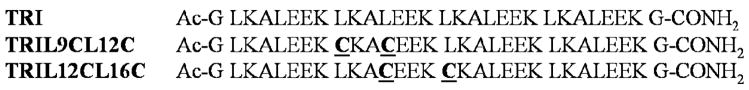Abstract
A de novo protein design strategy provides a powerful tool to elucidate how heavy metals interact with proteins. Cysteine derivatives of the TRI peptide family (Ac-G(LKALEEK)4G-NH2) have been shown to bind heavy metals in an unusual trigonal geometry. Our present objective was to design binding sites in α-helical scaffolds that are able to form higher coordination number complexes with Cd(II) and Hg(II). Herein, we evaluate the binding of Cd(II) and Hg(II) to double cysteine substituted TRI peptides lacking intervening leucines between sulfurs in the heptads. We compare a -Cysd-X-X-X-Cysa- binding motif found in TRIL12CL16C to the more common -Cysa-X-X-Cysd- sequence of native proteins found in TRIL9CL12C. Compared to TRI, these substitutions destabilize the helical aggregates, leading to mixtures of two and three stranded bundles. The three stranded coiled coils are stabilized by the addition of metals. TRIL9CL12C forms distorted tetrahedral complexes with both Cd(II) and Hg(II), as supported by UV-vis, CD, 113Cd NMR, 199Hg NMR and 111mCd PAC spectroscopy. Additionally, these signatures are very similar to those found for heavy metal substituted rubredoxin. These results suggest that in terms of Hg(II) binding, TRIL9CL12C can be considered as a good mimic of the metallochaperone HAH1, that has previously been shown to form protein dimers. TRIL12CL16C has limited ability to generate homoleptic tetrahedral complexes (Cd(SR)42−). These type of complexes were identified only for Hg(II). However, the spectroscopic signatures suggest a different geometry around the metal ion, demonstrating that effective metal sequestration into the hydrophobic interior of the bundle requires more than simply adding two sulfur residues in adjacent layers of the peptide core. Thus, proper design of metal binding sites must also consider the orientation of cysteine sidechains in a vs d positions of the heptads.
Introduction
In the past few years the rate of research in the field of metallopeptide design has seen a tremendous jump.1–5 This is because one can selectively isolate specific aspects of protein structure to study fundamental properties of metal coordination chemistry. Many structural motifs such as α-helical bundles,5–10 β-sheet proteins,5,11,12 α/β-proteins5,13,14 and fibrous proteins5,13 have been exploited as scaffolds for de novo protein design. Two very common properties of metalloproteins are that they often bind metals in α-helical tertiary structures and that the sequence C-X-X-C is utilized as a pseudochelating agent to anchor metals to a structural site. These two structural features, the α-helix and the C-X-X-C motif, are often associated since this linear sequence will place two cysteine residues on the same face of an α-helix, making chelation possible. It has been recognized for many years that helical peptides can be prepared by repeating a seven amino acid sequence, termed a heptad, multiple times in a linear sequence (e.g., G(LKALEEK)4G). If the first residue (designated a) and the fourth residue (designated d) are always a hydrophobe such as leucine or isoleucine, and the remaining residues are hydrophilic, one can obtain α-helices that orient the a and d positions into the hydrophobic core of an aggregate that may be a two, three or four stranded coiled coil. Thus, the common Ca-X-X-Cd motif provides the simplest way to place two cysteine residues as chelating ligands. However, this is not the only way to prepare a helix with cysteines in an adjacent layer. An alternative approach would be to exploit the sequence defga′. This would yield the sequence Cd-X-X-X-Ca. Despite the fact that this sequence also places cysteines in adjacent layers of a helical bundle, one rarely sees this as a metal binding motif in proteins. A systematic comparison of the relative metal binding preferences of these motifs is a perfect example of the use of de novo protein design for understanding metal/protein interactions.
Our group has focused on investigating primarily three-stranded α-helical coiled coil peptides. Since our first report with TRIL16C,7 we have investigated a number of peptides that belong to the TRI peptide family. The primary structures of these peptides consist of four repeats of the heptad sequence (Ac-G(LKALEEK)4G-NH2) that fold into coiled coil structures by association of peptidic strands. According to size exclusion chromatography and analytical sedimentation equilibrium experiment results, pH dependent aggregation state preferences were observed for that system.15 Two-stranded coiled coils that exist at pH < 5 associate with another peptide unit to form three-stranded coiled coils when the pH exceeds 6. The transition depends on the protonation state of the glutamic acid residues that occupy the helical interface positions in the coiled coil structure of the peptide. Metal binding sites have previously been introduced by substitution of one or two of the Leu residues by Cys, and these binding sites are excellent targets for soft metal ions such as Hg(II) or Cd(II). We were able to design peptides that bind Cd(II) in either S3 or S3O configurations by introducing bulky penicillamine (Pen) residues to the TRI peptide directing it to a Cd(SR)3− complex formation or substituting an N-terminally located leucine residue with alanine to form a cavity that allows for a pure S3O coordination mode of Cd(II) bound to cysteine residues.16 We have also reported the doubly substituted TRI peptide derivatives containing intervening leucines that show heterochromic properties and are able bind two Cd(II) ions, one with S3 and another with S3O environments.17,18
Because of our deep understanding for the design of thiol rich metal binding sites within a moderately simple peptidic scaffold, we felt it was appropriate to assess the metal binding features of two related dicysteine substituted sequences. Therefore, we used the doubly substituted variants of TRI peptides, TRIL9CL12C and TRIL12CL16C to generate -Ca-X-X-Cd- or –Cd-X-X-X-Ca- binding motifs, respectively (Scheme 1). Peptidic systems similar to the -Ca-X-X-Cd- sequence have been investigated by Ogawa et al.19,20 However, their studies differed from our interests in two important ways. First, they examined a designed peptide sequence that was unable to fold into a defined three-dimensional structure in the absence of metal. This allowed them to follow protein folding as had been done with Baby peptides.3 Second, these authors only examined the common -Ca-X-X-Cd-sequence, but did not compare the chemistry of this sequence to that of the lesser known sequence Cd-X-X-X-Ca.
Scheme 1.
Sequence of the TRI peptide and its derivatives.
We felt that the application of TRI peptide derivatives that form coiled coils both in the presence and absence of metal ions may provide new details on metal ion interaction with the proteins and address why the sequence Cd-X-X-X-Ca is so rarely seen in helical structures. Since there are a number of metalloproteins where -C-X-X-C- binding motif is present, taking rubredoxin or Cu(I)-chaperones as examples, the results we obtain may allow further insight into understanding the chemistry of these proteins. In the case of rubredoxin analogues, one can ask several simple but interesting questions. Can one prepare equivalent MS4 binding sites with either Ca-X-X-Cd-or Cd-X-X-X-Ca? Is the ability to form an MS4 site not only sequence dependent but also metal dependent? If one can prepare these MS4 sites in helical aggregates, will the spectroscopic features of the center mimic that of metal substituted rubredoxin which is found in an entirely different protein fold (the “α/β-sandwich” fold)? In this article, we will describe the structures of the Hg(II) and Cd(II) sites of these designed peptides and then answer these intriguing questions.
Experimental section
Fmoc-protected amino acids and the MBHA rink amide resin were purchased from Novabiochem; N-hydroxybenzotriazole (HOBT) and 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) were bought from Anaspec Inc.; diisopropylethylamine (DIEA), acetic anhydride, and pyridine were purchased from Aldrich; and piperidine was supplied by Sigma. N-Methylmorphopyridinone (NMP) was from Fisher Scientific.
Peptide Synthesis and Purification
Peptides were synthesized on an Applied Biosystems 433A automated peptide synthesizer, with Fmoc-protected amino acids by use of the FastMoc activation protocol (Applied Biosystems). Rink amide MBHA resin was used as the solid support so that the resulting peptides would be amidated at the C-terminus. The N-terminus was acetylated on the column with a solution of 4% (v/v) acetic anhydride, 4.3% pyridine, and 91.7% N,N dimethylformamide (DMF). Cleavage from the resin was performed for 90 min in a 90% trifluoroethanol (TFA) solution containing 5% anisole, 3% thioanisole, and 2% ethanedithiol as free radical scavengers.
After precipitation with cold ether, the peptide was redissolved in water and lyophilized to obtain a fluffy off-white powder. The solid was redissolved in 10% acetic acid and purified by reversed-phase HPLC on a Waters 600 Semiprep HPLC with a preparative C18 column (Vydac protein & peptide) with a linear gradient of 0.1% TFA in water to 0.1% TFA in 9:1 CH3CN/H2O over 50 min. The identity of the peptides was verified by ES (electrospray) mass spectrometry (expected MW = 3401.05, found 3410.52), and the purity was determined by analytical HPLC to be greater than 90%. The peptide concentration of these solutions was determined by quantitation of the cysteine thiols by Ellman’s test,21 which uses the product of the reaction of the aliphatic thiols with dithionitrobenzoate (DTNB) as a spectroscopic probe.
Analytical centrifugation
In order to determine oligomerization status, we performed sedimentation equilibrium experiments on the following samples: TRIL12CL16C with Hg(II) in CHES; pH 9.4 further considered as sample 1, TRIL12CL16C with Cd(II) in TrisHCl; pH 8.5 (sample 2), TRIL12CL16C in phosphate; pH 8.5 (sample 3), TRIL9CL12C with Cd(II) in 50 mM TRIS; pH 8.5 and 6 mM TCEP (sample 4), TRIL9CL12C in 50 mM phosphate; pH 8.5 (sample 5), TRIL9CL12C Hg(II) in 50 mM CHES; pH 9.4 6 mM TCEP (sample 6).
All sedimentation experiments were performed with a Beckman Optima XL-I at the Center for Analytical Ultracentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center at San Antonio. Sedimentation equilibrium and Monte Carlo analyses were performed with UltraScan22 version 9.2. Hydrodynamic corrections for buffer conditions were made according to data published by Laue et al,23 and as implemented in UltraScan. All samples were measured at 3 loading concentrations and sedimented to equilibrium at 5 speeds (45, 48.7, 52.5, 56.2, and 60 krpm) in the AN-60-TI rotor. All samples were measured in 6-channel titanium centerpieces (Nanolytics, Gesellschaft für Kolloidanalytik mbH, Am Mühlenberg 11, 14476 Potsdam, Germany), and scanned at equilibrium at the following wavelengths: Sample 1: 301 nm, Sample 2: 263 nm, Sample 3: 236 nm, Sample 4: 263 nm, Sample 5: 230 nm, Sample 6: 290 nm. This resulted in measurements in the concentration range of 0–4 mM for Sample 1, 0–0.6 mM for Sample 2, 0–0.7 mM for Sample 3, 0–0.6 mM for Sample 4, 0–0.2 mM for Sample 5, and 0.06–0.125 mM for Sample 6. We used the following extinction coefficients for the concentration conversion: Sample 1: 5,840 OD301/(Mol cm), Sample 2: 4,430 OD263/(Mol cm), Sample 3: 4,660 OD236/(Mol cm), Sample 4: 7,040 OD263/(Mol cm), Sample 5: 3,509 OD230/(Mol cm), Sample 6: 8,710 OD290/(Mol cm). The molar extinction coefficients were based on the measured molar absorptivity of each sample at its respective wavelengths. The partial specific volume of each peptide was estimated from peptide sequence according to the method by Durchschlag,24 and as implemented in UltraScan was found to be 0.760 ccm/g for both TRIL9CL12C and TRIL12CL16C. All equilibrium data were fitted to multiple models. The most appropriate model was chosen based on visual inspection of the residual run patterns, and based on the best statistics. 95% confidence intervals were determined by Monte Carlo analysis using the Linux cluster at the UTHSCSA Bioinformatics Core Facility.
