Summary
Aberrations in chromatin dynamics play a fundamental role in tumorigenesis, yet relatively little is known of the molecular mechanisms linking histone lysine methylation to neoplastic disease. ING4 (Inhibitor of Growth 4) is a native subunit of an HBO1 histone acetyltransferase (HAT) complex and a tumor suppressor protein. Here we show a critical role for the specific read-out of histone H3 trimethylated at lysine 4 (H3K4me3) by the ING4 PHD finger in mediating ING4 gene expression and tumor suppressor functions. The interaction between ING4 and H3K4me3 augments the acetylation activity of HBO1 on H3 tails, and drives H3 acetylation at ING4 target promoters to effect a DNA damage-dependent gene expression program. Further, ING4 facilitates apoptosis in response to genotoxic stress and inhibits anchorage-independent cell growth, and these functions are dependent on ING4 interactions with H3K4me3. Together, our results demonstrate a mechanism for brokering crosstalk between H3K4 methylation and histone H3 acetylation, and reveal a new molecular link between chromatin modulation and tumor suppressor mechanisms.
Introduction
ING4, a member of the ING family of type II tumor suppressor proteins, harbors several anti-cancer activities such as inhibiting angiogenesis and cell proliferation and promoting cell death and contact inhibition (Doyon et al., 2006; Garkavtsev et al., 2004; Kim, 2005; Kim et al., 2004; Ozer and Bruick, 2005; Shen et al., 2007; Shiseki et al., 2003). Multiple tumor cells and tissue types contain mutations within the ING4 gene, exhibit reduced ING4 expression levels, or display aberrant ING4 sub-cellular localization (Li et al., 2008; Shi and Gozani, 2005). At the molecular level, ING4 is thought to link HBO1 HAT activity to tumor suppression; however the specific mechanism by which this occurs remains obscure (Doyon et al., 2006; Iizuka et al., 2008; Shi and Gozani, 2005). HBO1 regulates S phase progression and is responsible for the bulk of acetylation on histone H4; further, ING4 is required for HBO1 to acetylate H4 and H2A on chromatin substrates (Doyon et al., 2006; Iizuka et al., 2008). Thus, one possibility is that an HBO1-ING4 complex couples histone acetylation and proper chromatin modulation to DNA replication, and that this function is important for preventing cellular transformation.
In addition to being a stable subunit of an HBO1 complex, ING4 has also been reported to repress transcription of the NF-kB and hypoxia response pathways (Colla et al., 2007; Garkavtsev et al., 2004; Ozer and Bruick, 2005). Because ING4 is associated with HAT activity, which is generally linked to transcriptional activation, the ability of ING4 to act as a repressor in these pathways may not be through histone regulation. In this regard, ING4 has been shown to directly interact with the RelA subunit of NF-kB and with Egln1/HPH2 (Hypoxia inducible factor Prolyl Hyrdoxylase 2), a negative regulator of HIF1-α (Hypoxia Inducible Factor) (Garkavtsev et al., 2004; Ozer and Bruick, 2005). However, how the binding of ING4 to these proteins mediates transcriptional repression at the molecular level is not known. One possibility is that ING4 directs HBO1 to acetylate non-histone substrates in these pathways, and that the consequence of such acetylation events is to inhibit the activity of the targeted proteins. Regardless of the molecular mechanism, the ability of ING4 to bind RelA is important for its activity to suppress angiogenesis and prevent tumor growth in mice with glioblastoma xenografts (Garkavtsev et al., 2004). Further, the role of ING4 in suppressing HIF target genes is possibly important for the ability of tumors to survive and grow under the hypoxic conditions common to the tumor microenvironment (Ozer and Bruick, 2005; Ozer et al., 2005). Thus, while there are clear links between ING4 and inhibiting the development and progression of cancers, the specific molecular mode of action underlying ING4 activity in tumor suppression remains unclear (Ozer et al., 2005).
H3K4me3 and histone acetylation are both implicated in gene activation and these marks are often enriched near transcription start sites (Bannister and Kouzarides, 2004; Sims and Reinberg, 2006). In this context, the binding of the S. cerevisiae protein Yng1 to H3K4me3 regulates stabilization of NuA3 HAT activity at target genes (Taverna et al., 2006). Whether a similar mechanism exists in mammalian gene regulation is not known. Here we have performed biochemical, structural, genomic, and functional analyses to investigate the biological consequence of methylation sensing by ING4 (Shi et al., 2006). We find that H3K4me3-recognition by an ING4-HBO1 complex drives acetylation on H3 at a set of genes in response to genotoxic stress. Further, we show that the ability of ING4 to prevent anchorage-independent growth is critically dependent on H3K4me3-recognition. Taken together, these data demonstrate a new mechanism by which crosstalk between distinct histone modifications can influence gene activation, possibly resulting in tumor suppression.
