takes advantage of the fact that a transcription factor fused to the yeast Gal4DBD (DNA-binding domain) can recognize the upstream activating sequence (UAS) and drive expression of a reporter. In our experiments, a postnatal day 1 (P1) rat cortical cDNA library was fused to Gal4DBD and broken down into 200 pools. Each pool was transfected along with a UAS-CAT (chloramphenicol acetyltransferase) reporter into embryonic day 18 (E18) cortical cultures. Pools containing a putative calcium-regulated transactivator were identified by comparing the number of CAT-positive neurons in unstimulated and KCl-stimulated wells (Fig. 1 A and B). The cDNA that conferred KCl-dependent transactivation in 1 of the pools was found to encode an HMG box-containing protein. A partial sequence of the gene has previously been posted with the National Center for Biotechnology Information (NCBI) as TNRC9 and CAGF9 (17) and renamed TOX3 on the basis of its homology to the HMG box protein, TOX (NCBI Reference Sequence). We isolated and sequenced a full-length clone of TOX3 from a P1 rat cortical cDNA library. The coding region of TOX3 is 1,734 nucleotides long, which translates into 578 aa residues (supporting information Fig. S1). The sequence has been deposited to GenBank (accession no. EU194254).
Fig. 1.
Characterization of TOX3-mediated transcription. (A and B) Immunocytochemistry of rat E18 neurons transfected with the pool containing Gal4.TOX3. The images show CAT-immunoreactivity in unstimulated (A) and 50 mM KCl-stimulated (B) cells transfected with the Gal4.TOX3-containing pool together with UAS-CAT. The arrows point to the CAT-positive cells. (C) Relative CAT activity in rat E18 cortical neuronal cultures transfected with UAS-CAT and either Gal4.TOX3 or vector on DIV3 and assayed for CAT activity on DIV 5. Cultures were treated either with 50 mM KCl or PBS on DIV4 for 16 h. *, P < 0.05. (D) Diagrammatic representation of the various deletion constructs of TOX3. (E) Relative CAT activity in rat E18 cortical neuronal cultures transfected with full-length Gal4.TOX3 (FL), Gal4.TOX3 base pairs 1–870, Gal4.TOX3 base pairs 1–711, or Gal4.TOX3 base pairs 712-1734 and UAS-CAT, stimulated as indicated and assayed for CAT activity on DIV5. (F) Expression of the deletion constructs shown by Western blot. Equivalent amounts of whole-cell lysates of HEK293 cells transfected with full-length Gal4.TOX3 (FL) or Gal4.TOX3 base pairs 1–870, 1–711, or 712-1734 were probed with anti-Gal4 antibody. (G) Northern blot of RNA isolated from rat cortices and cerebella at different developmental ages and hybridized with a probe to TOX3. The same blots were stripped and reprobed with GAPDH. Error bars represent +SEM.
Structurally, TOX3 contains an HMG box near the middle of the protein and has a glutamine-rich domain near the C terminus (Fig. 1D). Sequence comparison shows that rat TOX3 is closely related to mouse and human TOX3, although homologues in zebrafish and yeast can also be identified (Fig. S1 B and C). The sequence similarity of TOX3 with other TOX family HMG box proteins is shown in Fig. S1D. The HMG box of TOX family proteins is highly conserved and is closely related to the HMG box of the mouse HMGB1 gene (Fig. S1D).
To characterize calcium activation of TOX3-mediated transcription, we transfected E18 cortical cultures with Gal4.TOX3 and UAS-CAT and examined depolarization-induced activation of the UAS-CAT reporter. Membrane depolarization with elevated extracellular KCl leads to an influx of calcium via voltage-gated calcium channels and has been shown to induce calcium-dependent transcription (10). Stimulation with 50 mM KCl to depolarize the neurons did not lead to reporter activity in the absence of Gal4.TOX3 (Fig. 1C). In contrast, transfection of Gal4.TOX3 led to a significant increase in reporter activity, which was further increased by KCl stimulation (Fig. 1C). KCl stimulation did not lead to an increase in Gal4.TOX3 levels (Fig. S2D). In addition, stimulation with the neurotransmitter glutamate, or increasing activity in culture by blocking GABAergic signaling with bicuculline, led to an increase in Gal4.TOX3-mediated transcription (Fig. S2 B and C). These observations indicate that membrane depolarization regulates TOX3-mediated transcription.