Electronic spectroscopy
Cd(II) binding titrations were performed by adding aliquots of 2mM CdCl2 to solution containing 50mM TRIS*HCl buffer, pH 8.5 containing 60μM TRIL12CL16C or TRIL9CL12C, respectively and monitored on Carey 100 Bio UV-vis spectrophotometer. Hg(II) titration were performed by adding aliquots of 2mM HgCl2 to 60μM peptide in CHES buffer, pH 9.4.
CD Spectroscopy
CD titration experiments were performed on an Aviv 62DS CD spectrophotometer with the temperature regulated at 298 K. UV CD spectra for 40 μM peptide in double distilled water solutions were collected from 280 to 195 nm every 0.5 nm. The path length of quartz cells used was 2 mm. The pH of the solution was adjusted using small aliquots of 1M KOH and HCl. The molar ellipticity, [Θ], is given in units of degrees centimeter2 decimole−1 per residue. Spectra for Cd(II) complexes were collected at pH 8.5 from 300 to 225 nm every 1 nm. Spectra for Hg(II) complexes were collected at pH 9.4 from 320 to 230 nm every 1 nm. Due to dynode saturation the spectra were not recorded at lower wavelengths. All metal titrations were performed on 90 μM peptide solutions in the presence of 50 mM of the appropriate buffer using 1 cm stain-free quartz cells. Signal averaging was 1s for all systems. The molar ellipticity, [Θ], is given in units of degrees centimeter2 decimole−1.
NMR Spectroscopy
113Cd and 199Hg NMR spectra were collected at room temperature on a Varian Inova 500MHz spectrometer (110.92 MHz for 113Cd and 89.45 MHz for 199Hg) equipped with a 5mm tunable broadband probe. The 0.1M Cd(ClO4)2 in D2O and 0.1M Hg(ClO4)2 in D2O were used as external references.
a) 113Cd NMR
A spectral width of 847 ppm (93 897 Hz) was sampled using 5μs 90° pulse and 0.05 s acquisition time without delays between scans. Samples were prepared by dissolving 25–30mg of lyophilized and degassed peptide in 600 μL of 20% D2O/H2O under Ar flow followed by the addition of desired amount of 250 mM Cd(NO3)2 solution (95% of isotopically enriched 113CdO obtained from Oak Ridge National Laboratory, Oak Ridge, TN) and the adjustment of pH with KOH and HCl solutions. The resulting samples contained 9–12mM peptide. All scans were collected for 2 hours. All data were processed using the software Mestre C v2.3a. All the induction decays were zero-filled to double the amount of original points and were treated with an exponential function with the line broadening of 100 Hz, unless otherwise noted.
b) 199Hg NMR
All 199Hg NMR spectra were externally reference to 0.1 M Hg(ClO4)2 in 0.1 M HCl04/D2O solution at −2250ppm.25 A spectral width of 1340ppm (119 940 Hz) was sampled using 5.0μs 90° pulse and 0.01 s acquisition time with the delay between scans of 0.005 s. Samples were prepared as described above using 125mM stock solution of Hg(NO3)2 prepared from 91% isotopically enriched 199HgO purchased from Oak Ridge National Laboratory, Oak Ridge, TN). Scans were collected for 3 hours. The data were processed using the same software and procedure as was described for 113CdNMR.
Perturbed Angular Correlation spectroscopy
All perturbed angular correlation (PAC) experiments were performed with a setup using six detectors at a temperature of 1 ± 2 °C, which was controlled by a Peltier element. The radioactive cadmium was produced on the day of the experiment at the University Hospital cyclotron in Copenhagen and extracted as described previously,26 except for the HPLC separation of zinc and cadmium. This procedure may lead to zinc contamination of the sample, but the level of contamination should not interfere with the experiment. The 111mCd solution (10–40 μL) was mixed with nonradioactive cadmium acetate and TRIS buffer. The TRI12CL16C or TRIL9CL12C peptide was then added (dissolved in ion-exchanged water), and the sample was left to equilibrate for 10 min to allow for metal binding. Finally, sucrose was added to produce a 55% w/w solution, and the pH of the solution was adjusted with H2SO4 or KOH. To measure the pH, a small volume of sample was removed from the solution to avoid chloride contamination of the sample. The pH reported in table 2 was measured at room temperature the following day. The pH of solutions buffered by TRIS is temperature dependent. Because of the pH dependence on the temperature of TRIS solutions, the pH of the solution at 1 °C was calculated using pH (1 °C) = 0.964 [pH(25 °C)] +0.86.26 The samples were either used immediately after preparation or left on ice for up to 2 hours until the measurement was started. All buffers were purged with Ar and treated so as to lower metal contamination. The final volume of the samples ranged between 0.05 and 0.5 mL with concentrations of 300 μM peptide and 20 mM TRIS. All fits were carried out with 300–400 data points, disregarding the 3–5 first points due to systematic errors in these. The analytical expression for the perturbation function is known, and five parameters are fitted to the data40: ω0 is a measure of the nuclear quadrupole interaction strength; the asymmetry parameter, η, refers to the symmetry of the structure at the site of the PAC probe. In an axially symmetric structure η is 0, and the largest value of this parameter, namely 1, is found, for example, in a tetrahedral complex with 2A and 2B ligands; the amplitude of the signal, A. If more than one structure at the site of the PAC probe is present in the sample, the relative amplitudes will in general directly reflect the relative population of the different sites. The lineshape of the Fourier transformed data can be affected by structural variations at the site of the PAC probe from one molecule to the next, reflected in the relative peak width Δω0/ω0; dynamics similarly affects the PAC signal, and is reflected in the rotational correlation time τc. In summary, five parameters (A, η, νQ (or ω0), δ and τc) are determined for each structure present at the site of the PAC probe in the sample.
Table 2.
Spectroscopic values for Hg(II)/TRILXC complexes.
| Hg Coordination Mode | λ(Δε)a | pKa | RHgS (Å)b | δ (199Hg ppm) |
|---|---|---|---|---|
| Linear 2-coordinate | 240 (2700)c | - | 2.32c | −844d |
| Trigonal 3-coordinate a site | 247 (19200)e | - | 2.44f | −185d |
| 265 (11900)e | ||||
| 295 (5800)e | ||||
| Trigonal 3-coordinate d site | 230 (21300)g | - | 2.44f | −316g |
| 247 (15000)g | ||||
| 297 (5500)g | ||||
| Linear 2-coordinate within a 3-stranded coiled coil | 247 (2000)i | 7.6±0.2f | - | −908d |
| 8.5±0.2h | ||||
| Tetrahedral 4-coordinate | 230 (8100) | - | - | −500 |
| TRIL12CL16C | 289 (7100) | |||
| TRIL9CL12C | 252 (6300) | - | - | −308 ppm |
| 289 (6800) | −400 ppm |
λ given in nm and Δε given as M−1 cm−1.
Average Hg-S EXAFS bond lengths.
Data for TRIL16C from Reference 15.
Data for TRIL9C from Reference 44
Data for TRIL16C from Reference 29.
Data for the a site peptide TRIL9C from Reference 62.
Data for TRIL12C from Reference 44.
Data for the d site peptide TRIL12C from Reference 29.
Data for TRIL12C from Reference 63.
Results
Analytical centrifugation
In the analysis of the equilibrium data of our peptides in the presence and absence of metals, we found various oligomers present. However, using an ideal reversible self-association model for a monomer-dimer, monomer-trimer, or monomer-dimer-trimer system did not produce the statistically most favorable result, suggesting that reversibility of the process is either very slow or not mass-action driven. To characterize the propensity of the samples to oligomerize we chose the following approach: First, we fitted the data with a continuous molecular weight distribution model with 100 species ranging between 1 and 20 kDalton. We then performed a Monte Carlo analysis to these fits.27 Since lower molecular weight species tend to correlate with baseline signals, we cropped the resulting distributions between 3 kDa and 20 kDa and determined weight and number averages. These averages provide a trend of oligomerization potential for each sample. For all cases, no signal was reported for species larger than trimer (10.08 kDa). Next, we fitted the same data to a fixed molecular weight distribution for only molecular weights corresponding to monomer, dimer and trimer and report the same averages (Figure S1, supporting information). The fact that AUC data could not be fit to a reversible model indicates that equilibria are probably too slow to be established when disturbed from the equilibrium point during the speed changes in the equilibrium experiment. Such a situation can be modeled well with the fixed molecular weight distribution model,28 and still provide a good estimation of the relative concentrations of the oligomeric species. Indeed, some of the systems could be fit well by reversible models but in the interest of comparing among all models we found the less constrained model of a fixed molecular weight distribution more appropriate. On each plot, the relative number concentration of each oligomeric form is shown. Variances, averages and oligomerization potential are summarized in Table 1.
Table 1.
Fitting results for analytical centrifugation data for samples 1–6. Shown are weight average (Mw), number average (Mn), and z-average molecular weight (Mz), and the residual mean square deviations (RMSD) for the Monte Carlo analysis of the continuous distribution, and a 3-component discrete molecular weight distribution analysis. No signal of any species larger than a trimer (~10.1 kDalton) was detected in any sample.
| Sample: | Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | Sample 6 |
|---|---|---|---|---|---|---|
| RMSD | 7.621×10−3 | 7.973×10−3 | 6.996×10−3 | 6.568×10−3 | 4.941×10−3 | 6.076×10−3 |
| Mw (cont.) | 7,224 | 7,372 | 6,973 | 7,723 | 6,634 | 8,636 |
| Mn (cont.) | 6,857 | 7,275 | 6,661 | 7,346 | 6,332 | 8,426 |
| Mz (cont.) | 7,601 | 7,456 | 7,293 | 8,006 | 6,897 | 8,678 |
| RMSD | 7.898×10−3 | 8.287×10−3 | 7.057×10−3 | 6.757×10−3 | 5.005×10−3 | 6.548×10−3 |
| Mw (discr.) | 7,561 | 7,561 | 6,699 | 7,214 | 6,272 | 7,155 |
| Mn (discr.) | 6,694 | 6,694 | 6.064 | 6,278 | 5,336 | 5,972 |
| Mz (discr.) | 8,232 | 8,232 | 7,225 | 7,970 | 7,160 | 8,131 |
Electronic spectroscopy
Electronic spectroscopy was examined in our studies since formation of both Cd(II) and Hg(II) complexes of TRI peptide derivatives is accompanied by LMCT (ligand to metal charge transfer) transitions within the UV energy range. A summary of the spectral parameters for these and related peptides is given in Tables 2 and 3.
Table 3.