Results
The ING4 PHD finger binds selectively to H3K4me3
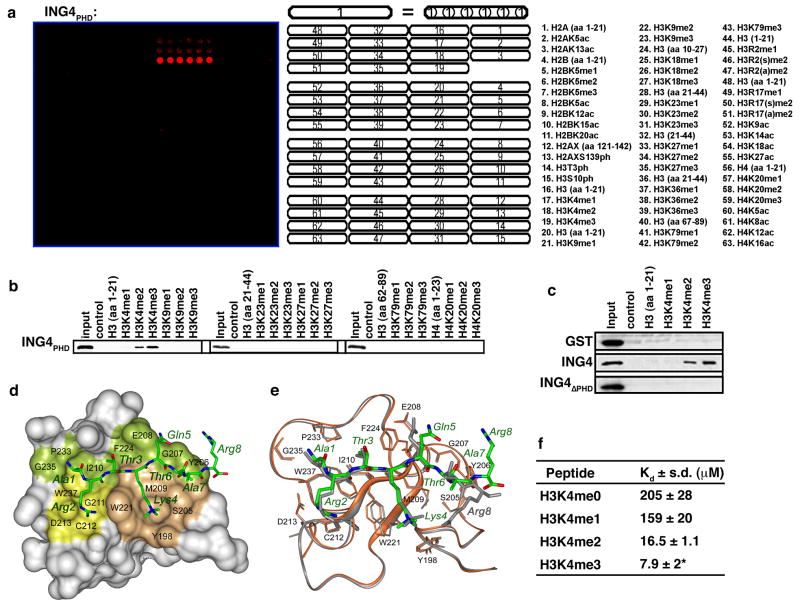
To determine the specificity of the ING4 PHD finger for H3K4me3 versus other methylation states and sites on histones, we performed an in vitro screen with recombinant ING4 PHD finger (ING4PHD: aa 195–241) on peptide arrays containing 50 distinct modified and unmodified histone peptides (Matthews et al., 2007). As shown in Figure 1a, ING4PHD binds strongest to H3K4me3 peptides, followed by H3K4me2 and H3K4me1, and is unable to recognize any other peptide present on the array. This result was also observed in peptide pull-down assays with ING4PHD (Fig. 1b). On longer exposures, binding to H3K4me1 was also observed (e.g. Fig. 2g; data not shown). Full-length ING4 recognizes methylated H3K4 peptides, but an ING4 derivative lacking the PHD finger, ING4ΔPHD (aa 1–194), does not, demonstrating that the PHD finger of ING4 is necessary and sufficient for the H3K4me peptide binding activity of ING4 (Fig. 1c).
Figure 1. The ING4 PHD finger binds specifically to H3K4me3.
(A) ING4PHD preferentially binds H3K4me3 peptides. Microarrays spotted with the indicated histone peptides (as in (Matthews et al., 2007)) were probed with glutathione S-transferase (GST) fused to ING4195–241 (ING4PHD). Red spots indicate positive binding. H3, histone H3; H4, histone H4; me, methylation; ac, acetylation; ph, phosphorylation; s, symmetric; a, asymmetric. (B) Western analysis of histone peptide pulldowns with GST-ING4PHD and the indicated biotinylated peptides. (C) Full-length ING4, but not ING4 PHD, recognizes H3K4me3. Histone peptide pulldowns as in (B) with the indicated protein. (D) 1.8 Å crystal structure of the ING4PHD-H3K4me3 complex. The PHD finger is shown as a solid surface with the binding site residues colored and labeled. H3K4 and H3R2 binding grooves are in brown and yellow. The histone peptide is shown as ball-and-stick model with C, O and N atoms colored green, red and blue, respectively. (E) Superimposition of the backbone structures of the ING4 (brown) and ING2 (gray) PHD fingers bound to H3K4me3 (green and gray stick models, respectively). (F) ING4PHD binds with highest affinity to H3K4me3. Tryptophan fluorescence was used to determine disassociation constants (Kds) for the interaction between the ING4PHD and the indicated peptides. *H3K4me3 Kd was previously determined (Pena et al., 2006).
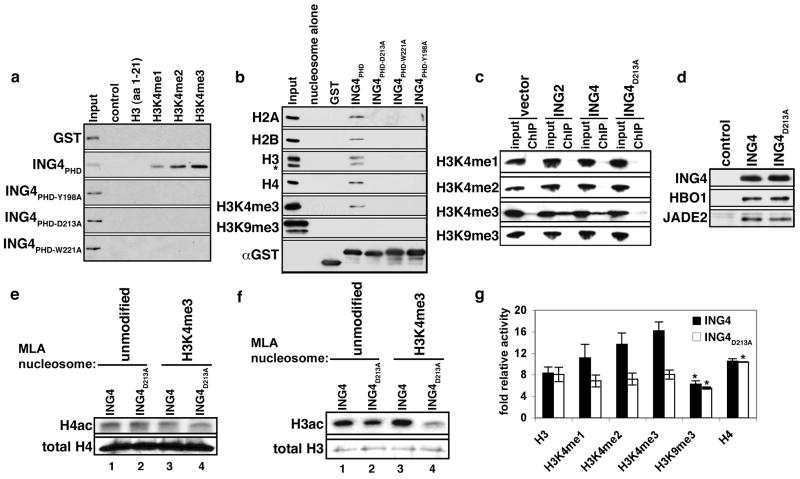
Figure 2. ING4PHD binding to H3K4me3 promotes HBO1 acetylation of histone H3.
(A) Identification of residues in the ING4 PHD finger critical for H3K4me3 binding. Western blot of histone peptide pull down assays with the indicated GST-fusion proteins and biotinylated peptides. (B) ING4PHD binding to polynucleosomes is abrogated by substitution of critical residues in the H3K4me3-binding surface. The indicated recombinant proteins were incubated with purified polynucleosomes and binding determined by Western analysis with antibodies against the four core histones and H3K4me3 and H3K9me3 antibodies. (C) The ING4 interaction with H3K4me3 occurs at chromatin in vivo and requires an intact PHD finger. Western analysis of wild-type or mutant Flag–ING4 protein–protein ChIPs. ING2 is used as a positive control. Input represents 5% of total. (D) Western analysis of affinity purified Flag-ING4 and Flag-ING4D213A complexes with the indicated antibodies. Control, empty vector IP. (E)–(F) Histone acetylation by HBO1 in wild-type, but not mutant, ING4 complexes is increased by binding to H3K4me. Autoradiograms of in vitro HAT reactions by ING4 complexes with the indicated MLA nucleosomes. Western analysis of histones is shown as a loading control. (G) Quantitation of HAT activity of ING4 and ING4D213A complexes on the indicated histone peptides from three independent experiments, except for *, which indicate two independent experiments. Error bars indicate the S.E.M.