To identify the domains of TOX3 that mediate activity-dependent transcription, we generated the deletion constructs shown in Fig. 1D. Gal4 fusions of these constructs were cotransfected into E18 cortical neurons with UAS-CAT, and transactivation of the reporter after KCl stimulation was measured using CAT assays (Fig. 1E). We found that the constructs lacking the C terminus retained their basal transactivation activity but lost their calcium responsiveness. In contrast, constructs lacking the N terminus were transcriptionally inactive (Fig. 1E). Western blot analysis confirmed that each of the constructs was expressed (Fig. 1F). These observations suggest that multiple domains of TOX3 contribute to transcriptional activation. The N terminus is absolutely necessary for transcription; the C terminus is not required for basal transactivation but is required for calcium responsiveness.
We carried out Northern blot analysis to determine the expression pattern of TOX3 in the developing brain. TOX3 is expressed in the cortex and the cerebellum as 2 separate isoforms, 5 kb and 3 kb (Fig. 1G). In the cortex, the 5-kb isoform was expressed embryonically, and its expression remained stable through adulthood. However, expression of the 3-kb isoform was tightly regulated during development. The 3-kb isoform was already expressed at high levels at E18 and declined to baseline levels between P7 and P14. In the cerebellum, the 5-kb isoform was first detected at P0 and remained at high levels thereafter. The expression of the 3-kb isoform in the cerebellum peaked between P7 and P14. The developmental regulation of TOX3 isoforms suggests that the different isoforms might serve distinct functions in the developing and adult brain. It is not known whether both of the isoforms encode the same protein or represent splice variants.
Contribution of TOX3 to Depolarization-Induced c-Fos Expression.
Among the principal targets of calcium signaling in neurons are the immediate early genes (1). The prototypical immediate early gene is c-fos, which is rapidly induced upon calcium influx (10). To determine whether TOX3 contributes to calcium activation of the c-fos promoter, we examined the effect of inhibiting TOX3 on the expression of c-fos. We designed 2 RNAi constructs targeting different regions of TOX3 (see Materials and Methods). The TOX3 RNAi constructs were very effective in knocking down expression of cotransfected TOX3 (Fig. 2A). To examine the contribution of TOX3 to depolarization-induced expression of c-fos, we knocked down TOX3 by cotransfecting the TOX3 RNAi constructs along with a pBos.eGFP plasmid into rat E18 neuronal culture on day in vitro 3 (DIV3). When the neurons were depolarized with 50 mM KCl for 2 h on DIV4, the enhancement of the endogenous c-fos expression was clearly detected by immunofluorescence (Fig. 2B). However, KCl-induced c-fos expression was greatly reduced in the TOX3 knockdown cells (Fig. 2C). The effect of RNAi-mediated knockdown of c-Fos expression could be reversed by cotransfecting an RNAi-insensitive TOX3 construct carrying same-sense mutation in TOX3. These observations indicate that endogenous TOX3 contributes significantly to depolarization-induced expression of c-fos in neurons.
Fig. 2.
Calcium-dependent c-Fos expression is regulated by TOX3. Western blot of whole-cell lysates of HEK293 cells transfected with either Myc.TOX3 and scrambled RNAi (Scr.RNAi), TOX3 RNAi#1, or RNAi#2, or Myc.TOX3R and scrambled RNAi or RNAi#1. The blot was probed with anti-Myc and anti-α-tubulin antibodies. (B) E18 neuronal cultures were cotransfected with pBos.eGFP and scrambled RNAi (scrRNAi) or TOX3 RNAi#1 on DIV3. Cells were fixed on DIV4 after stimulation with KCl for 2 h and stained with c-Fos antibody. Arrows indicate TOX3 knockdown cells. (Scale bar, 50 μm.) (C) Quantification of the c-Fos staining of E18 neuronal cultures cotransfected with either pBos.eGFP and scrambled RNAi (scrRNAi), RNAi#1, or RNAi#2, or Myc.TOX3R and RNAi#1. *, P < 0.05.
Association of TOX3 with CREB and CBP.