Spectroscopic values for Cd(II)/TRILXC complexes.
| System | UV-Visa | 113Cd NMRb | 111mCd PAC | ||
|---|---|---|---|---|---|
| λmax (nm) | Δε | Δ (ppm) | ω0 (rad/ns) | η | |
| TRI L16Cc | 232 | 22600 | 625 | 0.337 0.438 |
0.23 0.20 |
| TRI L12AL16Cd | 230 | 21800 | 574 | 0.341 | 0.141 |
| TRI L16Pend | 227 | 28900 | 684 | 0.454 | 0.02 |
| TRIL12CL16C | 222 sh 238 |
16250 14920 |
560 | 0.380(1) 0.270(14) |
0.486(5) 0.50(7) |
| TRIL9CL12C | 227 sh 240 |
20200 17800 |
650 680 |
0.14(4) | 1(1) |
| CmtRe | 240 | ≈16000 | 480 | 0.291(1) | 0.18(1) |
Cd(II) complexes of TRIL12CL16C and TRIL9CL12C
Titration of Cd(II) into a buffered solution of TRIl12CL16C results in a linear increase of the extinction coefficients at 238 nm and at 220 nm up to 0.5 equivalents of metal ion vs. peptide monomer. (Figure 1a) This linear dependency might reflect the formation of single complex species, i.e. a dimer, that requires simultaneous deprotonation of two to three protons. The observed 238 nm maximum absorption band is consistent with the energy already reported by Kharenko et al for C16C19-GGY peptide that contains a -Ca-X-X-Cd- binding motif.20 In addition, the extinction coefficient of 14920 M−1 cm−1 suggests formation of a species involving three rather than four cysteine residues in the coordination sphere of the Cd(II) ion.29 A similar extinction coefficient has been reported for the Cd(II) bound CmtR metalloregulatory protein that is proposed to bind Cd(II) ion within trigonal thiolate environment.30 The pH titration carried out at a 0.5 Cd(II) to TRIL12CL16C ratio gives some idea about the apparent log K value of 4.05 which is much lower than the values previously reported for other TRI peptides forming three-stranded coiled coils with Cd(II).18,29
Figure 1.
Difference spectra of titrations of Cd(II) into the solution of 50mM TRIS buffer pH 8.5 containing 60 μM TRIL12CL16C (a) and 60 μM TRIL9CL12C (b).
Titration of CdCl2 into a buffered solution of TRIL9CL12C at pH 8.5 is shown in Figure 1b. Although the spectroscopic properties of complexes formed with that system are very similar, the mode of Cd(II) binding is different. We still observe the coincidence of two bands at 222 nm and 241 nm. A linear binding dependence is observed until the metal concentration reaches 33 percent of the peptide molar content. This indicates that at this point we may expect formation of complexes within a three-stranded, rather than two-stranded, coiled coil. Addition of subsequent aliquots of CdCl2 leads to the formation of another complex species that reaches its maximum concentration at 0.6 equivalents of metal ion versus peptide. This species exhibits maximum absorption at 227 nm and a shoulder at around 245 nm. A similar absorption spectrum has been reported for Cd(II) substituted rubredoxin.31 Since we are dealing with a mixture of two forms of peptide association, it is hard to calculate extinction coefficients that refer to the types of complexes formed in solution. However, the ratio between maximum absorption bands at 227 nm and 245 nm appears to be comparable and indicates that the geometries of the complexes formed are almost identical. In addition, spectroscopic features suggest that the coordination properties of TRIL9CL12C are different than those of TRIL12CL16C, where Cd(SR)42− complex formation is not observed.
Hg(II) complexes of TRIL12CL16C and TRIL9CL12C
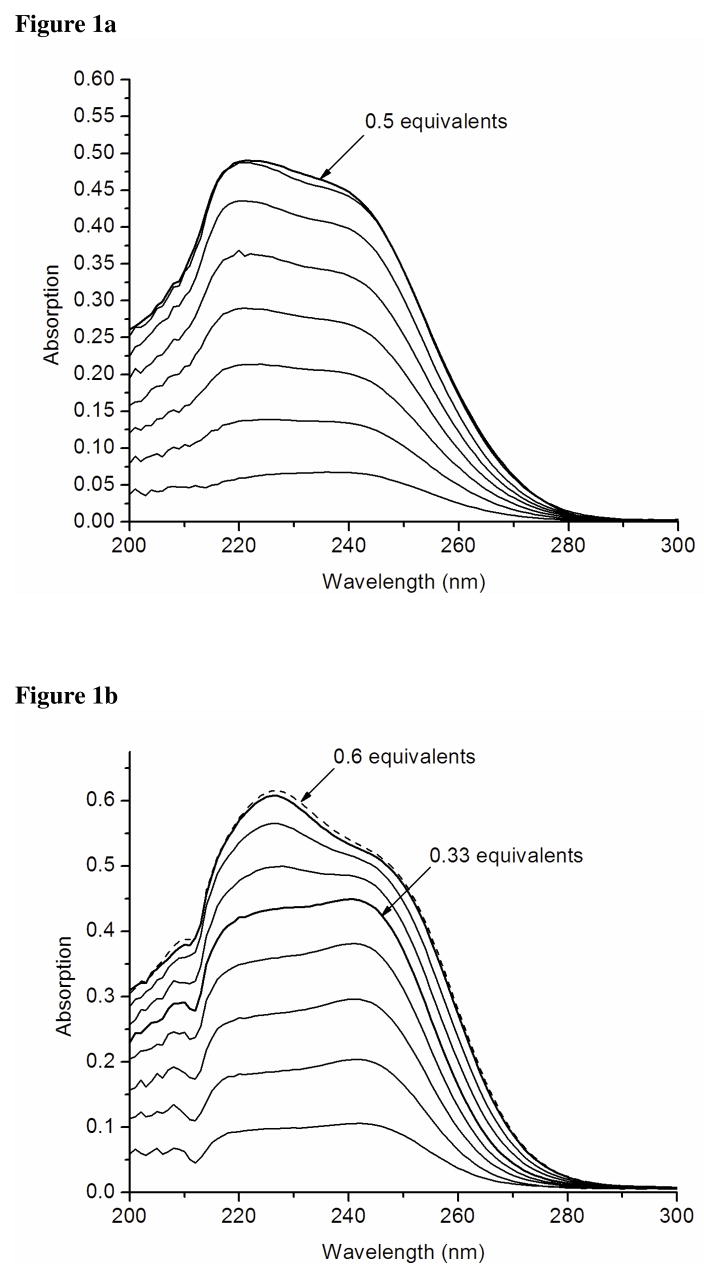
Titration of a buffered solution of TRIL12CL16C with HgCl2 was monitored by electronic spectroscopy (Figure 2a). Addition of subsequent aliquots of Hg(II) result in the formation of complexes that have two absorption maxima at 232 and 289 nm. The latter may be considered a chemical signature of a distorted tetrahedral Hg(II) complex bound to four cysteine thiolate sulfur atoms.32 The band at 289 nm reaches its maximum at 0.5 equivalents of peptide and decays when additional aliquots of HgCl2 are titrated. This suggests that new complex species with higher numbers of Hg(II) ions bound within the two-stranded coiled coil framework are formed.
Figure 2.
Difference spectra of titrations of Hg(II) into the solution of 50mM CHES buffer pH 9.4 containing 60 μM TRIL12CL16C (a) and 60 μM TRIL9CL12C (b).
Titration of HgCl2 into a buffered solution of TRIL9CL12C leads to formation of different complexes than were observed for TRIL12CL16C. This is mainly illustrated by the presence of the absorption band at 250 nm which was not observed for the Cd-X-X-X-Ca system (Figure 2b). This band was previously assigned to trigonal Hg(II) complexes formed within a three-stranded coiled coil.7 Analogous to the spectrum reported for Hg(II) substituted rubredoxin,32 the 284 nm λmax yields a maximum extinction coefficient at a 0.5 ratio of [Hg(II)]/[TRIL9CL12C]. Therefore, one expects Hg(SR)42− complexes with a geometry similar to that present in the mercury(II) substituted rubredoxin.
Circular Dichroism
Protein secondary structures have specific circular dichroism (CD) signatures in the far UV region. For example, α-helical peptides and proteins display spectra with two minima at 208 nm due to an α-helix (π−π* parallel) contribution and at 222 nm due to an α-helix (n−π*) contribution.33 To elucidate the relationship between pH and the secondary structure of TRIL12CL16C and TRIL9CL12C, far UV circular dichroism spectra were recorded in the absence of metal at different pH conditions. Since TRIL12CL16C and TRIL9CL12C correspond to Cd-X-X-X-Ca and Ca-X-X-Cd substituted peptides, respectively, we expected that their metal complexes exhibit different properties in CD spectroscopy. For that reason, titration experiments of Cd(II) and Hg(II) to buffered peptide solutions were completed.
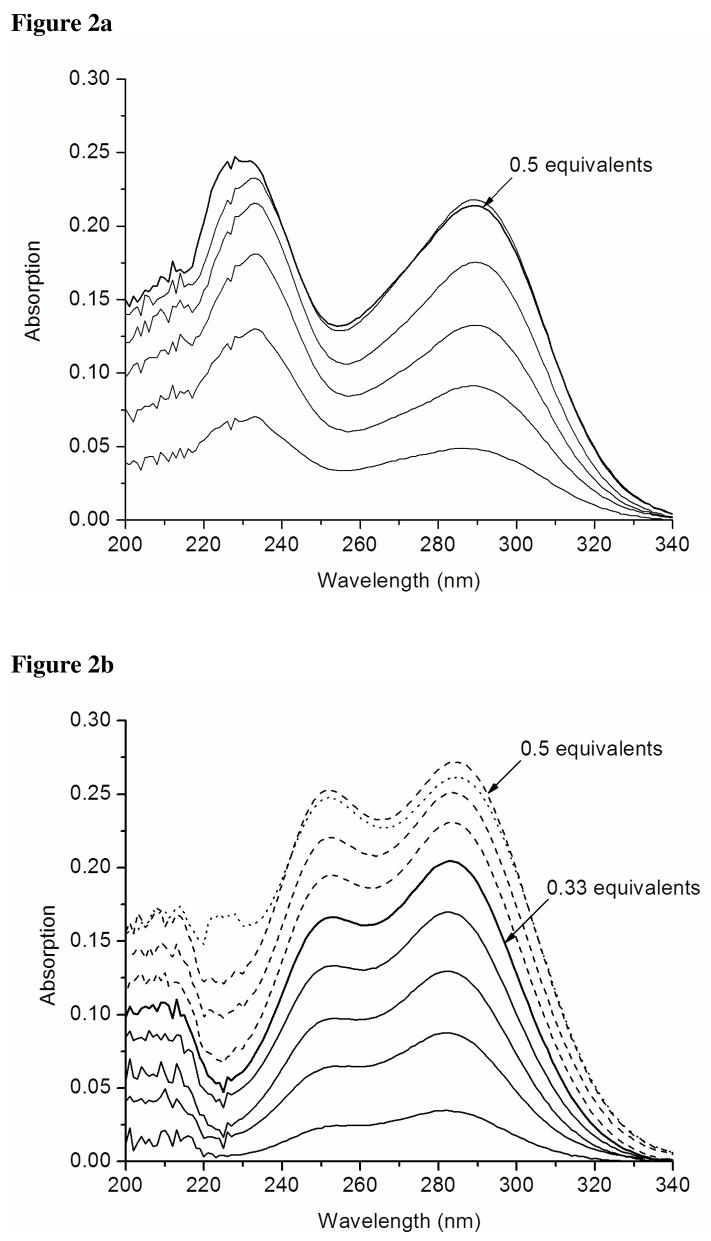
Structural properties of peptides in absence of metal
CD spectra showing the pH dependence of secondary structure of TRIL12CL16C in solution are shown on Figure 3a. The coexistence of two bands at 208 nm and 222 nm is characteristic for α-helical structures and the ellipticity ratio between those signals (≈ 1.05) falls within the range indicating the presence of coiled coils.8 This relationship is observed in acidic and neutral solutions; however, when the pH reaches 9.5 we observe a decreased ellipticity accompanied by a decrease in the [Θ222]/[Θ209] ratio. This perturbation is a consequence of peptide unfolding due to deprotonation of the Nε-amine groups of Lys residues that are no longer able to stabilize the coiled coil structure through the salt-bridges they form with glutamate residues.