Structural basis of H3K4me3-recognition by the ING4 PHD finger
To elucidate the molecular basis of the interaction between ING4 and H3K4me3, we determined the crystal structure of the ING4PHD-H3K4me3 complex at 1.8 Å resolution, and refined it to an Rwork of 19.9% and Rfree of 21.1% (Table 1). The protein structure consists of long loops stabilized by two zinc binding clusters and an antiparallel β sheet, which superimposes well with the structure of the ING2PHD-H3K4me3 complex (Figs. 1d and 1e) (Pena et al., 2006). In ING4, the H3K4me3 peptide adopts an extended conformation and forms characteristic β-sheet hydrogen bonds with the protein. The K4me3 sidechain is restrained by hydrophobic and cation-π interactions within a groove formed by Y198, S205, M209, and W221. The long sidechain of H3R2 extends into an adjacent groove and makes electrostatic interactions with D213 (Fig 1d). The side chains of H3R2 and H3Q5 in the ING4PHD complex are displaced 4.5 Å and 2 Å in comparison to how these residues are coordinated by ING2PHD (Figs. 1d, 1e). In addition, the ING4PHD-H3K4me3 complex has fewer hydrogen bonding contacts than the corresponding ING2PHD complex – possibly accounting for the weaker binding observed for the H3K4me3 interaction with ING4 (Kd 7.9 μM) versus ING2 (1.5 μM) (Fig. 1f; (Pena et al., 2006)). These measurements also indicate that the binding affinity of ING4PHD for the H3 tail increases concomitant with the number of methyl groups marking K4 (Fig. 1f). These results are in agreement with our array and pull-down data, but contrast with a study concluding that the extent of methylation on H3K4 did not alter affinity (Palacios et al., 2006). However, a latter study by the same group observed binding affinities similar to those reported here (Palacios et al., 2008). Based on our data, we conclude that in vitro, the ING4PHD finger preferentially binds to H3K4me3, similar to what has been observed for several H3K4me-binding PHD fingers (e.g. ING2, RAG2, BPTF, Taf3, Yng1 and others (Li et al., 2006; Martin et al., 2006; Matthews et al., 2007; Pena et al., 2006; Shi et al., 2006; Shi et al., 2007a; Taverna et al., 2006; Vermeulen et al., 2007; Wysocka et al., 2006); data not shown)).
Table 1.
Data collection and refinement statistics of the H3K4me3-bound ING4 PHD finger.
| Data Collection | ||||
|---|---|---|---|---|
| Zn MAD | ||||
| Space group | P43, a=b=68.16, c=27.96Å, α=β=γ=90°, two molecules per A.U. | |||
| peak | inflection | remote | ||
| Resolution (Å) | 48-1.8 | 48-2.0 | 48-2.05 | |
| Wavelength (Å) | 1.282 | 1.283 | 1.257 | |
| Redundancy 1 | 6.69 (3.92) | 7.22 (7.22) | 7.21 (7.09) | |
| Completeness (%) | 98.8 (90.1) | 99.9 (100.0) | 99.8 (100.0) | |
| Rmerge 2 | 0.089 (0.244) | 0.133 (0.312) | 0.106 (0.319) | |
| I/σ(I) | 13 (2.2) | 8.3 (1.8) | 10.2 (2.1) | |
| Refinement Statistics (|F|>0) | ||||
| Resolution (Å) | 44-1.8 | |||
| Rworking, % | 19.92 | |||
| Rfree, % 3 | 21.08 | |||
| Number of protein atoms | 973 | |||
| Number of non-protein atoms | 131 water molecules and 4 zinc ions | |||
| R.m.s.d. from ideal bond length (Å) | 0.004 | |||
| R.m.s.d. from ideal bond angle (°) | 1.621 | |||
| Ramachandran statistics | Most favored 83 residues | Additionally allowed 10 residues | Generously allowed 2 residues | Disallowed 4 2 residues |
Numbers in parenthesis represent values for the highest resolution bin.
Rmerge = Σ| Iobs - Iavg | / ΣIavg.
Rfree was calculated with 7.2% of reflections.
Residue Glu237 in chains A and C are clearly in a conformation which Phi/Psi values fall into the disallowed region of the Ramachandran plot.
The residues essential for specific recognition of H3K4me3 in ING2 are conserved in ING4 (Fig. 1e and Supplementary Fig. 1). As expected, individual substitutions at ING4 residues Y198A, D213A, and W221A abrogated binding of the ING4PHD to the H3K4me peptides (Fig. 2a). We confirmed that the interaction between ING4PHD and H3K4me3 occurs in the context of nucleosomes (Fig. 2b), and that mutations of critical residues on the H3K4me3 recognition surface largely abolished this association. (Fig. 2b). Finally, ING4 wild-type, but not mutant proteins, interacted with H3K4me3 in vivo by protein-protein ChIP (Fig. 2c). Thus, in vivo, an intact PHD finger is required for the interaction of ING4 with H3K4me3.
