Abstract
Activation-induced cytidine deaminase (AID) introduces DNA cleavage in the Ig gene locus to initiate somatic hypermutation (SHM) and class switch recombination (CSR) in B cells. The DNA deamination model assumes that AID deaminates cytidine (C) on DNA and generates uridine (U), resulting in DNA cleavage after removal of U by uracil DNA glycosylase (UNG). Although UNG deficiency reduces CSR efficiency to one tenth, we reported that catalytically inactive mutants of UNG were fully proficient in CSR and that several mutants at noncatalytic sites lost CSR activity, indicating that enzymatic activity of UNG is not required for CSR. In this report we show that CSR activity by many UNG mutants critically depends on its N-terminal domain, irrespective of their enzymatic activities. Dissociation of the catalytic and CSR activity was also found in another UNG family member, SMUG1, and its mutants. We also show that Ugi, a specific peptide inhibitor of UNG, inhibits CSR without reducing DNA cleavage of the S (switch) region, confirming dispensability of UNG in DNA cleavage in CSR. It is therefore likely that UNG is involved in a repair step after DNA cleavage in CSR. Furthermore, requirement of the N terminus but not enzymatic activity of UNG mutants for CSR indicates that the UNG protein structure is critical. The present findings support our earlier proposal that CSR depends on a noncanonical function of the UNG protein (e.g., as a scaffold for repair enzymes) that might be required for the recombination reaction after DNA cleavage.
Keywords: N-terminal region deletion, point mutants, SMUG1
In response to antigen stimulation, the Ig locus of B cells undergoes 2 types of DNA modification: class switch recombination (CSR) and somatic hypermutation (SHM) (1). CSR and SHM diversify antibodies in 2 different modes: SHM introduces point mutations in the recombined V(D)J region, whereas CSR changes the heavy-chain constant (CH) region by looping-out deletion of upstream CH genes, which is mediated by DNA cleavage and relegation of 2 different switch (S) regions located 5′ to each CH gene. CSR results in switching Ig isotype from IgM to IgG, IgA, or IgE, keeping the same antigen specificity but altering effector functions of the expressed antibody.
AID has been shown to be essential to both CSR and SHM (2, 3), which are initiated by introducing DNA breaks in V and/or S regions (4–6). However, the molecular mechanism of DNA cleavage by AID has been a subject of controversy. The RNA editing model postulates that AID edits an unknown mRNA to generate endonuclease or its cofactor (7). On the other hand, the DNA deamination model (8–10) proposes that AID deaminates cytidine (C) to uridine (U) in S regions, generating U/G mismatches, which are recognized by the base excision or mismatch repair pathway. The majority of Us are proposed to be processed by uracil DNA glycosylase (UNG) and an apurinic/apyrimidic endonuclease, generating single-stranded nicks and staggered double-strand breaks (DSBs) in CSR.
According to the DNA deamination model, the U-removal activity of UNG was assumed to be critical for DSB generation. In fact, UNG-deficient B cells show only 10% CSR compared with WT B cells. However, we have provided lines of evidence that the U-removal activity of UNG is not required for CSR, although the UNG protein is involved in CSR. First, series of point mutations in the well-established catalytic sites did not abolish CSR activity of mouse (m)UNG (11). Second, mutations in the WxxF motif, which is known to be required for interaction of UNG with Vpr, an HIV encoded protein, strongly reduced CSR activity without loss of U-removal activity (12). Third, IgM+ hybridoma generated from stimulated UNG−/−Msh2−/− B cells clearly accumulated mutations or small deletions, footprints of DNA cleavage in the S region (12). Fourth, Ugi, a specific inhibitor of UNG, did not block generation of γH2AX focus formation, the hallmark of DSBs in S regions, although Ugi severely damaged class switching (11). These results suggest the possibility that UNG participates in CSR after DNA cleavage by its noncanonical function.
However, an alternative interpretation was recently proposed, claiming that barely detectable residual catalytic activities of mUNG, which presumably range between 0.05% and 0.5% of WT, as in the case of hUNG (13), were sufficient for generating DSBs in S regions (14). It is therefore necessary to verify the contribution of residual catalytic activities because the DNA deamination model absolutely relies on the U-removal activity of UNG for CSR and SHM.
