Abstract
The synthesis of protein-based polymers with controlled conformational properties and functional group placement offers many opportunities for the design of advanced materials. In this work, protein engineering methods have been used to produce repetitive alanine-rich protein polymers with the sequence [(AAAQ)5-(AAAE)(AAAQ)5]x (x = 2 and 6); these macromolecules may mimic architectural features of certain alanine-rich helical sequences found in natural proteins. Various proteins from this family can be readily expressed and purified from Escherichia coli. Circular dichroic spectroscopy (CD) characterization demonstrates that the purified proteins are highly helical under a variety of conditions. Thermal analysis of [(AAAQ)5(AAAE)-(AAAQ)5]2 via differential scanning calorimetry (DSC) and CD indicates that the protein undergoes a reversible helix–coil transition at approximately 45 °C and that the protein conformation can be manipulated at elevated temperatures depending on solution conditions. The demonstrated conformational properties of these artificial proteins suggest that they may be excellent candidates for elucidating structure–function relationships in biopolymers for nanotechnology and biological applications.
Introduction
The central importance of macromolecular structure in controlling material function has fueled the development of a variety of polymer synthetic methods that have been useful for producing macromolecules with defined molecular architectures. Among these, protein engineering strategies have been increasingly employed for producing protein polymers that exhibit precise molecular weights and monomer sequence. In natural proteins, this precise control of molecular weight and amino acid sequence permits the adoption of thermodynamically stable conformations that mediate the assembly of proteins into explicit hierarchical structures (such as those in collagen, silk, and elastin) or impart the catalytic functions of enzymes. This level of control in artificial proteins has resulted in the production of a variety of protein-based materials that exhibit interesting crystalline, liquid crystalline, assembly, mechanical, and biological properties.1–14 Specifically, the molecular weight control of the methods has proven critical for the production of novel smectic liquid crystalline phases from solutions of biosynthetically derived poly(benzyl-α,L-glutamates),3 as well as the assembly of elastin-like block copolymers into nanoparticles of controlled size and properties.9,15 The sequence control of the method provides additional opportunities for controlling macromolecular folding, assembly, and function and has been used to produce proteins with β-sheet-like structures reminiscent of those of silk,1,5,6,12,16,17 with α-helical structures of coiled-coil proteins,4 and with controlled densities of functional groups to control cross-linking and mechanical properties.7,18,19
Outside of investigations that employ polyglutamate3 or the leucine zipper motif for control of macromolecular assembly,4,20 there have been few reports of the production of repetitive artificial α-helical proteins for materials applications, despite the prominence of helical motifs in controlling protein assembly and the presentation of functional groups in natural proteins. Adoption of an appropriate helical conformation by the protein involucrin is hypothesized to be an important determinant in controlling formation of the corneocyte protein envelope in the epidermis,21 and the controlled presentation of hydroxyl groups over short length scales in alanine-rich, α-helical, type I antifreeze proteins is critical for their ability to inhibit ice crystal growth in arctic fish.22,23 The production of artificial proteins that mimic the sequences and conformational properties of these proteins may therefore find use in a variety of materials applications, such as in controlling assembly and/or for the multivalent presentation of functional groups.
We have employed protein engineering strategies for the synthesis of a family of alanine-rich helical proteins of the general sequence [(AAAQ)y(AAAE)(AAAQ)y]x; the amino acids in the sequence have been chosen owing to the high helical propensity of alanine (A) and the helical tendencies of glutamine (Q) and glutamic acid (E).24,25 The number of glutamic acid residues in the helical proteins, and their density in the sequence, can be manipulated by simple variations of the x and y values in this amino acid sequence, which provides opportunities to tailor these polymeric architectures for specific applications. The design, synthesis, and characterization of the conformational properties of one family of these protein polymers, in which y=5, are described.
Materials and Methods
Materials
A mutagenized version of the pET-19b plasmid (altered via site-directed mutagenesis to eliminate a SapI site26) was a gift of N. L. Goeden-Wood and J. D. Keasling. The restriction endonuclease Eam1104I was obtained from MBI Fermentas (Hanover, MD), while all other restriction endonucleases were obtained from New England Biolabs (Beverly, MA) or Invitrogen (Carlsbad, CA). Synthetic oligonucleotides were obtained from Sigma Genosys (The Woodlands, TX). Oligonucleotide and plasmid purification kits and nickel-chelated Sepharose resin were obtained from Qiagen (Valencia, CA). General reagents for protein expression and purification were obtained from Sigma (St. Louis, MO) and Fisher Scientific (Fairlawn, NJ).