ING4PHD recognition of H3K4me3 promotes HBO1 acetylation at histone H3
Previously, it was demonstrated that ING4 is required for HBO1 HAT activity on free histones and chromatin (Doyon et al., 2006). We therefore investigated the role of the ING4PHD on HBO1 HAT activity. Introduction of a point mutation that abrogates H3K4me3 binding (ING4D213A, see Fig. 2) did not alter the composition of purified ING4-HBO1 complex compared to the wild-type (WT) complex (Fig. 2d; data not shown), and therefore this mutation could be used to selectively investigate the importance of the PHD finger-H3K4me3 interaction in HAT assays. In order to test for potential crosstalk between H3K4me3 and H4 acetylation (mediated by HBO1) we utilized nucleosome substrates, allowing H3 and H4 proteins to be physically coupled within the same physiologically-relevant molecular entity. MLA (methyl-lysine analog) chemistry was employed to generate nucleosomes that are exclusively trimethylated at H3K4 and not otherwise modified (Simon et al., 2007). As shown in Figure 2e, acetylation of H4 by HBO1 occurs irrespective of the methylation status of H3K4. In addition, a PHD finger competent for H3K4me3-binding is not required for H4 acetylation – providing evidence that the ING4D213A-HBO1 complex is functionally intact (Fig. 2e). In contrast, the ability of the mutant complex to acetylate H3 was slightly compromised on unmodified nucleosome compared to WT complex (Fig. 2f, lanes 1 and 2). Moreover, K4me3 enhanced acetylation of H3 by the WT complex, but inhibited the activity of the mutant complex (Fig. 2f). Thus, in the context of nucleosomes, ING4 PHD finger recognition of K4me3 promotes in vitro HBO1 HAT activity on H3, but does not affect H4 acetylation.
To further investigate the interplay between HBO1 activity and H3K4 methylation, in vitro HAT assays were performed on histone peptides containing different levels of methylation at H3K4 as well as H3K9me3 peptides and H4 peptides. As shown in Figure 2g, ING4-HBO1 acetylation on H3 peptides was augmented commensurate with increasing levels of H3K4 methylation, and requires an intact ING4 PHD finger. In contrast, H3K9me3 did not promote HBO1 activity on H3 peptides, and the integrity of the ING4 PHD finger had no influence on the acetylation of unmodified H4 and H3K9me3 peptides (Fig. 2g). Taken together, our data indicate that in vitro the ING4 PHD finger is not intrinsically required for HBO1 acetylation of histone H4, but rather serves to sense the methylation state of H3K4 to facilitate HBO1 HAT activity on H3. These results are consistent with an independent study demonstrating a role for H3K4me3 recognition by the ING4 PHD finger in switching the substrate preference of HBO1 from H4 to H3 (J. Cote, personal communications).
H3K4me3-recognition required for ING4 cellular functions
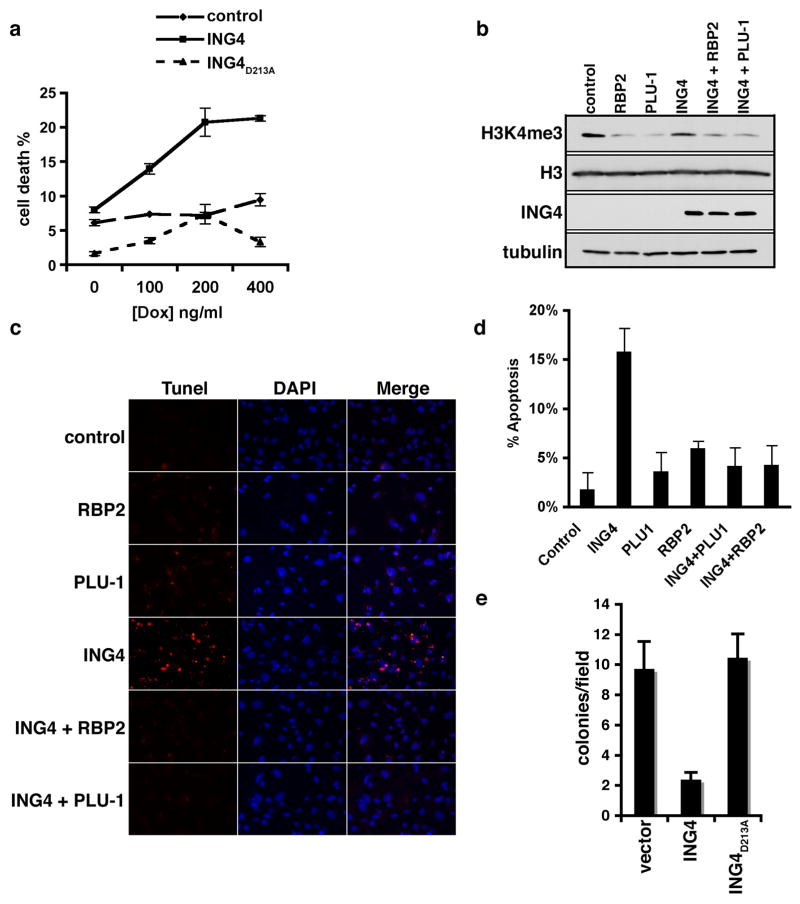
Next we explored whether H3K4me3 recognition plays a role in ING4-mediated cell death due to DNA damage (Shiseki et al., 2003). The sensitivity of HT1080 cells expressing flag-tagged ING4, flag-ING4D213A, or vector alone to low levels of the radiomimetic doxorubicin was determined (Fig. 3a). Cells expressing ING4 were more susceptible to DNA damage when compared to control cells or ING4D213A expressing cells (Fig. 3a). Next, ING4 genotoxic stress response activity was tested under conditions in which endogenous H3K4me3 levels were directly decreased via independent expression of two distinct H3K4me3-demethylases, RBP2 and PLU-1(Christensen et al., 2007; Iwase et al., 2007; Klose et al., 2007; Yamane et al., 2007). Expression of both enzymes decreased total H3K4me3 levels (Fig. 3b) and also abrogated ING4-dependent apoptosis (Fig. 3c and 3d). These data suggest that ING4 requires access to H3K4me3 in order to promote cell death.