Figure 3.
Far-UV circular dichroism spectra of pH titrations of TRIL12CL16C (a) and TRIL9CL12C (b). Changes in ellipticity at 209 nm and 222nm are plotted as a function of pH in the inset.
The pH titration of TRIL9CL12C monitored by circular dichroism (Figure 3b) is similar to that observed for TRIL12CL16C. The negative ellipticity maxima remain at 209nm and 222nm, indicating an α-helical fold; however, the ratio [Θ222]/[Θ209] is almost unaffected across the entire pH range. The plot of ellipticity fluctuation at both wavelengths vs. pH (Figure 3b inset) demonstrates a similar pH dependent destabilization of the peptide self-assembly. Our analytical ultracentrifugation data suggest the presence of two forms of TRIL9CL12C in solution under these conditions.
Comparison of TRIL9CLl2C to similar systems with a -Ca-X-X-Cd- binding motif may possibly explain the reason why peptides with two adjacent leucine layers substituted by cysteine residues tend to form two-stranded instead of three-stranded coiled coils. C16C19-GGY does not fold by itself into a coiled coil, needing a metal cofactor to form the coiled-coil.20 The unusual behavior is reminiscent of the BABY peptides (one heptad shorter than TRI) which also required metals to nucleate the protein fold.34 It is interesting to note that this behavior is not simply the result of replacing leucines with sterically less bulky residues in adjacent a and d layers as TRIL12AL16C and TRIL16CL19A peptides both yield exclusively well folded three stranded coiled coils under the same conditions as these dicysteine substituted systems. Clearly, the replacement of cysteines for leucines is somewhat special, leading to a significant helix destabilization which cannot be easily accommodated within the hydrophobic layers of the core of the coiled coil scaffold.
Cd(II) complexes of TRIL12CL16C and TRIL9CL12C
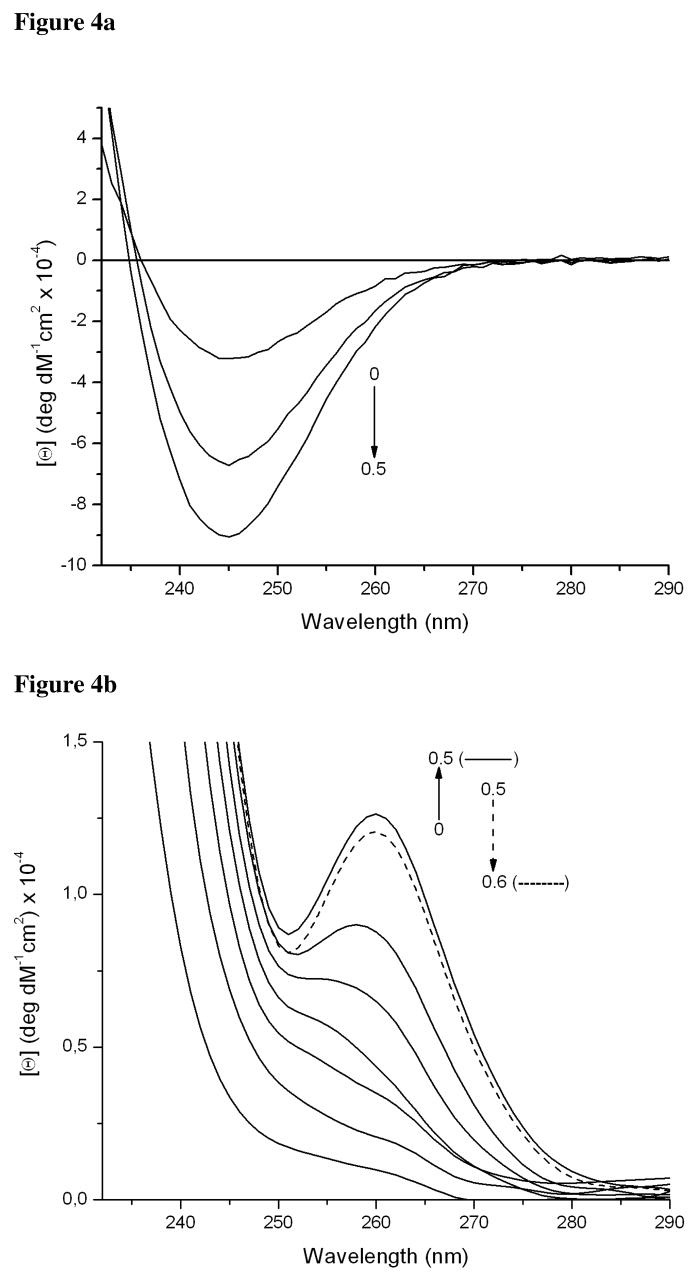
When we titrate CdCl2 into a buffered solution of 90 μM TRIL12CL16C we observe a linear increase of negative ellipticity at 245 nm up to the point where one reaches a 0.5 molar ratio of Cd(II) to peptide (Figure 4a). This inflection point is presumptive evidence for the formation of a complex within a two-stranded coiled coil, supporting results obtained in sedimentation equilibrium experiments (vide supra).
Figure 4.
Circular dichroism difference spectra of pH titrations of Cd(II) into the solutions of 90 μM TRIL9CL12C in 50mM TRIS buffer pH 8.5 (a) and 90 μM TRIL9CL12C in 50mM TRIS buffer pH 8.5 (b).
Although the electronic spectra associated with complex formation upon titration of CdCl2 into a buffered solution of TRIL9CL12C are similar to those for TRI12CL16C, the circular dichroism spectra are markedly different (Figure 4 a and b). Formation of a Cd(TRIL9CL12C)n adduct does not lead to formation of a negative band at 245 nm. Instead, a positive band appears that changes its energy from 257 nm to 260 nm when the CdCl2 concentration reaches half the peptide concentration.
Hg(II) complexes of TRIL12CL16C and TRIL9CL12C
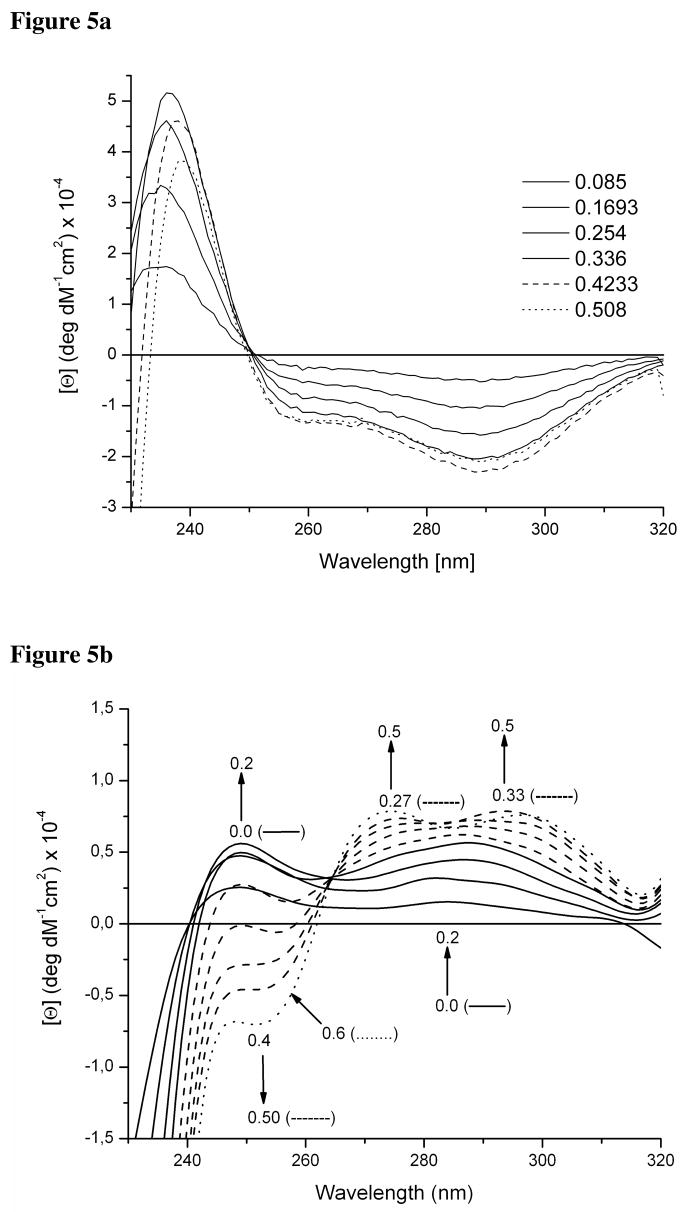
Titration of HgCl2 into a buffered solution of TRIL12CL16C results in the appearance of a negative ellipticity band at 289 nm and a shoulder around 257 nm (Figure 5a). Although the pattern of transitions (+,−,−) is not the same, the energy of the transitions might be considered as a signature of Hg(SR)42− species of Hg(II) substituted rubredoxin.32 Therefore, this indicates the formation of a distorted tetrahedral Hg(II) thiolate complex that reaches its maximum concentration at a 0.5 [Hg(II)]/[peptide] ratio. Titration of subsequent aliquots of HgCl2 into the system causes the negative band to lose its intensity.
Figure 5.
Circular dichroism difference spectra of titrations of Hg(II) into the solution of 50mM CHES buffer pH 9.4 containing 90 μM TRIL12CL16C (a) and 90 μM TRIL9CL12C (b).
Spectra for the titration of HgCl2 into a buffered solution of TRIL9CL12C indicate the presence of different species (Figure 5b). At very low concentration of Hg(II), the formation of a trigonal Hg(SR)3− is observed. However, this species is not similar to those formed for TRIL12C and TRIL16C as is corroborated by a different pattern of LMCT transitions.29 Although the transition at around 250 nm appears, its intensity is over 50 times lower than would be expected for a Hg(SR)3− system. When we increase the Hg(II) concentration, we are able to observe further changes leading to the appearance of two positive bands around 294 nm and 275 nm as well as a negative band at 253 nm. The band at 294 nm reaches its maximum intensity at a 0.5 [Hg]/[TRIL9CL12C] ratio, whereas the bands at 275 nm and 253 nm maximize at a 0.6 ratio. This observation suggests the presence of two complexes, pseudotetrahedral reflected by the transition at 294 nm, and distorted trigonal, reflected by the presence of the band at 275 nm. Correlating the data with the analytical centrifugation results obtained for TRIL9CL12C in the presence of Hg(II) suggests these two compounds correspond to two-stranded and three-stranded coiled coils. The pattern of transitions is very similar to those observed for mercury(II) substituted rubredoxin.32
NMR
Both 113Cd and 199Hg NMR are powerful tools that can provide detailed information on the structure of complexes formed in solution since there is a strong correlation between particular donor sets (coordination number, ligand type and coordination geometry) and the chemical shift.