To examine the effect of residual U-removal activities of catalytically defective mutants, Escherichia coli (e)UNG is most suitable because their reaction kinetics have been precisely determined (15, 16). Unexpectedly, 2 eUNG catalytic mutants showed totally different CSR activities. We found that the N-terminal region, which is missing from eUNG and not required for the catalytic activity, is critical to the CSR activity of many mUNG mutants at the catalytic sites as well as the WXXF motif. In addition, single-stranded monofunctional uracil DNA glycosylase1 (SMUG1) mutants at equivalent catalytic sites retained moderate U-removal activities but little CSR activity. Furthermore, we confirmed that Ugi did not block DNA cleavage in the S region by a direct detection method of cleaved ends. Taken together, the present study strengthens the idea that UNG is not involved in CSR as a U-removal catalyst.
Results
Differential CSR Activities by Catalytic Mutants of eUNG.
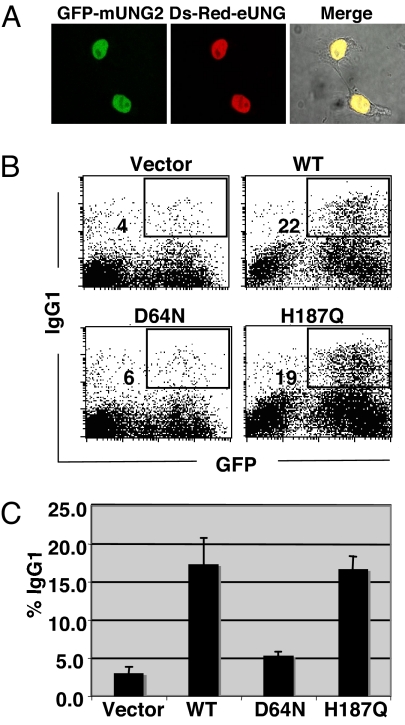
To verify the contribution of residual catalytic activities of UNG mutants in CSR, we took advantage of eUNG and its catalytic mutants, whose residual activities have been precisely measured with a highly sensitive UNG assay (15, 16). First, we confirmed the expression and localization status of eUNG in mammalian cells and showed that they are comparable with mUNG (Fig. 1A). Next we tested the CSR rescue ability of eUNG by retroviral transduction of eUNG-IRES-GFP reporter construct into UNG-deficient B cells. IgG1 switching efficiency was measured in the GFP-positive population expressing WT and mutant eUNGs. Fig. 1B shows typical FACS profiles obtained for eUNG WT and catalytic mutants D64N and H187Q, equivalent to D145N and H268L of mUNG, respectively. Unlike the mUNG mutants, which are highly proficient in CSR, one of the eUNG catalytic mutants (D64N) failed to rescue CSR, whereas the other mutant (H187Q) showed the same CSR activity as mUNG H268L mutant (11) (Fig. 1C). It is important to stress that both D64N and H187Q mutants of eUNG have 10−3 and 3 × 10−3–10−2 WT enzymatic activity on double-stranded DNA and single-stranded DNA, respectively (15). Nonetheless, only the H187Q mutant retained CSR activity. The result clearly indicates dissociation of CSR activity from the enzymatic activity of eUNG.
Fig. 1.
CSR rescue efficiency differs between catalytic mutants of E.coli UNG. (A) Immunofluorescence microscopy of nuclear isoform of mUNG (mUNG2) and eUNG in 293T cells. EGFP and DsRed were fused to the N terminus of mUNG2 and eUNG, respectively. (B) Flow cytometric (FACS) analysis of ung−/− spleen cells after retroviral transduction of WT and catalytic mutants of eUNG. CSR efficiency was measured as percentage of surface IgG1-expressing cells in GFP-positive (infected) cells and indicated next to the gate box. (C) CSR complementation efficiency by WT and mutant eUNG was plotted from 3 experiments.
N-Terminal Region Requirement for CSR by D145N and N204V mUNG Mutants.