Figure 3. The ING4-H3K4me3 interaction is required for ING4-mediated cell death and inhibition of anchorage-independent growth.
(A) Increased sensitivity of ING4 overexpressed cells to DNA damage-induced cell death requires an intact PHD finger. HT1080 cell viability ± Flag-ING4 and Flag-ING4D213A, in response to indicated concentrations of doxorubicin for 20 hr.. (B) Rbp2 or Plu-1 transfection in the presence or absence of ING4 overexpression reduces global H3K4me3 levels as determined by Western analysis of cell lysates. (C) Dependency on H3K4me3 for ING4-mediated apoptosis. Representative images of TUNEL assays performed on HT1080 cells transfected with the indicated plasmids and treated with 400 ng/ul doxorubicin. Control indicates empty vector controls. red: TMR red, blue: DAPI. (D) Quantification of (C), error bars represent s.e.m. from 3 independent fields. (E) ING4 inhibition of anchorage-independent cell growth requires an intact PHD finger. Soft agar assay of T47D cells stably expressing ING4, ING4D213A, or control vector. (A, B, D, and E) error bars indicate S.E.M. from four independent experiments.
ING4 was previously shown to be functionally deficient in the breast cancer T47D cell line, and ING4 complementation into these cells was found to inhibit growth in soft agar (Kim et al., 2004). Therefore, the role of H3K4me3-recognition in this anti-tumor associated activity was assessed. As expected, introduction of WT ING4 into T47D cells strongly inhibited growth in soft agar compared to control cells (Fig. 3e). In contrast, complementation with ING4D213A failed to inhibit soft agar growth (Fig. 3e), arguing that H3K4me3-recognition by ING4 plays a critical role in the ability of ING4 to prevent anchorage-independent growth.
ING4 occupancy at target genes is disrupted by abrogation of H3K4me3-binding
Based on the in vitro demonstration that H3K4me3 promotes ING4-HBO1 HAT activity and the in vivo observation that H3K4me3-recognition is critical for ING4 function, we postulated that ING4 binding to H3K4me3 at target genes leads to localized HAT activity and subsequent gene activation. To test this hypothesis, DNA purified by anti-Flag chromatin immunoprecipitation (ChIP) from Flag-ING4 or Flag-ING4D213A stable HT1080 cells, in the absence or presence of genotoxic stress, were hybridized to Nimblegen whole genome promoter tiling arrays. HT1080 cells were utilized due to low levels of endogenous ING4 protein (Supplementary Figure 2). 63 of the ~30,000 gene promoters examined on the arrays were strongly bound by ING4 under basal conditions, with an increase to 292 promoters upon DNA damage (see Supplementary Tables 1–4). Nimblescan, a standard Nimblegen peak-calling algorithm, was used to identify the ING4 binding sites. Briefly, the log2ratio of ChIP to input signal was calculated, and positive binding sites were determined using the following criteria: (1) a minimum of four consecutive probes where the log2 ratio is between 20–90% of a hypothetical maximum (mean log2ratio of the array + 6 standard deviations), (2) a false discovery rate <.2, and (3) an average log2 ratio of all the probes in a peak>1.5, which represents a 2.82 enrichment of ChIP over Input (see Supplementary information).
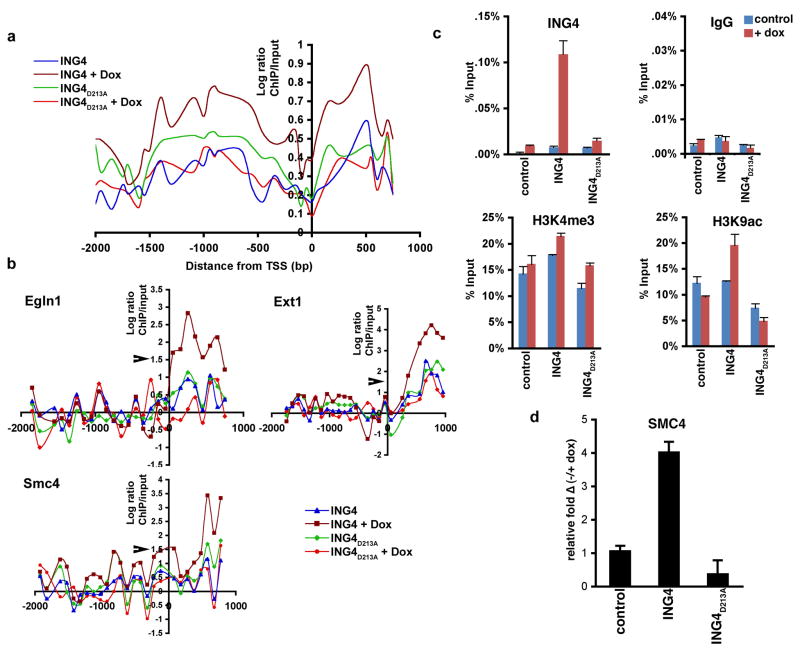
We next determined the average occupancy of WT and mutant ING4 proteins on DNA damage-dependent promoters. As shown in Figure 4a, with doxorubicin treatment, the wild-type ING4 protein is generally enriched on either side of the transcriptional start site (TSS). This is consistent with the absence of two nucleosomes at the TSS, and resembles the pattern of occupancy for H3K4me3 (Raisner et al., 2005). Induced occupancy of the mutant protein in response to DNA damage was largely absent compared to the wild-type protein (Fig. 4a), arguing that H3K4me3-recognition is important for ING4 stabilization at chromatin targets. Upon further analysis of ING4 promoter occupancy at the level of individual genes, two patterns emerged: one in which ING4 is enriched immediately downstream of the transcription start-site (Fig. 4b), and a second in which ING4 is enriched upstream of the transcription start-site (Supplementary Figure 3).