113Cd NMR
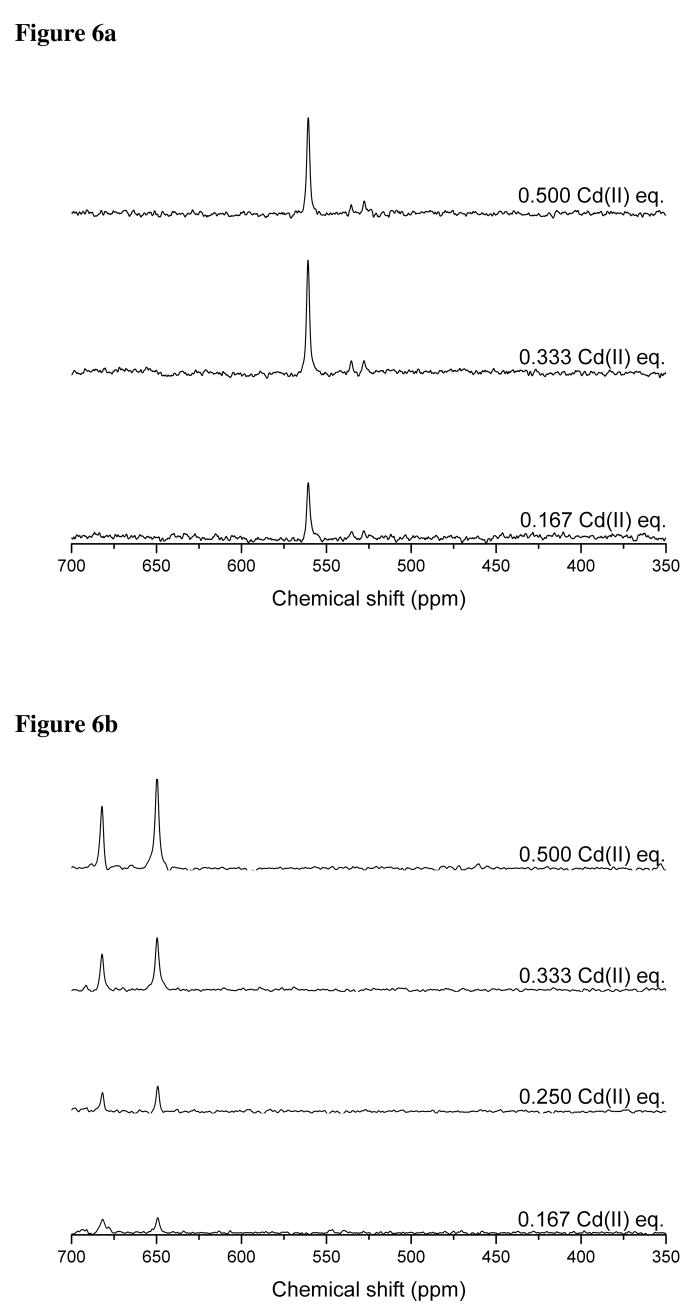
Titration of TRIL12CL16C with aliquots of 113Cd(II) at pH 8.5 indicates the formation of a single species that reaches its maximum concentration at a 2:1 ligand to metal ratio. This stoichiometry indirectly indicates that, in the presence of Cd(II), the peptide assembles into a two stranded coiled coil. The observed single resonance in the 113Cd NMR spectra (Figure 6a) with chemical shift of 560 ppm excludes formation of a homoleptic four-coordinate species involving four sulfur atoms in the coordination environment of the Cd(II). Moreover, the presence of trigonal Cd(SR)3− complexes, that were previously observed for mono- and disubstituted TRI peptides, is also unlikely.16,17 The observed chemical shift is in the range of values already reported for distorted tetrahedral complexes where an S3O coordination mode of Cd(II) is proposed.16 This assignment is also consistent with theoretically calculated values.35
Figure 6.
113Cd NMR spectra of subsequent titrations of 113Cd(II) into the solution containing 10 mM TRIL12CL16C (a) and 10 mM TRIL9CL12C (b) at pH 8.5.
The collected electronic spectra suggested that the coordination properties of TRIL9CL12C, which contains a -Ca-X-X-Cd- motif, are different than those found for TRIL12CL16C. This conclusion is further supported by titration experiments of TRIL9CL12C using 113Cd NMR spectroscopy (Figure 6b) which show the formation of two complexes that have very similar chemical shifts (650 ppm and 680 ppm). These chemical shift values are different from those found for Cd(II) bound to TRIL12CL16C; however, there are similarities in the binding of Cd(II) between TRIL12CL16C and TRIL9CL12C. For example, the Cd(II) to peptide ratio at which the amplitude of 113Cd NMR peaks reaches its maximum is the same. The fact that the amplitude is the highest at half saturation of the peptide with the metal indicates that the two-stranded coil-coil is the major species in solution, and is accompanied by a lesser concentration of three-stranded coiled coil. Reported chemical shifts signify formation of a Cd(II) species having three or four thiolate sulfur atoms bound to the metal ion. Furthermore, based on the chemical shift range, we can exclude the sole contribution of O or N ligands to the coordination sphere of Cd(II).35
199Hg NMR
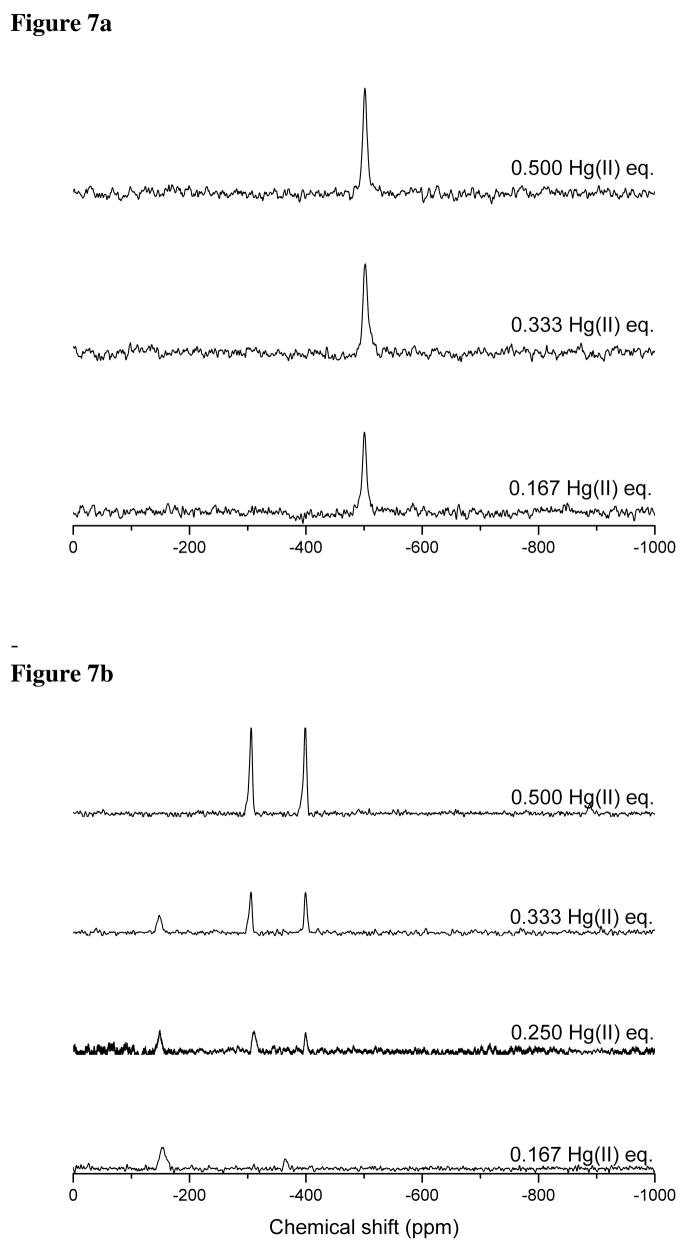
All 199Hg NMR titrations were carried out at pH 9.4 to ensure complete metal binding to our peptides. The spectra (Figure 7a) indicate that TRIL12CL16C forms a single species with Hg(II) in solution with a chemical shift of −500 ppm. Based on available literature data, this chemical shift is the signature of a Hg(II) ion bound to four thiolate sulfur atoms.36,37 It also suggests that the Hg(II) is distorted from an ideal tetrahedral geometry, since the observed resonance is shifted toward higher field in comparison to values found for Hg(II) substituted rubredoxin38 and those generated in situ.39
Figure 7.
199Hg NMR spectra of subsequent titrations of 113Hg(II) into the solution containing 10 mM TRIL12CL16C (a) and 10 mM TRIL9CL12C (b) at pH 9.4.
The 199Hg(II) NMR spectrum of a solution containing TRIL9CL12C is dependent on the mercury(II) to peptide ratio (Figure 7b). At very low metal/peptide ratios, i.e. 0.167/1, we are able to detect species with chemical shifts at −149 ppm and −365 ppm. These data correlate well with chemical shifts of previously described complexes of the TRI family peptides, where Hg(II) was bound within a three-stranded coiled coil scaffold to three sulfur atoms of a cysteine residues in a (−179 ppm) and d (−316 ppm) sites, respectively.7 Hence, at very low metal concentrations, a Hg(SR)3− coordination mode is the dominant structure for TRIL9CL12C complexes. The relative ratio of the −149 ppm and −365 ppm resonances is also consistent with the previous observation that Hg(II) prefers to form trigonal structures in a layers (TRI L9C) rather than d layers (TRI L12C). When we add subsequent aliquots of 199Hg(NO3)2, there is a reduction of the signal at −149 ppm and loss of the −365 ppm resonance. Two new species appear, having identical intensities, with chemical shifts of −308 ppm and −400 ppm. The amplitude of these peaks reaches maximum intensity at a 0.5 Hg(II)/TRIL9CL12C molar ratio. A further increase of Hg(II) concentration up to 1 equivalent of peptide results in the appearance of new species with chemical shifts of −876 ppm (indicating the formation of 2-coordinate linear Hg(II) complexes).
Perturbed Angular Correlation (PAC) spectroscopy
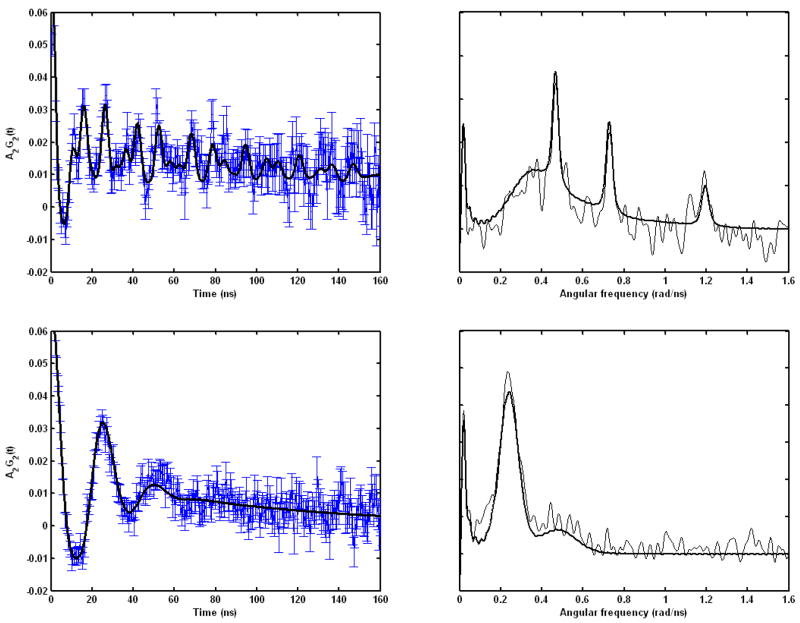
PAC spectroscopy was used to determine the cadmium coordination geometry within the coiled coils formed by TRIL12CL16C and TRIL9CL12C. A description of this technique is provided in Hemmingsen et al.40 and Matzapetakis et al.29 The measured and fitted perturbation functions and Fourier transforms of these spectra are shown in Figure 8, and in Table 4 the parameters fitted to the PAC data are presented.