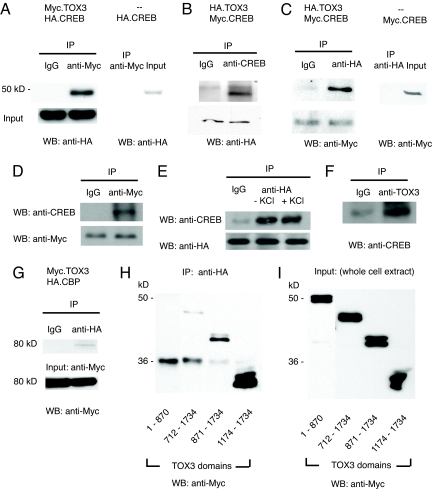
Previous studies have identified the CREB–CBP complex as a key regulator of calcium-dependent c-fos expression. We therefore decided to explore the possibility that TOX3 might influence calcium-dependent transcription by exerting an effect on CREB and/or CBP. To determine whether TOX3 and CREB were in a complex together, we cotransfected Myc.TOX3 and HA.CREB into HEK293T cells and evaluated whether the 2 proteins could be coimmunoprecipitated. Immunoprecipitation with anti-Myc antibodies followed by an anti-HA Western blot revealed that the 2 proteins are indeed in the same complex (Fig. 3A). The association of CREB with TOX3 was confirmed in experiments in which the tags in the transfected proteins were reversed (Fig. 3C). In these experiments, we could immunoprecipitate Myc.CREB and pull down HA.TOX3, or immunoprecipitate HA.TOX3 and pull down Myc.CREB (Fig. 3 B and C). To further examine the interaction of CREB and TOX3, we transfected neurons with Myc.TOX3 and could pull down endogenous CREB with anti-Myc immunoprecipitation (Fig. 3D). The association of transfected TOX3 and endogenous CREB in neuron was not affected by KCl stimulation (Fig. 3E). Immunoprecipitation of endogenous TOX3 in neurons also coprecipitated endogenous CREB (Fig. 3F). These experiments indicate that TOX3 interacts with CREB, and the interaction is not depolarization dependent.
Fig. 3.
TOX3 associates with CREB and CBP. HEK 293T cells were transfected with (A) Myc.TOX3 and HA.CREB or HA.CREB alone, or (B and C) HA.TOX3 and Myc.CREB or Myc.CREB alone. (A) Immunoprecipitation (IP) with anti-Myc antibody, Western blot (WB) with anti-HA antibody. (B) IP with anti-CREB antibody, WB with anti-HA antibody. (C) IP with anti-HA antibody, WB with anti-Myc antibody. (D) Cortical neurons were transfected with Myc.TOX3. IP performed with Myc antibody, WB with anti-CREB and anti-Myc antibodies. (E) Cortical neurons transfected with HA.TOX3 and treated with 50 mM KCl for 10 min before IP. IP with anti-HA antibody, WB with anti-CREB and anti-HA antibodies. (F) IP of the E18 cortical lysate with TOX3 antiserum and WB with anti-CREB antibody. (G) HEK 293T cells were cotransfected with Myc.TOX3 and HA.CBP. IP with anti-HA antibody and WB with anti-Myc antibody. (H and I) HEK 293T cells were cotransfected with Myc.TOX3 base pairs 1–870, 712-1734, 871-1734, or 1174–1734 and full-length CBP. (H) IP with anti-HA antibody and WB with anti-Myc antibody. (I) Western blot of the input with anti-Myc antibody.
We next investigated whether TOX3 could also associate with CBP by cotransfecting HEK293T cells with HA-tagged CBP and Myc-tagged TOX3. As shown in Fig. 3G, HA.CBP could be coprecipitated with Myc.TOX3, indicating that TOX3 and CBP are likely to be in the same complex also the degree of coprecipitation was generally less than TOX3 and CREB. To map the domain of TOX3 that mediates association with CBP, we transfected HEK293T cells with HA.CBP along with Myc-tagged deletion constructs of TOX3. These experiments showed that the N terminus (1–870) domain alone did not interact with CBP but that the C-terminal domain (1174–1734) could be coprecipitated with CBP. Thus, TOX3 seems to interact with CBP through its C terminus (Fig. 3 H and I).
Effect of TOX3 on CREB- and CRE-Mediated Transcription.
To determine whether TOX3 contributed to depolarization-induced activation of CREB- and CBP-mediated transcription, we transfected neurons with Gal4-CREB and UAS-CAT, along with a scrambled RNAi or a TOX3 RNAi. As shown in Fig. 4 A and B, KCl-induced transcription mediated both by Gal4-CREB and Gal4-CBP was attenuated by the TOX3 RNAi. Gal4-CREST-mediated transcription was not affected by TOX3 RNAi, indicating that TOX3 is not involved in all calcium-dependent transcription (Fig. 4C). The association of TOX3 with CREB and CBP suggested that TOX3 might influence transcription via CRE, because transcription via this element is regulated by the CREB–CBP complex. To explore this possibility, we examined the effect of TOX3 on the expression of a c-fos-CAT reporter. As shown in Fig. 4D, stimulation with 50 mM KCl leads to a significant increase in the expression of the c-fos-CAT reporter. Coexpression of TOX3 increased the c-fos-CAT reporter activity both at baseline and after stimulation (Fig. 4D).