Figure 4. ING4 occupancy at target genes is disrupted by abrogation of H3K4me3-recognition.
(A) Average occupancy of ING4 at the 292 doxorubicin-induced target promoters. Flag-ING4 or Flag-ING4D213A ChIP-chip ± doxorubicin on high resolution whole genome promoter tiling arrays, average occupancy calculated every 50 bp along the promoters. (B) ING4 and ING4D213A occupancy ± doxorubicin at the indicated target promoters. Log ratios >1.5 denoting significant ChIP peaks is indicated with arrow (see Supplementary Info). (C) ING4 occupancy correlates with H3 acetylation at target promoters. Realtime PCR of ChIP assays on the Smc4 promoter with the indicated antibodies and cell lines. IgG is used as the negative control. Values = ChIP/input percent. Error bars indicate s.e.m. (D) Realtime PCR analysis of smc4 transcript levels in control, ING4 and ING4D213A cells with DNA damage. Values shown represent the relative change of transcript in the indicated samples with DNA damage as compared to untreated wild-type cells. Note that ING4D213A inhibits baseline expression ofsmc4. Error bars indicate s.e.m.
ChIP assays on three DNA damage-dependent ING4 target genes were performed to determine whether ING4 occupancy correlates with H3K4me3 and H3 acetylation markings. In response to doxorubucin treatment, binding of WT ING4 to the Smc4 (Fig. 4c), Egln1 (Hph2), and Ext1 (Supplementary Figure 4) promoters is significantly higher than that observed for ING4D213A. Further, H3K9 acetylation at all three promoters is considerably higher in the wild-type cells versus mutant cells (Fig. 4c; Supplementary Figure 4), suggesting that the ING4-H3K4me3 interaction stabilizes HBO1 HAT activity at target genes. However, we were unable to determine HBO1 occupancy at these genes as commercial anti-HBO1 antibodies failed to work in our hands under ChIP conditions (data not shown). In all samples, H3K4me3 levels were modestly induced with doxorubicin treatment at the Smc4 (Fig. 4c) Egln1,and Ext1 promoters (Supplementary Figure 4). We note that due to low endogenous ING4 protein levels in HT1080 cells, the response of control cells was largely not observed (Fig. 4c; Supplementary Fig. 2). Finally, mRNA levels for Egln1, Ext1 and Smc4 were greatly induced by DNA damage in the ING4 cells, but not in the ING4D213A cells (Fig. 4d; Supplementary Figure 5)). Based on these data, we propose that the ING4PHD-H3K4me3 interaction is critical for proper occupancy of the ING4-HBO1 complex at target promoters, and results in H3 acetylation and transcriptional activation of the promoter’s cognate gene.
Discussion
Previously we demonstrated that ING4 contains a C-terminal PHD finger that binds to H3K4me3 (Shi et al., 2006). Here we have demonstrated that the recognition of H3K4me3 by ING4 regulates a new HBO1 function – H3 acetylation and gene expression in response to genotoxic stress. These functions also represent a new function associated with the H3K4me3 mark in metazoans. In this regard, a growing number of H3K4me3-binding proteins link this mark to remarkably diverse biological outcomes. For example, recognition by the TAF3 PHD finger bridges H3K4me3 to RNA polymerase-II-mediated transcription, whereas H3K4me3 recognition by the BPTF PHD finger stabilizes a chromatin-remodeling complex at promoters (Vermeulen et al., 2007; Wysocka et al., 2006). In contrast, ING2 recognition of H3K4me3 stabilizes the co-repressor Sin3a/HDAC1 complex at target promoters to acutely repress gene expression (Shi et al., 2006). H3K4me3 is also involved in non-transcription functions. For example, CHD1 couples H3K4me3 to the mRNA splicing machinery (Sims et al., 2007). Finally, H3K4me3-recognition by the PHD finger of RAG2 is critical for V(D)J recombination, and a mutation in RAG2 that specifically interferes with this activity is found in patients suffering from immunodeficiency syndromes (Matthews et al., 2007). Here we have demonstrated that the ability of ING4 to bind H3K4me3 is required to suppress growth of T47D breast cancer cells in soft agar (Fig. 3e), a common cellular manifestation observed in neoplastic transformation (Kim, 2005; Kim et al., 2004). Contact inhibition is also regulated by ING4, and the cell-adhesion related gene family was the most enriched gene set associated specifically with the doxorubicin-dependent ING4 target promoters (Supplementary Table 5). These findings further highlight the key role that protein domain recognition of histone methylation events play in physiology and in the prevention of pathologic states.