DNA Monomer and Cloning Plasmid Construction
All molecular biology procedures were conducted according to standard protocols27 and are not described in detail here. A DNA sequence was designed to encode the amino acid sequence [(AAAQ)5(AAAE)(AAAQ)5] flanked by Eam1104I restriction sites to permit removal of the monomer DNA sequence from a cloning plasmid. The Eam1104I restriction sites were also flanked by EcoRI and BamHI restriction sites for efficient cloning of the oligonucleotide into the cloning plasmid pUC19 via standard published protocols.27 Single-stranded oligonucleotides encoding the monomer sequence and flanking restriction sites (Figure 1A) were annealed, purified via 2% agarose gel electrophoresis, and phosphorylated via treatment with T4 polynucleotide kinase. The recipient cloning vector, pUC19, was digested with the restriction enzymes EcoRI and BamHI for 4 h at 37 °C, dephosphorylated via the addition of calf intestinal alkaline phosphatase (CIP), and purified via 1% agarose gel electrophoresis. The oligonucleotide and digested pUC19 were ligated at 25 °C with T4 DNA ligase. The ligation mixture was transformed into chemically competent cells of Escherichia coli strain DH5αF′, and the DNA of isolated transformants was purified via the Qiagen QIAquick miniprep kit and protocol. A plasmid that contained the appropriate DNA sequence was identified via sequencing analysis and designated as pUC19-KLK-C.
Figure 1.
DNA sequence of inserts used in plasmid construction. (A) Oligonucleotide sequence inserted into the cloning vector pUC19 to yield pUC19-KLK-C. (B) Oligonucleotide sequence inserted into the expression vector pET19b to yield pET19b-RF1.
Expression Plasmid Construction
The multiple cloning site of the mutagenized expression plasmid pET-19b26 was modified to permit insertion of the target protein sequence. A DNA linker was designed to contain terminal NcoI and BamHI restriction sites that permit insertion of the oligonucleotide linker into pET-19b. The linker DNA sequence was also designed to contain two internal SapI restriction sites (Figure 1B), which permits insertion of the DNA sequence of the target protein. The oligonucleotides encoding this linker sequence were annealed as described above, and ligated, via incubation with T4 DNA ligase (15 °C, 16 h), into pET-19b that had been digested with NcoI and BamHI in NEBuffer BamHI supplemented with 100 µg/mL BSA, dephosphorylated via treatment with CIP, and purified via 1% agarose gel electrophoresis. The ligation mixture was transformed into E. coli strain DH5αF′, and the sequence of the DNA of isolated transformants was determined. Plasmid DNA containing the correct linker DNA sequence was designated as pET19b-RF1.
To isolate the oligonucleotide sequence for insertion into the pET19b-RF1 expression plasmid, the plasmid pUC19-KLK-C was digested with the restriction enzyme Eam1104I (37 °C, 16 h), and the DNA fragment corresponding to the target protein (140 bp) was isolated via agarose gel electrophoresis. The purified monomer DNA was then ligated into the pET19b-RF1 plasmid that had been previously digested with SapI (37 °C, 16 h), dephosphorylated via treatment with CIP, and purified. The production of expression plasmids carrying artificial repetitive genes of varying lengths was confirmed via restriction digest analysis of the plasmids with the enzymes EcoRI and BglII (37 °C, 4 h) and subsequent analysis via agarose gel electrophoresis. The sequences of all genes were confirmed via dideoxy sequencing analysis. The plasmids containing the target protein DNA sequences were designated as pET19b-RF1-CX, where X denotes the number of [(AAAQ)5(AAAE)(AAAQ)5] repeats. The pET-19b-derived plasmids also encode an N-terminal decahistidine tag to permit protein purification via metal chelate affinity chromatography.