Fig. 4.
Effect of TOX3 on CRE-mediated transcription. (A) Relative CAT activity in rat E18 cortical neuronal cultures transfected with Gal4.CREB, UAS.CAT, and scrambled RNAi (scrRNAi) or TOX3 RNAi#1 on DIV3 and assayed for CAT activity on DIV5. Cultures were treated either with 50 mM KCl or PBS on DIV4 for 12 h. (B) Relative CAT activity in rat E18 cortical neuronal cultures transfected with Gal4.CBP, UAS.CAT, and scrambled RNAi (scrRNAi) or TOX3 RNAi#1 on DIV3 and assayed for CAT activity on DIV 5. Cultures were treated with either 50 mM KCl or PBS on DIV4 for 12 h. (C) Relative CAT activity in rat E18 cortical neuronal cultures transfected with Gal4.CREST, UAS.CAT, and scrambled RNAi (scrRNAi) or TOX3 RNAi#1 on DIV3 and assayed for CAT activity on DIV5. Cultures were treated with either 50 mM KCl or PBS on DIV4 for 12 h. (D) Relative CAT activity in rat E18 cortical neuronal cultures transfected with c-fos-CAT and either vector (pBos) or Myc.TOX3 on DIV3 and assayed for CAT activity on DIV5. Cultures were treated with either 50 mM KCl or PBS on DIV4 for 12 h. (E) Relative CAT activity in rat E18 cortical neuronal cultures transfected with m2G/foscat and vector (pBos) or Myc.TOX3 on DIV3 and assayed for CAT activity on DIV5. Cultures were treated with either 50 mM KCl or PBS on DIV4 for 12 h. (F) Relative CAT activity in rat E18 cortical neuronal cultures transfected with c-fos-CAT and either scrambled RNAi (RNAi.scr), TOX3 RNAi#1, RNAi#2, or Myc.TOX3R and RNAi#1 on DIV3 and assayed for CAT activity on DIV5. Cultures were treated with either 50 mM KCl or PBS on DIV4 for 12 h. *, P < 0.05.
To determine whether TOX3 acts at the CRE sites, we used the m2G/fosCAT reporter construct, which consisted of the c-fos promoter but with all 3 CRE sites mutated (18). As shown in Fig. 4E, this promoter construct is weakly calcium responsive. We found that Myc.TOX3 expression did not increase expression of the m2G/fosCAT reporter (Fig. 4E), suggesting that TOX3 exerts its effects via the CRE sites.
Finally, to determine whether endogenous TOX3 contributes to calcium activation of the c-fos promoter, we examined the effect of 2 different TOX3 RNAi constructs on KCl activation of c-fos-CAT. As shown in Fig. 4F, the induction of c-fos-CAT expression was completely blocked by both TOX3 RNAi constructs, indicating that TOX3 is required for depolarization-induced activation of the c-fos promoter. These observations suggest that TOX3 is a key regulator of CRE-mediated transcription and likely exerts its effect via interaction with the CREB–CBP complex.
Discussion
Our experiments identify TOX3 as a novel regulator of calcium-dependent transcription. The importance of TOX3 in calcium-dependent transcription is underscored by the observation that knockdown of TOX3 attenuates endogenous c-fos expression and completely blocks KCl-induced activation of the c-fos promoter. It has long been known that the CREB–CBP complex mediated transcription via CRE sites on calcium responsive promoters. We find that TOX3 interacts with this complex, suggesting that this interaction is important for the effects of TOX3 on mediating calcium-dependent gene expression. Calcium signaling regulates expression of a number of different genes, and calcium-dependent transcription has been implicated in a wide range of neuronal adaptive responses, such as activity-dependent dendritic development, long-term memory, and addiction [reviewed in Qiu and Ghosh (19)]. It will be interesting to determine whether these biologic responses depend on TOX3 function.
The effect of TOX3 on transcription depends on a functional interaction with CREB. This is supported by the observation that the ability of TOX3 to confer calcium responsiveness to the c-fos-CAT reporter is abolished when the CRE sites are removed. The fact that TOX3 associates with CBP via its C terminus, which is also required for calcium-dependent activation of Gal4-TOX3, suggests that the interaction with CBP is important for calcium-dependent activation of TOX3-mediated transcription. We should emphasize that although our experiments support the association of TOX3 with both CREB and CBP, we do not yet know whether all 3 proteins are in fact present in 1 complex and whether the interaction of TOX3 with CREB and CBP are direct. A definitive resolution of this will require purification of the endogenous complexes, as well as binding assay with purified proteins.