ING4 has been implicated in negatively regulating both the NF-kB pathway and the HIF pathway (Garkavtsev et al., 2004; Ozer and Bruick, 2005). However the role of the H3K4me3-recognition by the ING4 PHD finger in these functions is unclear. ING4 was previously demonstrated to repress RelA activity in an NF-kB-dependent luciferase reporter assay, but in a similar experiment we did not observe differential effects on RelA activity between wild-type and PHD finger mutant ING4 proteins (data not shown) (Garkavtsev et al., 2004).
In addition, ING4 influences the HIF pathway by directly binding to HPH2 (Ozer et al., 2005). Notably, our data indicate that ING4 up-regulates transcription of Egln1/HPH2 in response to doxorubicin, likely via directing HBO1 HAT activity to the Egln1 promoter (Fig. 4). Thus, as well as directly binding to HPH2, ING4 may repress the HIF pathway indirectly via transcriptional upregulation of the HIF inhibitor, HPH2. In summary, we have provided evidence that ING4 can regulate gene expression in response to DNA damage via mediating crosstalk between H3K4me3 and H3 acetylation, and that this activity may function to transduce ING4-dependent tumor suppressor signals.
Experimental Procedures
Plasmids, Antibodies, MLA modified nucleosomes and Peptides
Antibodies against the following proteins were used in this study: Rabbit anti-ING4 (Covance), H3K4me1, H3K4me2, H3K4me3, H3K9me3, JADE2 (Abcam), GST, HBO1 (Santa Cruz Biotechnology), and FLAG (M2 and M5), H3K9Ac, IgG (Sigma). ING4 cDNA was cloned into p3XFlag7.1 (Sigma), pGEX6P1 (Amersham) and pMSCV. MLA generated H3K4me3 and control nucleosomes were generated as previously described (Simon et al., 2007). Biotinylated-H3 and H4 unmodified and modified peptides were synthesized at the Yale W.M. Keck facility as previously described (Shi et al., 2006).
Interaction, Soft Agar, and DNA damage sensitivity assays
Interaction assays were performed essentially as described in (Shi et al., 2006) except that for nucleosome binding, bound proteins were visualized by Western analysis. For soft agar assays, 55,000 T47D cells stably expressing pMSCV-ING4, pMSCV-ING4D213A, or pMSCV vector control were plated onto 30 mm dishes in a 0.04% soft agar growth media (Invitrogen) on top of a 1% agar media. Cells were supplemented with fresh media every three days. Colonies were scored after 21 days. Cell death assays on HT1080 cells was determined by trypan blue exclusion (Invitrogen) as previously described (Shi et al., 2007b). Apoptosis was determined by performing TUNEL assays. Briefly, 48 hours after transfection and 20 hours after 400 ng/ml Doxorubicin treatment (Sigma), TUNEL assays were performed using the In Vitro Cell Death Detection Kit-TMR red (Roche). Cells were fixed with methanol at −20C for 20 minutes, stained with TUNEL reaction mixture as per manufacturer’s instructions, and mounted in ProLong Gold Antifade Reagent With Dapi (Invitrogen).
ING4-HBO1 complex purification and HAT assays
Protein complexes were purified from 25 confluent 150mm plates of 293T cells stably expressing Flag-ING4 and Flag-ING4D213A. Cells were sonicated on ice in buffer (50 mM TrisHCl, 250 mM NaCl, 0.5% Triton X100, 10% glycerol, PMSF, PI (Roche)) and supernatant incubated at 4°C overnight with anti-Flag(M2)-agarose beads (Sigma). Protein complexes were eluted using 3xFlag-peptide (Sigma). Nucleosome HAT assays: MLA nucleosomes were incubated with 5μL FLAG-purified ING4 complexes and 2 μL of 3.3 Ci/mmol [3H]acetyl Coenzyme A (acetyl-CoA) in HAT buffer (10mM Tris-Cl, pH7.5, 0.1mM EDTA, 10% glycerol, 1mM DTT) for one hour at 30°C and separated by SDS page for autoradiography. Peptide HAT assays: As above except that 2.0μg of biotinylated peptides were used instead of nucleosomes and the reaction was performed for 18h at 30°C. The peptides were purified using 30μL of 50% streptavidin beads, unincorporated radioactivity was removed by three 1mL PBS 0.1% Tween-20 washes, and C.P.M. was measured by scintillation.
ChIP-chip, ChIP, and mRNA expression Realtime-PCR
HT1080 cells stably expressing Flag-ING4 were treated with 400ng/ml Doxorubicin for 4h, then prepared as described previously for ChIP (Shi et al., 2006). RealTime-PCR reactions using 1.2 uL of unamplified ChIP DNA and gene expression quantification were performed as previously described using Taqman Gene Expression Assays (Applied Biosystems) (Shi et al., 2007b). ChIP-chip assays were performed on Two-Array Set HG18 whole genome promoter arrays (Roche NimbleGen, Inc. (Madison, WI)). These arrays tile from 3500 bp upstream to 750bp downstream of the transcriptional start site using 50–75mer oligos at a 100bp interval. ChIP DNA was amplified using the WGA2 Whole Genome Amplification Kit (Sigma) as previously described (O’Geen et al.,2006). All hybridization and data extraction were performed according to the standard NimbleGen protocol. Raw data is available for download at Gene Expression Omnibus (GEO), Accession number: GSE13412.