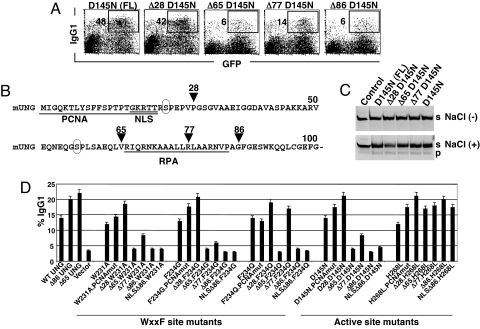
To explore the reason for the difference in CSR activity between D64N eUNG and D145N mUNG, which are mutated at the equivalent residue in the catalytic center on the 3D structure, we compared the primary amino acid sequences of eUNG and mUNG. The amino acid sequence of eUNG is 77 residues shorter in the N terminus than in mUNG, although their catalytic domains are highly conserved (Fig. S1). We generated the N-terminally truncated D145N mutant of mUNG to determine whether the N-terminal region of mUNG is important for CSR. Comparison between D145N and Δ86D145N mutants of mUNG revealed that the CSR activity by D145N mutant entirely depended on the N-terminal region (Fig. 2A). The results are incompatible with the idea that CSR is mediated by the residual catalytic activity of mUNG mutants because the N-terminal region is known to be dispensable for catalysis (13), thus divergent between nuclear and mitochondrial isoforms of UNG and also among the species (17). Because the N terminus of mUNG possesses the major nuclear localization motif, we verified the intracellular distribution of Δ86 mUNG and Δ86D145N mUNG by GFP fusion. Both appeared to be distributed in the cytoplasm and nucleus (not shown), and no obvious defect was observed at their expression levels (Fig. 2B).
Fig. 2.
N-terminal–dependent CSR activity of mUNG mutants. (A) Representative FACS profile showing IgG1 switching efficiency in ung−/− spleen cells 2 days after retroviral transduction of 2 types of catalytic mutants, with and without N terminus. Surface IgG1 expression in GFP-positive (infected) cells was indicated in the FACS profile. (B) Immunoblot analysis of mutant proteins and tubulin as a loading control. (C) N-terminal–dependent CSR restoration by WxxF site mutants of mUNG. Flow cytometric analysis of ung−/− spleen cells to IgG1 switching. WT and WxxF site mutants, with or without N terminus, were expressed by retroviral transduction. Percent IgG1 expression was calculated from infected GFP-positive cells and indicated next to the gated population. (D) A scatterplot showing the catalytic activity vs. CSR efficiency of various mutants at catalytic and WxxF sites. Enzyme assay and IgG1 switching were measured using the same population of retrovirus-infected cells. Relative percentages were calculated in reference to WT as 100%. Data are representative of 3 independent experiments.
To verify whether this was the case for other mutants as well, we applied the same approach and generated the N-terminally truncated form of the other catalytic mutants of mUNG (i.e., N204V and H268L). Δ86N204V shared the same features as Δ86D145N (Fig. 2 A and B). However, H268L mutant did not show any significant reduction in CSR activity in the absence of the N terminus (Figs. 2D and 3D), consistent with H187Q eUNG (Fig. 1C). The finding suggests that 2 key residues (D145 and N204) in the active pocket play a critical role in CSR in collaboration with the N terminus, which is unlikely to be catalytic function.
Fig. 3.
Identification of the N terminus sequence responsible for CSR. (A) Flow cytometry of ung−/− spleen cells after retroviral introduction of full-length (FL) and serial truncations of D145N mutant. Switching to IgG1 was examined 4 days after retroviral transduction and indicated in the respective FACS profile. (B) N-terminal amino acid sequence of mUNG. Arrowheads indicate positions of deletions. Phosphorylation sites conserved between mouse and human are indicated by circles, and other relevant motifs are underlined. (C) Denaturing polyacrylamide gel showing U-removal activity of ung−/− spleen cell extracts expressing different truncated mutants of D145N as indicated. Control cell extract was prepared from cells infected with retroviral vector alone. s, intact oligo-substrate; p, cleaved product. (D) Efficiency of IgG1 switching of 3 WxxF site mutants and 2 catalytic mutants that were subjected to serial N-terminal truncations as indicated. Mutants were introduced into ung−/− spleen cells by retroviral transduction, and percentage IgG1 switching was determined from infected GFP-positive population.
WxxF Mutants Require N Terminus for CSR.