TOX3 is an HMG box protein, which are nonhistone chromosomal proteins that have been demonstrated to bend DNA by interacting with the minor grooves of the DNA helix (6). For example, LEF, an HMG box transcription factor involved in mediating the effects of Wnt signaling, is thought to activate transcription via its DNA-bending activity (20). The putative DNA-bending activity of TOX3 might facilitate CRE-mediated transcription by bringing the CREB–CBP complex close to the transcription start site. The DNA-bending activity of HMG box proteins, together with the interaction with sequence-specific transcription factors as demonstrated for TOX3, might allow HMG box proteins to mediate stimulus-dependent transcription via specific regulatory elements.
Materials and Methods
Cell Culture and Transfection.
Timed-pregnant Long Evans rats were purchased from Charles River. Animals were housed and handled according to animal protocols approved by the University of California, San Diego Animal Care Program. Cortical neurons from E18 rat embryos were cultured as previously described (21). Briefly, E18 rat cortical neurons were plated at 1 × 106 cells/mL on coverslips coated with poly-D-lysine and laminin. On DIV3, neurons were transfected with DNA using lipofectamine 2000 (Invitrogen). HEK293T cells were transfected with Fugene6 (Invitrogen) according to the manufacturer's instructions.
Constructs.
A partial sequence of TOX3 was initially cloned out from the P1 rat cDNA library through the Transactivator Trap screen. Subsequently, primers were designed to clone the full length of TOX3 from E18 rat brain cDNA by PCR and cloned into the polylinker of the pBos vector (22). The PCR primers were 20 bases long, each homologous to the 5′ end beginning at the start codon and to the 3′ end with the stop codon of the NCBI Reference Sequence predicted sequence. The RNA interference construct was made using NCBI BLAST by first selecting the antisense sequence consisting of 21 nucleotides in the coding region of TOX3, according to the selection criteria described by Ui-Tei et al. (23). Oligonucleotides containing the sense and antisense sequence with a loop in between were synthesized and annealed for cloning into pSilencer1 (Ambion). The scrambled RNAi sequence was: sense, GACGATGTTCCAGTGCTGA; anti-sense, TCAGCACTGGAACATCGTC. The TOX3 RNAi#1 sequence was: sense, GTGTTACCGCAGGTCAGTATT; anti-sense, AATACTGACCTGCGGTAACAC. The TOX3 RNAi#2 sequence was: sense, CACGTCACTGTCTCAACAT; anti-sense, ATGTTGAGACAGTGACGTG. PfuUltra (Stratagene) was used to clone out the TOX3 fragments into pBos. The rescue construct, pBos.myc.TOX3R, for RNAi#1, was constructed by using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene), for which 4 nucleotides were changed, without changing the amino acids. The following constructs have been described previously: HA-CBP (11), c-fos-CAT (18), and m2G/foscat (18).
CAT Reporter Assay.
Plasmids were transfected into E18 rat neuronal culture on DIV3 by lipofectamine 2000 (Invitrogen). Cells were treated with 50 mM KCl or PBS for 12 h on DIV4 to observe transactivation induced by the influx of calcium. Cells were collected at DIV5 with Tris-NaCl-EDTA (TNE) buffer, followed by lysis via repetitive freeze–thaw cycles. Lysates were collected and incubated with 14C-chloramphenicol for detection of the CAT activity. The mixtures were then loaded on TLC plates and allowed to migrate in a glass chamber equilibrated with 5% methanol and 95% chloroform. The TLC plate was exposed to a phosphorimager cassette overnight. The signals were detected by PhosphoImager (Amersham-Pharmacia) and ImageQuant (Molecular Dynamics).
Immunoprecipitation.
HEK 293T cells or rat E18 cortical neurons were transfected with different constructs by lipofectamine at 50% confluency. Cells were collected by nonionic detergent lysis buffer or modified RIPA buffer and subsequently incubated overnight with agarose A/G beads (Santa Cruz Biotechnology) that had already been preincubated with antibodies. The beads were washed and resuspended in 1× SDS buffer and loaded on a 10% SDS gel after boiling. The protein was then transferred to nitrocellulose paper (Invitrogen) for Western blot. The blot was detected by horseradish peroxidase at 1:5,000 dilution in blocking buffer.