Figure 8.
111mCd PAC data for the Cd(II) complexes of TRIL12CL16C (upper) and TRIL9CL12C (lower) at pH 8.5. The experimental perturbation function and the fit (full line) and the Fourier transform of the experimental data (thin line) and of the fit (thick line) are shown on the left and the right, respectively.
Table 4.
Parameters fitted to PAC-data. The numbers in parenthesis are the standard deviations of the fitted parameters.
| Peptide | [Cd(II)]/[peptide] | pH | ω0 (rad/ns) | η | Δω0/ω0 ×100 | 1/τc us−1 | A ×100 | χr2 |
|---|---|---|---|---|---|---|---|---|
| TRIL12CL16C | 0.5 | 8.5 | 0.380(1) 0.270(14) |
0.486(5) 0.50(7) |
0.8(5) 28(4) |
11(3) 2.4(7) |
2.9(3) 5.4(8) |
1.04 |
| TRIL9CL12C | 0.5 | 8.5 | 0.14(4) | 1(1) | 17(1) | 11(1) | 6.9(2) | 1.07 |
The results obtained for Cd(II) complexes of TRIL12CL16C reveal one clear and well defined nuclear quadrupole interaction (NQI1) with a relatively high ω0 of 0.380 rad/ns and an intermediate η. PAC data recorded previously for the TRIL12AL16C peptide gave a signal in the same spectral region, with ω0 of 0.340 rad/ns and low η, which was interpreted as an S3O coordination geometry.16 The higher value of η for TRIL12CL16C indicates that the structure is more distorted from a perfect tetrahedron, and this may also account for the higher ω0, although other interpretations are possible. The relatively small frequency spread Δω0/ω0 supports the existence of a well defined rigid coordination geometry. However, this species only accounts for roughly half the total amplitude of the PAC signal. Thus, there is one or more NQIs (and thereby coordination geometries) present. In the Fourier transformed data a broad signal is visible at frequencies from just below the most prominent peak (the latter belongs to NQI1). This signal has a peculiar shape that might indicate that it consists of several NQIs. It is, however, possible to fit it with just one NQI (NQI2) with a very high frequency spread, and this fit is presented in table 4. It should be noted that the high frequency spread probably makes the fitted amplitude of this signal too high. Thus, the relative amplitudes for this experiment are not as reliable as the standard deviations presented in the table.
Spectroscopic data recorded for Cd(II) complexes of TRIL9CL12C at a 1:2 [Cd(II)]/[peptide] ratio can be fitted with just one NQI with relatively low ω0 and large frequency spread (Δω0/ω0). Most importantly, the NQI is clearly different than those obtained for TRIL12CL16C. The large frequency spread indicates qualitatively that the there is a variation in structure from one peptide-Cd(II) complex to the next, i.e. a flexible metal ion binding site or even two or more binding sites with similar coordination geometries. Nevertheless, the low frequency of the PAC signal excludes the presence of S3 type complexes and indicates an S4 complex species as the best candidate.
Discussion
Recent studies have shown that the interaction of Cd(II) with proteins and peptides that naturally bind Zn(II) can provide valuable information about the toxicological roles of the former metal. Cd(II) can also be used to mimic the interaction of proteins with their natural cofactors in case direct analysis is challenging. Similar statement may apply to Hg(II) interaction with proteins that play a role in Cu(I) metabolism. Cd(II) and Hg(II) has been used to model the interaction of Fe(II) with rubredoxin, a protein involved in electron transfer.31,38,41–43
Cd(II) and Hg(II) have distinct geometrical preferences when they bind to sulfur. For Cd(II) the most common coordination environment is tetrahedral or octahedral, whereas Hg(II) prefers a linear binding mode. Our group has shown previously that peptidic systems can be used to alter the coordination environment of Cd(II) and Hg(II) by enforcing an unusual coordination mode. In the last few years we have focused on the peptidic system designed based on TRI peptide sequence. Direct substitution of Leu residues with Cys have allowed us to construct thiolate-rich metal binding sites that were able to sequester Cd(II) forming a mixture of Cd(SR)3− and Cd(SR)3O− complexes. We were able to design peptides that specifically bind Cd(II) either with fully S3 or S3O configurations by introducing bulky Pen residues in the Leu position or substituting Leu layer right above Cys with the less bulky Ala, respectively.16,17 These designed systems were able to bind Hg(II) with an unusual trigonal environment.7,34,44 Recently we have been interested in the design of peptidic systems that would allow for formation of homoleptic M(SR)42− complexes, with both Cd(II) and Hg(II).
We and others have used a de novo protein design strategy to examine the fundamental factors that are important for understanding the structure and function of metalloproteins. Despite the fact that a number of peptidic systems have been designed to mimic almost all structural motifs found in the majority of proteins (e.g., α-helices,8,45–47 β-sheets,48–50 and α-helical coiled-coils51–53), the α-helical coiled coils have become the main targets for metalloprotein mimics. This preference is due to a better understanding of the structural requirements for α-helical coil and bundle formation (although a few examples of structural motifs other than α-helices are known where soft metal binding sites are created by introduction of Cys or Pen residues into peptide sequence54–56). There are numerous examples of helical constructs containing hemes, binuclear metal clusters and monothiolate sites for soft metal ions like Cd(II), Hg(II), Pb(II), Bi(III), As(III)16–18,20,57–59 as well as first row transition metals like Cu(I).19 Recently helical peptides that contain the C-X-X-C motif have been studied.
Ogawa et al have studied this sequence in their design of the C16C19-GGy peptide, that in the presence of Cd(II) assembles exclusively into two-stranded coiled coils, thus forming a Cd(SR)42− species.20 One should realize that for C16C19-GGy metal binding is absolutely essential for coiled coil assembly. Addition of additional aliquots of Cd(II), up to 0.5 equivalents, resulted in an increase of the charge transfer band at 238 nm indicating metal binding. This is accompanied by a substantial change in the CD spectra, which reveals the formation of α-helical structure.20 The peptidic dimer sequesters one Cd(II) engaging four thiolates on metal binding. Ogawa et al also used the same peptidic system to generate four-stranded coiled coils capable of encapsulating a Cu4S4 cluster in the presence of Cu(I) ions.19
A different approach has been applied by Gibney at al, who utilizes protein maquettes, a synthetic analogs of natural metalloproteins, to examine the consequences of using noncoded amino acids.55 The comparison of Co(II) binding properties of cysteine and corresponding penicillamine derivatives indicates that the steric hindrance, introduced by the presence of methyl substituents, determines the chemical nature of the complexes formed in solution. IGA, the cysteine containing maquette, forms pseudotetrahedral Co(SR)42− complexes, whereas IGA-Pen forms CoS3O complexes. Interestingly, substitution of penicillamine for cysteine in the TRI peptides (e.g., TRI L16C to TRI L16Pen) causes a decreased Cd(II) coordination number.16
It is well known that C-X-X-C sequences are formidable metal binding motifs. Our aim in this study was to design Ca-X-X-Cd and Cd-X-X-X-Ca sequences incorporated into the TRI architecture that binds Cd(II) and Hg(II) ions with high affinity. Our objective is to compare the chemistry of metal binding to cysteines in a to d (TRI L9CL12C) versus d to a layers (TRI L12CL16C). Therefore, we synthesized TRIL9CL12C and TRIL12CL16C, which consist of two subsequent leucines residues substituted with Cys residues, in an effort towards achieving homoleptic MS4 complexes.
As the C16C19-GGy system warned, disruption of two adjacent hydrophobic layers can markedly impact the stability and folding propensity of a designed peptide. Therefore, it was important that we assess the impact of changing successive hydrophobic layers to cysteines in the TRI peptides. According to analytical centrifugation data, oligomerization state preferences of disubstituted TRI peptides with no intervening leucines depend on the presence or absence of metals. Comparison of molecular weight histograms (supplementary material, Figure S1) for TRIL12CL16C, TRIL9CL12C and their Cd(II) and Hg(II) complexes reveals a difference in oligomerization state preference due to metal binding. Unlike C16C19-GGy which require metals to fold, both TRI peptides preferentially associate into two-stranded coiled coils in the absence of metals. However, this is a change in association from apoTRI L12C and other monocysteine substituted apoTRI peptides that form three-stranded coiled coils. This tendency is more evident for TRIL12CL16C than TRIL9CL12C, which possesses some limited ability to form three-stranded coiled coils. Metal binding changes these preferences, especially in the case of Hg(II) complexes of TRIL9CL12C, where the metal induces the transition from the two-stranded coiled coil to the three-stranded coiled coil. This transition is also observed for Cd(II) complexes of this TRI peptide derivative, however, the two-stranded coiled coil still remains the dominant species in solution. The preferred aggregation state of TRIL12CL16C, regardless of the presence of metal ions, is a two-stranded coiled coil. Although both Cd(II) and Hg(II) increase the relative concentration of three-stranded coiled coil, we do not observe a noticeable change of association preferences.
We have shown using monosubstituted TRI peptides that the stability of the metal peptide complex correlates directly with the peptide self association affinity.60 The present system is more complex as metal binding shifts the equilibrium between 2- and 3- stranded coiled coils. None-the-less, one would expect that there would be enhanced metal binding to disubstituted peptides as the self association affinity increases.
The perturbations to the preferred aggregation states compared to TRI, all monosubstituted TRI and some disubstituted TRI peptides are probably related to the presence of two adjacent layers of cysteine residues that disrupt the net hydrophobic interactions causing attenuation of association forces between strands. The minor aggregation state difference between TRIL12CL16C and TRIL9CL12C is related to location of substituted residues in the primary structure of the peptide. In the former system, substitutions are made in the center of the primary structure, whereas in the latter they are located closer to the N-terminus. Hence, we suppose that substitution of leucines within the central part of the peptide results in much wider disruption of hydrophobic interactions, consequently leading to formation of two-stranded coiled coils that are not strong enough to associate with a third strand at a higher pH range.15 That allows the formation of wider cavities with a more hydrophilic environment around the coordinating metal ion. Such circumstances favor deprotonation of Cys at lower pH. The relatively low value of the apparent pKa of single Cd(II) ion binding observed for the complexes of both peptidic systems supports this hypothesis.
Both TRIL12CL16C and TRIL9CL12C display unusual coordination properties towards soft metal ions like Cd(II) or Hg(II) as compared to other mono- and di-substituted TRI peptides. Formation of thiol rich environments is a consequence of the association of two or three strands, providing a variety of possible anchoring sites for Cd(II) and Hg(II) ions. Therefore, the structure of the complexes depends on the metal ion concentration with respect to the peptide.