Previously, we also showed that the Vpr binding site of UNG, namely the WxxF motif, is critical for CSR because mutants of this site lose the CSR restoration property in UNG−/− B cells (12), although these mutants retain easily detectable U-removing activity. Because previous WxxF mutants were created on the N-terminally truncated UNG, the current observation prompted us to examine whether the N terminus of UNG can support the CSR function of the WxxF mutants. To our surprise, W231A, F234G, and F234Q mutants in the presence of the N-terminal region showed as strong CSR rescue function as WT (Fig. 2C). However, W231K mutant remained inactive in the presence or absence of the N-terminal region (Fig. 2D). The results again indicate that the selected residues within the protein interaction motif of UNG cooperate with the N-terminal region and play a critical role in CSR through a pathway independent of the U-removing activity. We plotted the relative activities of CSR and enzymatic activities of each mutant with or without the N terminus (Fig. 2D). This plot suggests that CSR activities of many UNG mutants correlate better with the presence of the N terminus rather than their enzymatic activities.
A Limited Region of UNG N Terminus Influences CSR.
Next we wanted to narrow down the critical region in the N terminus required for the restoration of CSR by the catalytic mutant D145N (Fig. 3 A and B). Studies on serial N-terminal deletion of mutant D145N showed that the deletion of the initial 28 aa, including the proliferating cell nuclear antigen (PCNA) binding site, did not affect the CSR restoration property. Deletion at residue 65 almost completely abolished CSR activity. Although further deletion up to residue 77 slightly enhanced CSR, it dropped to the basal level by truncation at residue 86. We further examined the enzymatic activity of various deletion forms of D145N in the presence and absence of 10 mM NaCl. We confirmed that all truncation mutants carried a similar level (detectable or undetectable depending on assay conditions) of U-removal activity, indicating that residual activities of mUNG do not correlate with CSR function (Fig. 3C). To confirm the essential region of the N terminus for CSR, we made similar deletion mutants of H268L, W231A, F234G, and F234Q. The H268L mutant was active regardless of the absence or presence of the N-terminal region. It is clear that the deletion at residue 65 led to almost complete abolition of CSR for W231A and F234G. F234Q retained CSR activity by truncation at residue 77 but lost the activity by truncation at residue 86 (Fig. 3D). Apparently residues 28–65 seem to be critical for the CSR function of D145N, W231A, and F234G, whereas residues 28–65 and residues 77–86 were involved for the CSR rescue function of the F234Q mutant. Conservation of CSR activity by deletion of 1–28 aa and additional internal deletions within 28–65 aa ruled out possible involvement of 2 phosphorylation sites (17), which were conserved between mouse and human UNG (Fig. 3B). The presence of the replication protein A (RPA) binding site seems unnecessary for CSR activity. The loss of nuclear localization signal (NLS) is not responsible for reduction of CSR activity because exogenous NLS addition to Δ86 mutants did not restore CSR activity (Fig. 3D).
Comparison of Catalytic Mutants of UNG and SMUG1.
If simple U-removal activity is required for S region cleavage, any U-removing enzyme, including SMUG1, is supposed to compensate the CSR function in UNG deficiency. It has been shown that SMUG1 is inefficient in rescuing CSR by transgenic expression in vivo (18). However, retroviral delivery of SMUG1 could fully restore CSR of UNG-deficient B cells (14). Because we revealed several novel features of UNG mutants, we examined whether UNG equivalent catalytic mutants of SMUG1 can restore CSR activity. We tested mouse (m)SMUG1 mutants that are structurally equivalent (19, 20) to mUNG mutants (Fig. S2), as summarized in Table 1. FACS profiles show that all mutants of mSMUG1 failed to restore CSR, although WT mSMUG1 could rescue CSR in place of mUNG (Fig. 4A). We next examined the U-removal activity of mSMUG1 and its mutants. Interestingly, most of the mSMUG1 mutants retained much more U-removal activity compared with equivalent UNG mutants (Fig. 4B), in agreement with the previous report (21). All catalytic mutants of mSMUG1, despite having a range of residual U-removal activities, are unable to rescue CSR. Most striking in contrast are G87Y and G87V, which have none and 58% of the U-removal activity of WT, respectively (21), despite their total loss of CSR activity. Loss of CSR rescue activity was also observed for conserved WxxF site mutants of mSMUG1 (data not shown). The results indicate that the role of U-removing activity in CSR remained in question and that a noncanonical function of mUNG and mSMUG1 is more likely to be involved in CSR.
Table 1.