Protein Expression and Purification
The plasmids pET19b-RF1-CX were used to transform chemically competent cells of E. coli strain BL21(DE3)pLysS. Protein expression of [(AAAQ)5(AAAE)(AAAQ)5]x, with x = 2 and 6, was conducted via standard methods employing chemical induction (isopropyl β-d-thiogalactopyranoside, IPTG, final concentration 0.4 mM) of cultures of the appropriate expression host [500 mL, supplemented with ampicillin (200µg/ mL) and chloramphenicol (35µg/mL)]. Cells were harvested 4 h after induction via centrifugation (6000 rpm, 10 min), the supernatant was decanted, and the cell pellets were resuspended in 8 M urea at pH 8.0 (1 g of cells/4 mL of buffer) and frozen at −20 °C. Protein expression was monitored via sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) of samples with normalized OD600 and visualized via Coomassie blue staining.
The target protein was purified from the cell lysate via immobilized metal chelate affinity chromatography at 4 °C with stepwise pH gradient elution under denaturing conditions (Qiagen). Eluted protein was dialyzed batchwise against distilled water at 4 °C for 5 days. The dialysate was lyophilized to yield 15–20 mg of the purified target protein [(AAAQ)5(AAAE)(AAAQ)5]2 per liter of culture. Protein yields decreased with the molecular weight of the protein; a yield of 5–10 mg for the protein [(AAAQ)5(AAAE)-(AAAQ)5]6 was obtained from 1 L cultures. The purity of the proteins was monitored via SDS–PAGE and confirmed via reverse-phase high-performance liquid chromatography (HPLC) and amino acid analysis. The molecular weight of the purified protein was confirmed via matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry, the conformational properties of the proteins were characterized via circular dichroic spectroscopy (CD), and thermally induced conformational changes were monitored via differential scanning calorimetry (DSC).
General Characterization
DNA sequencing of resulting plasmids was performed by the College of Agriculture sequencing facility at the University of Delaware. Analysis of the amino acid composition of purified protein was performed by the Molecular Analysis Facility at the University of Iowa (Iowa City, IA).
HPLC
Protein purity was determined via reverse-phase HPLC with a Delta600 HPLC (Waters, Milford, MA) equipped with a Symmetry300 C18 analytical column. Samples were made at a concentration of approximately 100 µM in deionized water. A linear gradient experiment was run from 5% to 70% solvent B over 49 min (1 mL/min) after a 100 µL injection of the sample, where solvent A is 0.1% trifluoroacetic acid (TFA) in water and solvent B is 0.1% TFA in acetonitrile. Data were recorded at 218, 254, and 280 nm to observe protein elution.
MALDI-TOF
MALDI-TOF analysis of purified protein was performed at the Mass Spectrometry Facility in the Department of Chemistry and Biochemistry at the University of Delaware on a Biflex III (Bruker, Billerica, MA). The samples were prepared in a 3,5-dimethoxy-4-hydroxy-cinnamic acid matrix with the calibration mixture of bovine insulin (MW=5734.59), thioredoxin from E. coli (MW=11 647.48), and horse apomyoglobin (MW=16 952.56). Data were recorded with the OmniFLEX program and subsequently analyzed in the XmassOmni program.
Circular Dichroism
Circular dichroic spectra were recorded on an Aviv 215 spectrophotometer (Proterion Corp., Piscataway, NJ) in a 1-mm path length quartz cuvette in the single-cell mount setup. Background scans of buffers (10 mM phosphate, pH 2.3; 10 mM phosphate and 150 mM NaCl, pH 2.3) were recorded and manually subtracted from the sample scans. Samples were made at a concentration of approximately 10–20 µM via dilution, with the appropriate buffer, of a 100 µM stock solution of protein in 10 mM phosphate buffer, pH 2.3. Protein concentrations were confirmed via amino acid analysis. The samples (400µL) were loaded into a 1-mm path length quartz cuvette. Data points for the wavelength-dependent CD spectra were recorded at every nanometer with a 1 nm bandwidth and a 10 s averaging time for each data point. Data points for the temperature scans were recorded at 222 nm, at 1 °C intervals with an equilibration time of 1 min. The molar ellipticity, [θ]MRW (degrees centimeter2 decimole−1), was calculated via use of the molecular weight of the protein and cell path length.