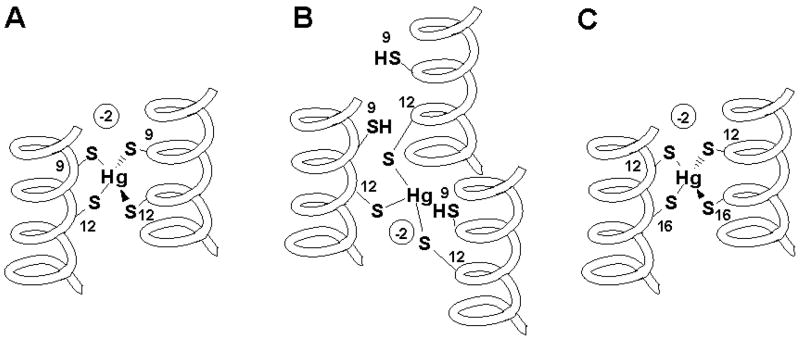
TRIL12CL16C binds Cd(II) within a two-stranded coiled coil scaffold to form a Cd(SR)3O complex (Scheme 2a) reaching its maximum concentration at 0.5 Cd(II) equivalents. This assignment is confirmed by the spectroscopic signatures of this chromophore including the 560 ppm 113Cd NMR chemical shift and the signal with ω0 = 0.380 rad/ns in the PAC spectra. Furthermore, the CD and UV-vis spectra are consistent with previously characterized S3O species formed in related systems. The PAC and NMR parameters support the existence of a distorted S3O donor set. Analysis of the PAC data indicates the presence of a second species, that is likely undergoing extremely fast water exchange, which is not detected by 113Cd NMR. We believe this is likely a Cd(SR)2O2 species; however, definitive assignment of this structure will require additional study.
Scheme 2.
Schematic representation of Cd(II) complexes of TRIL12CL16C (A) and TRIL9CL12C (B,C).
It is particularly interesting to compare the spectral parameters of TRIL12CL16C with Cd(II) that of the DNA binding repressor protein CmtR which can be isolated from Mycobacterium tuberculosis. Comparison of the spectroscopic signatures of Cd(II)(TRIL12CL16C)2 to Cd(II) complexes formed CmtR regulatory protein, where formation of trigonal thiolate Cd(II) species within –C-X-X-X-C- sequence has been proposed, reveals some similarity.30 CmtR is arranged in dimers in both apo and Cd-bound form.61 Binding of Cd(II) on the dimer interface, engaging Cys-57, Cys-61 from one subunit and Cys-102 from the other, reduces the conformational heterogeneity and locks the two subunits into a better defined reciprocal orientation. As a consequence, CmtR looses its ability to associate with DNA. Although the orientation of donor atoms is not identical, Cys-61 is at the end of the helix and Cys-102 is in a flexible loop, the first coordination sphere environments appear closely similar. Most important, the designed peptide with the C-X-X-X-C sequence matches much more closely the physical parameters of CmtR (also containing a C-X-X-X-C sequence) than does the more common C-X-X-C motif found in TRIL9CL12C. As it has been synthetically challenging to prepare stable Cd(II)S3 type structures in aqueous solution, one may conclude that TRIL12CL16C may be considered the most accurate structural model of the Cd(II) coordination environment of CdCmtR.
TRIL9CL12C (-Ca-X-X-Cd-) reveals different Cd(II) binding properties than TRIL12CL16C (–Cd-X-X-X-Ca-). There are two major differences between these peptides. The first difference is the relative aggregation state preferences discussed above. The second perturbation is the orientation of the cysteines from one layer to the next. Based on molecular models of a-helices, in the -Ca-X-X-Cd- peptide, the cysteines are closer to one another and the α-carbon position of the cysteine in the a position is rotated sixty degrees out of phase with respect to the d site cysteine. In contrast, the additional amino acid inserted into the -Cd-X-X-X-Ca- sequence extends the distance between layers and leads to a 40° rotation of the respective cysteine α-carbons. Both of these factors conspire against an optimal tetrahedral binding site. Thus, TRIL12CL16C yields an S3O structure. The coordination chemistry of TRIL9CL12C is dominated by having an optimal tetrahedral type binding site and the relative presence of two versus three-stranded coiled coils available for metal binding. Because the initial equilibrium contains three stranded coiled coils, the first aliquots of Cd(II) ion bind to four of six thiolate sulfurs (Scheme 2b) giving a first inflection point at 0.33 ratio metal to peptide ratio. However, with an increasing concentration of CdCl2 the aggregation state equilibrium shifts towards the two-stranded coiled coil scaffold (Scheme 2c). In this case, the available binding site transforms into the rubredoxin like spectra at 0.6 Cd(II) equivalents with the maximum absorption bands at 227 nm and 245 nm. The change of peptide aggregation state does not modify the nature of the complexes. The two signals in 113Cd NMR at 650 and 680 ppm, which were observed when the first aliquot of Cd(II) was added to the peptide solution, do not shift. Structural assignment based solely on 113Cd NMR can be ambiguous in this case, since these chemical shifts values can originate from either trigonal planar or tetrahedral complexes. Therefore, we analyzed the complexes by PAC spectroscopy. The PAC data reveal that both complexes are different forms of the same distorted tetrahedral complex engaging four thiolate sulfur atoms within the primary coordination sphere of the Cd(II) ion. The difference in chemical shift derives from small geometric changes in the coordination environment resulting from donor orientation in the two-stranded or three-stranded coiled coils.
The same structural and aggregation state considerations that were mentioned above define the binding of Hg(II) to the peptides. Furthermore, one must consider the binding preferences for metals within an individual layer as Hg(II) prefers lower coordination number complexes. In general, Hg(II) prefers forming linear, two-coordinate complexes in d layers, but can be forced into trigonal structures at high pH. In contrast, trigonal planar Hg(II) complexes in a layers form easily and at lower pH.
Combining these structural preferences provides an explanation for the Hg(II) binding behavior of both disubstituted peptides. The apoTRIL12CL16C exists predominantly as two-stranded coiled coils and forms simpler complex equilibria than apoTRIL9CL12C which exist as a mixture of two and three-stranded coiled coils. This is apparent at low Hg(II)/peptide ratio, at which trigonal complexes of TRIL9CL12C are formed. The presence of two peaks at −149 ppm and −365 ppm in 199Hg NMR spectra indicates formation of trigonal complex species involving mainly a and d cysteine residues, respectively. For comparison, the 199Hg NMR shifts for trigonal Hg(II) in the a layer peptide Hg(TRIL16C)3− and the d layer peptide Hg(TRIL12C)3− are −179 ppm and −316 ppm, respectively. As expected, the signal amplitude indicates that the trigonal complex formed in the a layer is the major species in solution.29 A further increase of Hg(II) concentration changes the complex speciation. Two maxima at 0.5 and at 0.6 equivalents of Hg(II) are observed in the circular dichroism spectra. This may suggest that two independent transition processes take place and that two independent complex species exist in solution. UV absorption bands at 284 nm and 250 nm also may indicate formation of two types of complexes in solution, pseudotetrahedral and trigonal, respectively. For the former species, the geometry around the metal ion is similar to that reported for Hg(II) substituted rubredoxin (Scheme 3a). The latter one corresponds to Hg(SR)3− complexes similar to those found for monosubstituted derivatives of TRI peptides (Scheme 3b). The existence of two species is confirmed by the presence of two peaks in the 199Hg NMR spectra with chemical shifts of −400 ppm and −308 ppm. When the concentration of Hg(II) exceeds a 0.5 metal to peptide ratio, formation of linear HgS2 complexes may be realized with the characteristic 199Hg NMR chemical shift at −876 ppm.
Scheme 3.
Schematic representation of Hg(II) complexes of TRIL9CL12C (A,B) and TRIL12CL16C (C).
Different coordination properties were observed for TRIL12CL16C, and are the consequence of the altered orientation of the thiolates, as well as the peptide framework consisting predominantly of a two-stranded coiled coil. Subsequent additions of HgCl2 lead to formation of a distorted tetrahedral complex using four thiolate sulfurs in metal binding. This is reflected by the presence of a band at 289 nm both in the electronic and circular dichroism spectra, reaching its maximum intensity at 0.5 equivalent of Hg(II) vs. TRIL12CL16C. It is interesting that the observed ellipticity of the -Cd-X-X-X-Ca- sequence has the opposite sign to that observed for the TRIL9CL12C system (vide supra). In addition, the difference between the two systems can be observed in their electronic spectra (the 250 nm band is absent for TRIL12CL16C). Therefore, we conclude that the coordination geometry around the metal ion is different to that of Hg(II) substituted rubredoxin (Scheme 3c). In addition a signal at −500 ppm in the 199Hg NMR spectra indicates the presence of a Hg(SR)42− species.36,37
Importantly, we are able to obtain nearly identical metal based spectroscopic signatures to that of metal substituted rubredoxin with TRIL9CL12C, despite the fact that our models are based on an α-helical framework and rubredoxin consists predominantly of a β-sheet fold with the metal binding site formed within a peptidic loop. Thus, we can conclude that the spectroscopic signatures of Hg(II) and Cd(II) substituted rubredoxins are primarily a consequence of the metal first coordination sphere and not the protein fold.
Conclusions
In this report, we have compared the peptide association affinities and metal binding propensities of designed peptides containing the common C-X-X-C binding motif and the less common C-X-X-X-C sequence. Both peptides represent opposite configurations of binding thiolates within a two-stranded coiled coil framework, a followed by d for TRIL9CL12C, compared to d followed by a for TRIL12CL16C. Our attempt to synthesize the homoleptic pseudo-tetrahedral complexes of Cd(II) was successful in the case of TRIL9CL12C that incorporates the -C-X-X-C- sequence while TRIL12CL16C, incorporating the uncommon -C-X-X-X-C- sequence, was unable to form homoleptic complexes with Cd(II). In this case, formation of a Cd(SR)3O− complex was observed. This dissimilarity is due to differences in the orientation of the binding thiolate ligands and the distances of the sulfurs within the hydrophobic core. In contrast, when considering Hg(II) complexation, both double substituted peptides are able to form Hg(SR)42− complexes. For TRIL9CL12C, however, the simultaneous formation of an Hg(SR)3− species is also observed. The different metal binding behavior of these peptides is due to their different self association affinities with respect to the formation of three-stranded coiled coils. The major factor that determines the degree of destabilization is the location of the substitutions. In general, the closer to the center, the weaker is the peptide ability to bind the third strand. Therefore, one may observe limited ability of TRIL9CL12C with N-terminal side substitution to associate with a third strand at basic pH, whereas no three-stranded coiled coil formation occurs for TRIL12CL16C. In both cases, the peptides ability to form three-stranded coiled coils is recovered by the binding of metal ions. Therefore, they may be considered as good models for the interaction of these metal ions with native proteins having tetrathiolate metal binding sites. As a matter of fact, although the tertiary structure of the TRI peptides is different from that of rubredoxin, their Cd(II) and Hg(II) complexes have very similar spectroscopic signatures.
Supplementary Material
Listings of Molecular weight histograms representing the molecular weight distribution between monomer, dimer and trimer for TRIL12CL16C, TRIL9CL12C, and their Cd(II) and Hg(II) complexes. This material is available free of charge via the Internet at http://pubs.acs.org.”
Acknowledgments
The authors thank Dr. A.F.A. Peacock for assistance in assemblying the data in tables 2 and 3. Vincent L. Pecoraro would like to thank the National Institute of Health (Grant R01 ES012236) for support of this work. Borries Demeler thanks the National Institute of Health (Grant R01RR022200-01A1), National Science Foundation (Grant TG-MCB070038), National Cancer Institute (CA54174) and UTHSCSA ERC for supporting the Center for Analytical Ultracentrifugation of Macromolecular Assemblies. Lars Hemmingsen thanks The Danish research council for Nature and Universe and EURONS for support of this work, and CERN/ISOLDE for beam time (IS448).