Generation of TOX3 Antiserum.
Full-length rat TOX3 cDNA was amplified and cloned into pQE70 (Novagen) for expression of His-tagged TOX3 in bacteria. After isopropyl-β-D-thiogalactopyranoside induction, the TOX3 recombinant protein was collected in the inclusion body from the bacteria by treating with 8 M urea, and subsequently was purified by passing through an Ni-NTA agarose column (Qiagen). The eluted protein was resolved on a 10% SDS-polyacrylamide gel. After staining with Coomassie brilliant blue, the TOX3 band was excised and injected into rabbits to produce anti-TOX3 antiserum.
Immunocytochemistry.
Cells were grown on glass coverslips (North Carolina Biological) pretreated with Radiacwash. When the cells were ready to be fixed, they were incubated in 4% paraformaldehyde/4% sucrose for 20 min and then washed with PBS 3 times. Subsequently the coverslips were blocked with 3% BSA and 0.3% Triton X-100 for 30 min. They were incubated with primary antibody for 2 h and then secondary antibody for 1 h at room temperature. The coverslips were mounted with Fluoromount (EMS). Dilutions of antibody used were: c-fos (EMD, rabbit polyclonal 1:2,000); and eGFP (Abcam, goat polyclonal 1:3,000).
Northern Blot.
Cortices were dissected out from E18, P0, P7, P14, P21, and adult rats. Total RNA was prepared with TRIZOL (Invitrogen). Ten to twenty nanograms of RNA were resolved on a 20% formaldehyde agarose gel. Probe hybridizing to the first 700 nucleotides of TOX3 was labeled with 32P (Perkin-Elmer) using Ready-To-Go DNA Labeling Beads (-dCTP) (GE Healthcare). The blot was exposed to x-ray film (Kodak) for 2–5 days and then developed.
Supplementary Material
Acknowledgments.
We thank David Ginty and Amir Kashani for constructs; Marc Montminy and members of the Ghosh Laboratory for comments and discussion; and Stephen LeSourd for manuscript preparation. This work was funded by a grant from the National Institute of Mental Health (MH068578). S.H.Y was supported by a National Institute on Aging K12 training grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. EU194254 (rat TOX3)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0805555106/DCSupplemental.
References
- 1.West AE, et al. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci USA. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aizawa H, et al. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- 3.Ince-Dunn G, et al. Regulation of thalamocortical patterning and synaptic maturation by NeuroD2. Neuron. 2006;49:683–695. doi: 10.1016/j.neuron.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 4.Kashani AH, et al. Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections. J Neurosci. 2006;26:8398–8408. doi: 10.1523/JNEUROSCI.0618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson B, et al. TOX: An HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol. 2002;3:272–280. doi: 10.1038/ni767. [DOI] [PubMed] [Google Scholar]
- 6.Travers AA. Priming the nucleosome: A role for HMGB proteins? EMBO Rep. 2003;4:131–136. doi: 10.1038/sj.embor.embor741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 9.Ginty DD, et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 10.Sheng M, Thompson MA, Greenberg ME. CREB: A Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 11.Chrivia JC, et al. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 12.Hu SC, Chrivia J, Ghosh A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- 13.Chawla S, Hardingham GE, Quinn DR, Bading H. CBP: A signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- 14.Hardingham GE, Chawla S, Cruzalegui FH, Bading H. Control of recruitment and transcription-activating function of CBP determines gene regulation by NMDA receptors and L-type calcium channels. Neuron. 1999;22:789–798. doi: 10.1016/s0896-6273(00)80737-0. [DOI] [PubMed] [Google Scholar]
- 15.Impey S, et al. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 16.Kitsos CM, et al. Calmodulin-dependent protein kinase IV regulates hematopoietic stem cell maintenance. J Biol Chem. 2005;280:33101–33108. doi: 10.1074/jbc.M505208200. [DOI] [PubMed] [Google Scholar]
- 17.Margolis RL, et al. cDNAs with long CAG trinucleotide repeats from human brain. Hum Genet. 1997;100:114–122. doi: 10.1007/s004390050476. [DOI] [PubMed] [Google Scholar]
- 18.Ginty DD, Bonni A, Greenberg ME. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 19.Qiu Z, Ghosh A. A brief history of neuronal gene expression: Regulatory mechanisms and cellular consequences. Neuron. 2008;60:449–455. doi: 10.1016/j.neuron.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 20.Sheridan PL, et al. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 21.Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 22.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ui-Tei K, et al. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.