Differential Scanning Calorimetry
Calorimetric experiments were performed on a VP-DSC from Microcal (North-hampton, MA), equipped with sample cells with a volume of approximately 0.5 mL. Initially, both cells were filled with buffer (10 mM phosphate, pH 2.3, or 10 mM phosphate and 150 mM NaCl, pH 2.3) to determine the background, which was manually subtracted from data. Samples at a concentration near 100 µM, confirmed via amino acid analysis, were heated at a constant rate of 1 °C/min and cooled at the machine-determined rate of approximately 6 °C/min. The raw data, in the form of heat flow (millicalories per minute) were converted to excess heat capacity, Cp ex:
| (1) |
where q is the heat flow after background subtraction (millicalories per minute), M is the protein molecular weight (grams per mole), R is the heating rate (degrees Celsius per minute), c is the protein concentration (grams per milliliter), and V is the volume of the sample cell (milliliters). This Cp ex was converted to apparent excess heat capacity, Cp,app ex, by correcting the data via subtraction of the average value of Cp ex at low temperatures, where no calorimetric transitions are observed. The Cp,app ex curve was then integrated over the entire temperature range to yield the calorimetric enthalpy change, ΔHcal. Calorimetric data exhibiting significant slopes were corrected, via subtraction of a baseline generated from a polynomial fit, prior to numerical integration.
Results and Discussion
The goal of this work was the synthesis of artificial protein polymers with architectural features that may mimic those of select natural helical proteins, such as the alanine-rich antifreeze proteins, and that may be useful for determining structure–function relationships based on variations in functional group density and conformation in a family of artificial protein polymers. Chemically reactive groups may be modified with, for example, sugars, short peptides, or metal nanoparticles to produce macromolecules with application in biomaterials design, therapeutics development, and biomineralization. Given that our primary interest was in the design of flexible, helical proteins that carry a controlled and limited number of chemically reactive functional groups, we turned our attention to the production of alanine-rich sequences equipped with chemically reactive amino acid residues. In these studies, the protein sequence [(AAAQ)5(AAAE)(AAAQ)5]x was chosen for investigation, on the basis of the high helical propensity of alanine, the helix-forming tendencies of glutamine and glutamic acid, and the requirement that the polymer backbone be chemically neutral. Alanine has the highest helical propensity of the natural amino acids, and alanine-rich sequences as short as 16 amino acid residues form isolated α-helices in aqueous solution.25 The first de novo designed α-helical peptide reported by Marqusee and Baldwin in 198728 comprised an alanine-rich sequence that contained glutamic acid and lysine residues for solubility. Since these early reports, an enormous number of additional alanine-rich, α-helical peptides have been produced and characterized, as the alanine-rich template provides an excellent model for determining the α-helical propensities of guest amino acids, both natural and nonnatural. In investigations such as these, peptides with the sequence (AAAAQ)x have been widely studied because they are nonaggregating, completely neutral, and carry no stabilizing side-chain interactions.29,30
Synthetic genes encoding the protein sequence [(AAAQ)5-(AAAE)(AAAQ)5]x were therefore designed; the AAAQ sequence was employed instead of the AAAAQ motif in order to improve the solubility of the proteins in aqueous solutions by increasing the density of the polar, helix-forming glutamine (Q) residue in the sequence. The potential success for expression of proteins of this composition from bacterial hosts was suggested by the fact that the (AAAQ)x motif can be expressed successfully in E. coli.31 The choice of the simple amino acid sequences employed in these investigations was motivated by a number of factors. Although a variety of strategies have long been applied for helix stabilization, such as helix cap design, formation of salt bridges between residues at i and i + 4 positions,24,32–36 and the use of coiled-coil domains,37–42 these strategies employ side chains that introduce chemical functionality that is undesirable for the planned uses of the proteins described here. Accordingly, only the chemically reactive, helix-forming glutamic acid (E) was introduced in these sequences. The carboxylic acid side chain of E provides sites for chemical attachment of desired biologically or otherwise active moieties at a regular density in the sequence; the presence of Q-E side-chain interactions at the (i, i + 4) positions may provide some stabilization.33 The selective placement of a limited number of functional groups within the sequence offers the potential for isolating the specific novel macromolecular functions imparted by the functional groups. The choice of glutamic acid functionality over other chemically reactive functional groups such as lysine and cysteine was motivated by the ease of expression and purification of anionic protein polymers, relative to highly cationic and thiolated protein polymers, from bacterial expression hosts. The nominal distance between the functional groups in a completely R-helical conformation of this sequence was estimated on the basis of energy minimization calculations (see Supporting Information) to be approximately 65 Å, which serves as a starting point for making future comparisons of protein polymer sequences with varying densities of functional groups. The presentation of functional groups with this precise spacing is not expected in solution, as the known flexibility of the alanine-rich sequences, based on the rapid conversion between helix and random coil conformations,24,43 will preclude these macro-molecules from existing as a single continuous helix. Nevertheless, the regularity of the sequence and the density of functional groups will provide unique opportunities to investigate structure–function relationships in these polymeric materials.