Summary of CSR efficiency vs. enzymatic activity of SMUG1 and UNG mutants
| Catalytic sites | mSMUG1 | CSR(%) | U-removal activity* | mUNG | CSR(%) | U-removal activity† |
|---|---|---|---|---|---|---|
| WT | 100 | 100 | WT | 100 | 100 | |
| H2O coordination | N85A | 9 ± 0 | 7 | D145N | 118 ± 20 | 0.04 |
| Stabilization of transition state | H239L | 0 | 26 | H268L | 100 ± 10 | 0.32 |
| Substrate binding | N163D | 9 ± 0 | 32 | N204V | 82 ± 9 | 0.52 |
| N204D | 9 ± 0‡ | 0.04 | ||||
| Thymine expulsion | G87V | 0 | 58 | Y147A | 9 ± 0‡ | 0.05 |
| G87Y | 0 | nd |
Data of U-removal activity of SMUG1 and UNG are taken from the references indicated. Background switching of ung−/− spleen cells (vector) was subtracted from all. For clarity, numbering of mouse catalytic mutants was kept the same as human catalytic mutants. nd, not detectable.
*, Ref. 21.
‡, Ref. 24.
Fig. 4.
Active site mutants of UNG but not SMUG1 can restore CSR. (A) Representative FACS profile showing IgG1 switching efficiency of ung−/− spleen cells expressing mSMUG1 and mUNG. Percentages of IgG1 switched population were calculated from infected GFP-positive cells. WT or catalytic mutants are indicated above each plot. (B) Uracil-removing activity of mSMUG1 and mUNG (Top) and Western blot of expressed proteins (Bottom). WT and indicated mutants were expressed in ung−/− spleen cells, and the same batch of cells was used for enzyme assay and protein expression. S, substrate. P, product.
S Region Cleavage Without CSR in Ugi-Expressing Cells.
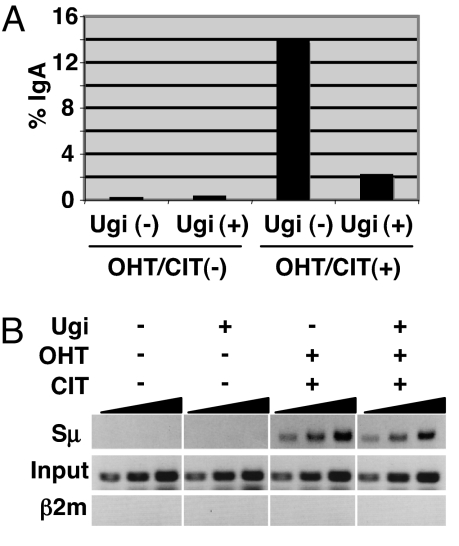
Ugi tightly binds to UNG and completely prevents UNG from interaction with DNA (22). Previously we generated a CH12F3–2–derived cell line (UAR), which expresses Ugi under the tetracycline-regulated promoter, and showed that DNA cleavage in the S region is not inhibited by Ugi using the γH2AX focus formation assay (11). However, this method detects DSB indirectly and in a broad area. We therefore re-examined the effect of Ugi on DNA cleavage directly by break-end labeling, as recently described (4, 23). Sμ DNA break ends were labeled in situ in CH12 cells by biotin-dUTP and terminal deoxyribonucleotide transferase (TdT) in the presence or absence of Ugi expression. After DNA isolation and restriction digestion, biotin-dUTP–labeled fragments were trapped by streptavidin beads and subjected to S region–specific PCR. Despite severe CSR inhibition by Ugi (Fig. 5A), Sμ-break signal can be readily detected in the biotin-dUTP–labeled DNA processed from stimulated cells, both in the presence or absence of Ugi, but not in the labeled DNA from nonstimulated cells (Fig. 5B). Detection of specific signals at the Sμ region but not at the β2m locus suggests that our established in situ end-labeling method detects breaks with specificity. The results are in agreement with previous DNA break assessment by γH2AX focus formation (11).
Fig. 5.
Detection of Sμ DNA break in Ugi-expressing cells by in situ labeling of DNA break end. (A) FACS data showing IgA switching efficiency of UAR cells 24 h after induction. (B) UAR cells were cultured in the presence and absence of tetracycline to obtain Ugi-suppressed and -expressed conditions, respectively. Both types of cells were either left untreated as control or stimulated by CIT and OHT. After an overnight induction, cells were harvested for DNA break assay as described in Materials and Methods. Pulled-down DNA labeled with biotin-dUTP at the cleaved ends was subjected to Sμ-specific PCR. Samples were 3-fold serially diluted.