Crystallization and Data Collection
The ING4 PHD finger (1.0 mM) was combined with H3K4me3 peptide (residues 1–12) in a 1:1.5 molar ratio prior to crystallization. Initial crystals of the complex were grown using the sitting drop vapor diffusion method at 18°C by mixing 1 ul of the protein-peptide solution with 1 ul of a well solution containing 90% 1.6 M sodium citrate tribasic dihydrate pH 6.5, and 10% of condition 6 from Hampton crystal screen 1 (0.2 M magnesium chloride hexahydrate, 0.1 M Tris HCL pH 8.5, and 30% (w/v) polyethylene glycol 4000). All crystals grew to ~ 0.05×0.05×0.3 mm3 in a tetragonal space group (P43) with unit cell parameters of a=68.16 Å, b=68.16 Å, c=27.96 Å. There are 2 similar molecules of the ING4:peptide complex per asymmetric unit (AB and CD; only the AB molecule complex is discussed in the text for clarity), with an estimated solvent content of 42%. Crystals were flash cooled in liquid nitrogen, and X-ray data were collected at 100K on a “NOIR-1” MBC system detector at beamline 4.2.2 at the Advanced Light Source (ALS) in Berkeley, CA. A complete Zn MAD dataset to 1.8Å was collected for peak, inflection and remote wavelengths. Data were processed with the DTREK. The coordinates have been deposited in the Protein Data Bank under accession number 2pnx.
Supplementary Material
Acknowledgments
We thank J. Côté for sharing unpublished results. We also thank K. Chua, X. Shi and C.L. Liu, R. Zhao, and P. Peña for helpful comments, and M. van der Woerd and J. Nix for X-ray data collection. This work was supported in part by grants from the NIH to O.G. (R01 GM079641-01A1), T.G.K., and H.Y.C. M.D.S. is supported by a Helen Hay Whitney Fellowship, A.J.K. by a Genetech Foundation Predoctoral Fellowship, and K.S.C. by an NIH NRSA Postdoctoral Fellowship. O.G. is a recipient of a Searle Scholar Award.
Footnotes
Additional information available in Supplementary Information
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bannister AJ, Kouzarides T. Histone methylation: recognizing the methyl mark. Methods Enzymol. 2004;376:269–288. doi: 10.1016/S0076-6879(03)76018-2. [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Colla S, Tagliaferri S, Morandi F, Lunghi P, Donofrio G, Martorana D, Mancini C, Lazzaretti M, Mazzera L, Ravanetti L, et al. The new tumor-suppressor gene inhibitor of growth family member 4 (ING4) regulates the production of proangiogenic molecules by myeloma cells and suppresses hypoxia-inducible factor-1 alpha (HIF-1alpha) activity: involvement in myeloma-induced angiogenesis. Blood. 2007;110:4464–4475. doi: 10.1182/blood-2007-02-074617. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, Barnett GH, Jain RK. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature. 2004;428:328–332. doi: 10.1038/nature02329. [DOI] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Iizuka M, Sarmento OF, Sekiya T, Scrable H, Allis CD, Smith MM. Hbo1 Links p53-dependent stress signaling to DNA replication licensing. Mol Cell Biol. 2008;28:140–153. doi: 10.1128/MCB.00662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Kim S. HuntING4 new tumor suppressors. Cell Cycle. 2005;4:516–517. doi: 10.4161/cc.4.4.1584. [DOI] [PubMed] [Google Scholar]
- Kim S, Chin K, Gray JW, Bishop JM. A screen for genes that suppress loss of contact inhibition: identification of ING4 as a candidate tumor suppressor gene in human cancer. Proc Natl Acad Sci U S A. 2004;101:16251–16256. doi: 10.1073/pnas.0407158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Martinka M, Li G. Role of ING4 in human melanoma cell migration, invasion, and patient survival. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn086. [DOI] [PubMed] [Google Scholar]
- Martin DG, Baetz K, Shi X, Walter KL, Macdonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. The Yng1p PHD Finger is a Methyl-Histone Binding Module that Recognizes Lysine 4 Methylated Histone H3. Mol Cell Biol. 2006 doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007 doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen, et al. Comparison of Sample Preparation Methods for Chip-chip assays. Biotechniques. 2006 doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer A, Bruick RK. Regulation of HIF by prolyl hydroxylases: recruitment of the candidate tumor suppressor protein ING4. Cell Cycle. 2005;4:1153–1156. doi: 10.4161/cc.4.9.2040. [DOI] [PubMed] [Google Scholar]
- Ozer A, Wu LC, Bruick RK. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF) Proc Natl Acad Sci U S A. 2005;102:7481–7486. doi: 10.1073/pnas.0502716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios A, Garcia P, Padro D, Lopez-Hernandez E, Martin I, Blanco FJ. Solution structure and NMR characterization of the binding to methylated histone tails of the plant homeodomain finger of the tumour suppressor ING4. FEBS Lett. 2006;580:6903–6908. doi: 10.1016/j.febslet.2006.11.055. [DOI] [PubMed] [Google Scholar]
- Palacios A, Munoz IG, Pantoja-Uceda D, Marcaida MJ, Torres D, Martin-Garcia JM, Luque I, Montoya G, Blanco FJ. Molecular basis of histone H3K4ME3 recognition by ING4. J Biol Chem. 2008 doi: 10.1074/jbc.M710020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Unoki M, Ythier D, Duperray A, Varticovski L, Kumamoto K, Pedeux R, Harris CC. Inhibitor of growth 4 suppresses cell spreading and cell migration by interacting with a novel binding partner, liprin alpha1. Cancer Res. 2007;67:2552–2558. doi: 10.1158/0008-5472.CAN-06-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Gozani O. The fellowships of the INGs. J Cell Biochem. 2005;96:1127–1136. doi: 10.1002/jcb.20625. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007a;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007b;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y, Appella E, Yokota J, Harris CC. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2373–2378. [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.