References
- 1.Lombardi A, Marasco D, Maglio O, Di Costanzo L, Nasti F, Pavone V. Proc Natl Acad Sci, U S A. 2000;97:11922–11927. doi: 10.1073/pnas.97.22.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nastri F, Lombardi A, Pavone V. Chem Rev. 2001;101:3165–3189. doi: 10.1021/cr000055j. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh D, Pecoraro VL. Inorg Chem. 2004;43:7902–7915. doi: 10.1021/ic048939z. [DOI] [PubMed] [Google Scholar]
- 4.Doerr AJ, McLendon GL. Inorg Chem. 2004;43:7916–7925. doi: 10.1021/ic0490573. [DOI] [PubMed] [Google Scholar]
- 5.DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. Annu Rev Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 6.Lombardi A, Summa CM, Geremia S, Randaccio L, Pavone V, DeGrado WF. Proc Natl Acad Sci, USA. 2000;97:6298–6305. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieckmann GR, McRorie DK, Tierney DL, Utschig LM, Singer CP, O’Halloran TV, Penner-Hahn JE, DeGrado WF, Pecoraro VL. J Am Chem Soc. 1997;119:6195–6196. [Google Scholar]
- 8.Hodges RS. Biochem Cell Biol. 1996;74:133–154. doi: 10.1139/o96-015. [DOI] [PubMed] [Google Scholar]
- 9.Gibney BR, Rabanal F, Skalicky JJ, Wand AJ, Dutton PL. J Am Chem Soc. 1999;121:4952–4960. [Google Scholar]
- 10.Lupas AN, Gruber M. In: Fibrous Proteins: Coiled-Coils, Collagen and Elastomers. David AD, Parry aJMS, editors. Vol. 70. Academic Press; 2005. pp. 37–38. [Google Scholar]
- 11.Venkatraman J, Naganagowda GA, Sudha R, Balaram P. Chem Commun. 2001:2660–2661. [Google Scholar]
- 12.Cox AAMM, Mayo KH. Biochem J. 2001;357:739–747. doi: 10.1042/0264-6021:3570739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodges R, Saund A, Chong P, St-Pierre S, Reid R. J Biol Chem. 1981;256:1214–1224. [PubMed] [Google Scholar]
- 14.Silverman JA, Balakrishnan R, Harbury PB. PNAS. 2001;98:3092–3097. doi: 10.1073/pnas.041613598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dieckmann GR, McRorie DK, Lear JD, Sharp KA, DeGrado WF, Pecoraro VL. J Mol Biol. 1998;280:897–912. doi: 10.1006/jmbi.1998.1891. [DOI] [PubMed] [Google Scholar]
- 16.Lee KH, Cabello C, Hemmingsen L, Marsh ENG, Pecoraro VL. Angew Chem, Int Ed. 2006;45:2864–2868. doi: 10.1002/anie.200504548. [DOI] [PubMed] [Google Scholar]
- 17.Iranzo O, Cabello C, Pecoraro VL. Angew Chem, Int Ed. 2007;46:6688–6691. doi: 10.1002/anie.200701729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matzapetakis M, Pecoraro VL. J Am Chem Soc. 2005;127:18229–18233. doi: 10.1021/ja055433m. [DOI] [PubMed] [Google Scholar]
- 19.Kharenko OA, Kennedy DC, Demeler B, Maroney MJ, Ogawa MY. J Am Chem Soc. 2005;127:7678–7679. doi: 10.1021/ja042757m. [DOI] [PubMed] [Google Scholar]
- 20.Kharenko OA, Ogawa MY. J Inorg Biochem. 2004;98:1971–1974. doi: 10.1016/j.jinorgbio.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 21.Ellman GL. Arch Biochem and Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 22.Demeler B. University of Texas Health Science Center; San Antonio, Dept. of Biochemistry: UltraScan 7.2 - An integrated data analysis software package for sedimentation experiments. http://www.ultrascan.uthscsa.edu. [Google Scholar]
- 23.Laue TMDSB, Ridgeway TM, Pelletier SL. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Horton JC, editor. Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
- 24.Durchschlag H. In: Thermodynamic Data for Biochemistry and Biotechnology. Hinz H-J, editor. Springer-Verlag; New York: 1986. pp. 45–128. [Google Scholar]
- 25.Wrackmeyer B, Contreras R. Annu Rep NMR Spectrosc. 1992;124:267–329. [Google Scholar]
- 26.Hemmingsen LBR, Bjerrum M, Adolph H, Zeppezauer M, Cedergren-Zeppezauer E. Eur J Biochem. 1996;241:546–551. doi: 10.1111/j.1432-1033.1996.00546.x. [DOI] [PubMed] [Google Scholar]
- 27.Demeler B, Brookes E. Colloid Polym Sci. 2008;286:129–137. [Google Scholar]
- 28.Demeler B. In: Modern Analytical Ultracentrifugation: Techniques and Methods. Scott DJ, Harding SE, Rowe AJ, editors. Royal Society of Chemistry; UK: 2005. pp. 210–229. [Google Scholar]
- 29.Matzapetakis M, Farrer BT, Weng TC, Hemmingsen L, Penner-Hahn JE, Pecoraro VL. J Am Chem Soc. 2002;124:8042–8054. doi: 10.1021/ja017520u. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Hemmingsen L, Giedroc DP. Biochemistry. 2005;44:8976–8988. doi: 10.1021/bi050094v. [DOI] [PubMed] [Google Scholar]
- 31.Henehan CJ, Poutney DL, Zerbe O, Vasak M. Protein Sci. 1993;2:1756–1764. doi: 10.1002/pro.5560021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faller P, Ctortecka B, Troger W, Butz T, Vasak M. J Biol Inorg Chem. 2000;5:393–401. doi: 10.1007/pl00010668. [DOI] [PubMed] [Google Scholar]
- 33.Chen YH, Yang JT, Chau KH. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 34.Farrer B, Pecoraro VL. Proc Natl Acad Sci, U S A. 2003;100:3760–3765. doi: 10.1073/pnas.0336055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemmingsen L, Olsen L, Antony J, Sauer SPA. J Biol Inorg Chem. 2004;9:591–599. doi: 10.1007/s00775-004-0553-0. [DOI] [PubMed] [Google Scholar]
- 36.Jalilehvand F, Leung BO, Izadifard M, Damian E. Inorg Chem. 2006;45:66–73. doi: 10.1021/ic0508932. [DOI] [PubMed] [Google Scholar]
- 37.Natan MJ, Millikan CF, Wright JG, O’Halloran TV. J Am Chem Soc. 1990;112:3255–3257. [Google Scholar]
- 38.Blake PB, Lee B, Summers MF, Park JB, Zhou ZH, Adams MWW. New J Chem. 1994;18:387–395. [Google Scholar]
- 39.Carson GK, Dean PAW. Inorg Chim Acta. 1982;66:157–161. [Google Scholar]
- 40.Hemmingsen L, Narcisz K, Danielsen E. Chem Rev. 2004;104:4027–4061. doi: 10.1021/cr030030v. [DOI] [PubMed] [Google Scholar]
- 41.Bonomi F, Iametti S, Kurtz DM, Jr, Richie KA, Ragg E. J Biol Inorg Chem. 1998;3:595–605. [Google Scholar]
- 42.Goodfellow BJ, Lima MJ, Ascenso C, Kennedy M, Sikkink R, Rusnak F, Moura I, Moura JJG. Inorg Chim Acta. 1998;273:279–287. [Google Scholar]
- 43.Lovenberg W, Sobel BE. Proc Natl Acad Sci, U S A. 1965;54:193–199. doi: 10.1073/pnas.54.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iranzo O, Thulstrup PV, Ryu SB, Hemmingsen L, Pecoraro VL. Chem Eur J. 2007;13:9178–9190. doi: 10.1002/chem.200701208. [DOI] [PubMed] [Google Scholar]
- 45.Regan L, DeGrado WF. Science. 1988;241:976–978. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- 46.Errington N, Iqbalsyah T, Doig AJ. In: Protein Design: Methods and Applications. Paz MLdl., editor. Vol. 340. Humana Press; 2006. pp. 3–26. [DOI] [PubMed] [Google Scholar]
- 47.Munoz V, Serrano L. Curr Opin Biotech. 1995;6:382–386. doi: 10.1016/0958-1669(95)80066-2. [DOI] [PubMed] [Google Scholar]
- 48.de la Paz ML, Lacroix E, Ramirez-Alvarado M, Serrano L. J Mol Biol. 2001;312:229–246. doi: 10.1006/jmbi.2001.4918. [DOI] [PubMed] [Google Scholar]
- 49.Yan YB, Erickson BW. Protein Sci. 1994;3:1069–1073. doi: 10.1002/pro.5560030709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pantoja-Uceda D, Santiveri CM, Jiménez MA. In: Protein Design: Methods and Applications. Guerois R, editor. Vol. 340. Humana Press Inc; 2006. pp. 27–52. [Google Scholar]
- 51.Baltzer L, Nilsson H, Nilsson J. Chem Rev. 2001;101:3153–3163. doi: 10.1021/cr0000473. [DOI] [PubMed] [Google Scholar]
- 52.Woolfson DN. In: Fibrous Proteins: Coiled-Coils, Collagen and Elastomers. David AD, Parry aJMS, editors. Vol. 70. Academic Press; 2005. pp. 79–112. [Google Scholar]
- 53.Mason JM, Arndt KM. ChemBioChem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 54.Kennedy ML, Petros AK, Gibney BR. J Inorg Biochem. 2004;98:727–732. doi: 10.1016/j.jinorgbio.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 55.Petros AK, Shaner SE, Costello AL, Tierney DL, Gibney BR. Inorg Chem. 2004;43:4793–4795. doi: 10.1021/ic0497679. [DOI] [PubMed] [Google Scholar]
- 56.Petros AK, Reddi AR, Kennedy ML, Hyslop AG, Gibney BR. Inorg Chem. 2006;45:9941–9958. doi: 10.1021/ic052190q. [DOI] [PubMed] [Google Scholar]
- 57.Li XQ, Suzuki K, Kanaori K, Tajima K, Kashiwada A, Hiroaki H, Kohda D, Tanaka T. Protein Sci. 2000;9:1327–1333. doi: 10.1110/ps.9.7.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matzapetakis M, Ghosh D, Weng TC, Penner-Hahn JE, Pecoraro VL. J Biol Inorg Chem. 2006;11:876–890. doi: 10.1007/s00775-006-0140-7. [DOI] [PubMed] [Google Scholar]
- 59.Touw DS, Nordman CE, Stuckey JA, Pecoraro VL. Proc Natl Acad Sci, U S A. 2007;104:11969–11974. doi: 10.1073/pnas.0701979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosh D, Lee KH, Demeler B, Pecoraro VL. Biochemistry. 2005;44:10732–10740. doi: 10.1021/bi0506674. [DOI] [PubMed] [Google Scholar]
- 61.Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Cavet JS, Dennilson C, Graham AI, Harvie DR, Robinson NJ. J Biol Chem. 2007;282:30181–30188. doi: 10.1074/jbc.M701119200. [DOI] [PubMed] [Google Scholar]
- 62.Farrer BT, Harris NP, Balchus KE, Pecoraro VL. Biochemistry. 2001;40:14696–14705. doi: 10.1021/bi015649a. [DOI] [PubMed] [Google Scholar]
- 63.Dieckmann GR. PhD thesis. University of Michigan; Ann Arbor, MI: 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Listings of Molecular weight histograms representing the molecular weight distribution between monomer, dimer and trimer for TRIL12CL16C, TRIL9CL12C, and their Cd(II) and Hg(II) complexes. This material is available free of charge via the Internet at http://pubs.acs.org.”