Discussion
According to the DNA deamination model (8–10), DNA cleavage in the S region depends on U-removal activity of mUNG. In the present study we have shown that 2 catalytic mutants of eUNG, whose residual activities have been precisely characterized to be at the level of 10−2–10−3 of WT (15), have totally different activities for CSR rescue in UNG-deficient mouse B cells. Because eUNG does not have the sequence corresponding to the N terminus of mUNG, we re-examined mUNG catalytic mutants and found that the presence of the N-terminal region of mUNG is critical to many of the catalytic as well as WxxF motif mutants. The results suggest that the N-terminal region, which is not required for U-removal activity, may be involved in CSR function of UNG. This clearly suggests that the U-removal activity is not critical but rather that the protein structure of UNG is more important to CSR.
Further evidence to support this conclusion was obtained by the analysis of mSMUG1 and its mutants. mSMUG1 mutants showing considerable levels of U-removal activity showed little CSR activity, although WT mSMUG1 can rescue CSR in UNG−/− B cells. mSMUG1 mutants were clearly far less active in CSR compared with equivalent mUNG mutants. This could be due to their structural difference, despite their similarity in catalytic function. Again, the finding supports our conclusion that the structure of the UNG protein is critical to CSR. In addition, we confirmed that UNG blocking by Ugi did not affect DNA cleavage in the S region by a direct DNA cleavage assay although Ugi blocks CSR severely, clearly indicating that UNG plays a role after DNA cleavage in CSR.
Taking all these results together, we confirm our previous claim that UNG is involved in CSR at a repair step after DNA cleavage. The N-terminus requirement is consistent with our previous proposal (11, 12, 24) that UNG serves as a scaffold for other proteins by interacting with unknown association protein(s) critical for CSR at several positions, such as D145 and N204 in the catalytic center, the WxxF motif, and the N-terminal region. As shown in Fig. 6, the N-terminal region requirement is minimal in WT UNG for interaction with the unknown association protein (F). However, the N-terminal region is critically required when either the catalytically active site or the WxxF site is mutated so that their interaction with F becomes weaker.
Fig. 6.
A scheme illustrating a possible mechanism for N-terminal–dependent CSR by catalytic and WxxF site mutants of UNG. UNG is assumed to interact with an unknown associated protein (F) or proteins for CSR. Del86 WT may interact with F at the catalytic and WxxF sites. Many mutants at these sites cannot interact with F without N terminus.
Materials and Methods
Mice and Cell Line.
UNG-deficient mice and UAR cells expressing Ugi and AIDER were described previously (11).
Retroviral Constructs and Class Switch Assay.
Genomic DNA of E. Coli-K12 strain was used to PCR-amplify eUNG. Catalytic mutants of eUNG were generated according to the instructions of the Quick Change Mutagenesis kit (Stratagene) and subcloned into retroviral vector pFB-IRES-GFP. SMUG1 was cloned by RT-PCR from CH12 cell RNA, and desired mutations were introduced by quick-change mutagenesis. WT and all mutants of SMUG1 and UNG were constructed in pFB-IRES-GFP vector. For the convenience of expression analysis a parallel set of mutants was produced by Flag-tag fusion. For intracellular localization study either DsRed or EGFP was fused at the N terminus of UNG.
Retroviral sap was prepared according to the standard method and Plat-E retroviral packaging line. Splenocytes were prepared from UNG−/− mice and preactivated for 1.5 days in the presence of lipopolysaccharide (50 μg/mL) and IL-4 (15 ng/mL). Either 2 days or 4 days after retroviral transduction, cells were prepared for FACS analysis by staining with biotinylated anti-IgG1 and allophycocyanin-labeled streptavidin.
U-Removal Assay.
The uracil DNA glycosylase assay in single-stranded DNA substrate was carried out using a 5′-FITC-labeled oligonucleotide of 30-mer with an internal single U residue. Reactions were performed at 37 °C using cleared spleen cell lysates and 10 pmol of substrate, as described previously (12). SMUG1 assay was performed using double-stranded oligo (14) with single U/G mismatch and as described by Pettersen et al. (21).
DNA Break Assay.
The CH12F3–2–derived UAR line cells were stimulated by both tamoxifen (OHT) and CIT (CD40L, IL-4, and TGF-β) as described previously (11), and switching efficiency was assayed by surface IgA staining. A modified Sμ DNA break assay (4, 23) was used to label the free ends of cleaved switch regions. UAR line cells were stimulated overnight. The next day, live cells were separated through Percoll gradient, followed by mild fixation and in situ DNA end-labeling by biotin-16-dUTP and TdT. Genomic DNA was prepared by phenol:chloroform extraction, and 20 μg of DNA was subjected to HindIII digestion. DNA fragments labeled with biotin-dUTP were captured by streptavidin magnetic beads (Promega) and subjected to 32 cycles of PCR (95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 S). The following primers were used for PCR of Sμ and β2microglobulin (β2m): Sμ, forward: 5′-GCTTCTAAAATGCGCTAAACTGAGGTGATT-3′; Sμ, reverse: 5′-GTTTAGCTCTATTCAACCTAG-3′; β2m, forward: 5′-GGTGACGACCTCCGGATCTG-3′; β2m, reverse: 5′-GCCGAGTAGCAGCCACTGAAA-3′.
Supplementary Material
Acknowledgments.
We thank Y. Shiraki and T. Kanda for preparation of the manuscript. This research was supported by a Grant-in-Aid for Specially Promoted Research (no. 17002015) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813252106/DCSupplemental.
References
- 1.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 4.Doi T, et al. The C-terminal region of AID is responsible for a novel function other than DNA cleavage in class switch recombination. PNAS. 2007 doi: 10.1073/pnas.0813253106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagaoka H, Ito S, Muramatsu M, Nakata M, Honjo T. DNA cleavage in immunoglobulin somatic hypermutation depends on de novo protein synthesis but not on uracil DNA glycosylase. Proc Natl Acad Sci USA. 2005;102:2022–2027. doi: 10.1073/pnas.0409491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen S, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honjo T, Nagaoka H, Shinkura R, Muramatsu M. AID to overcome the limitations of genomic information. Nat Immunol. 2005;6:655–661. doi: 10.1038/ni1218. [DOI] [PubMed] [Google Scholar]
- 8.Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 9.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 10.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 11.Begum NA, et al. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science. 2004;305:1160–1163. doi: 10.1126/science.1098444. [DOI] [PubMed] [Google Scholar]
- 12.Begum NA, et al. Requirement of non-canonical activity of uracil DNA glycosylase for class switch recombination. J Biol Chem. 2007;282:731–742. doi: 10.1074/jbc.M607439200. [DOI] [PubMed] [Google Scholar]
- 13.Mol CD, et al. Crystal structure and mutational analysis of human uracil-DNA glycosylase: Structural basis for specificity and catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 14.Di Noia JM, et al. Dependence of antibody gene diversification on uracil excision. J Exp Med. 2007;204:3209–3219. doi: 10.1084/jem.20071768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drohat AC, Jagadeesh J, Ferguson E, Stivers JT. Role of electrophilic and general base catalysis in the mechanism of Escherichia coli uracil DNA glycosylase. Biochemistry. 1999;38:11866–11875. doi: 10.1021/bi9910878. [DOI] [PubMed] [Google Scholar]
- 16.Drohat AC, et al. Heteronuclear NMR and crystallographic studies of wild-type and H187Q Escherichia coli uracil DNA glycosylase: Electrophilic catalysis of uracil expulsion by a neutral histidine 187. Biochemistry. 1999;38:11876–11886. doi: 10.1021/bi9910880. [DOI] [PubMed] [Google Scholar]
- 17.Hagen L, et al. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Noia JM, Rada C, Neuberger MS. SMUG1 is able to excise uracil from immunoglobulin genes: Insight into mutation versus repair. EMBO J. 2006;25:585–595. doi: 10.1038/sj.emboj.7600939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat Res. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 20.Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol Cell. 2003;11:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 21.Pettersen HS, et al. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35:3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mol CD, et al. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 1995;82:701–708. doi: 10.1016/0092-8674(95)90467-0. [DOI] [PubMed] [Google Scholar]
- 23.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 24.Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- 25.Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA—General mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.