Expression plasmids for helical proteins of varying molecular weights were produced via modified seamless cloning methods,26,44 as described in the Experimental Section. Expression plasmids of the correct sequence were transformed into the E. coli expression host BL21(DE3)pLysS to yield expression strains BL21(DE3)pLysS/pET19b-RF1-CX. Proteins of the sequence [(AAAQ)5(AAAE)(AAAQ)5]x, with x=2 and 6, were expressed from bacterial expression strains BL21(DE3)pLysS/pET19b-RF1-CX, with yields that decreased from 15 to 5 mg/L with increasing protein molecular weight. The target protein produced from these expression hosts carries a decahistidine fusion tag that facilitates protein purification via immobilized metal chelate chromatography. The target proteins are selectively eluted after endogenous host proteins have been washed from the resin; representative purification results obtained via SDS–PAGE analysis are shown in the Supporting Information. Amino acid analysis and HPLC analysis of the protein [(AAAQ)5(AAAE)(AAAQ)5]2, shown in the Supporting Information, indicate that the proteins purified in this manner are of at least 95% purity. MALDI-TOF mass spectrometric analysis of the protein yields single peaks with masses that are consistent with those theoretically predicted (Supporting Information). After purification, the fusion tag can be removed from the target protein via treatment with CNBr, although the short fusion tag (MGH10SSGHINM) has not been removed from proteins in these investigations, as it does not impact the conformation of the protein (see below).
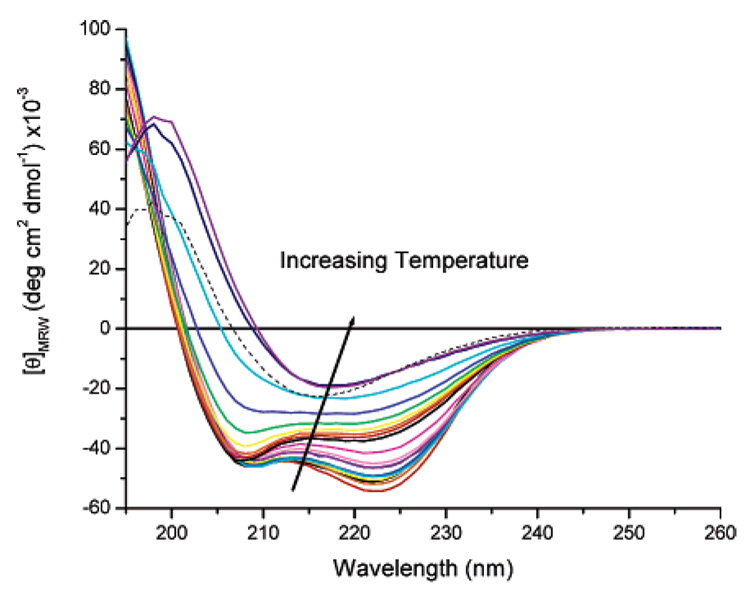
Circular dichroic spectroscopy studies were conducted to determine the conformation of these proteins under various conditions. Results are shown in Figure 2 for the protein [(AAAQ)5(AAAE)(AAAQ)5]2 at concentrations of approximately 25 µM in 10 mM phosphate buffer, pH 2.3, at 4 °C. A pH of 2.3 was used to ensure protonation of the glutamic acid residues, which would mimic the nonionic behavior of the protein after functionalization with neutral groups. The spectra of these proteins are not concentration-dependent in the concentration range employed for the CD measurements, suggesting that there is no significant change in aggregation state observed for the proteins under these conditions. As shown in Figure 2, the spectra observed exhibit a maximum at 192 nm and two minima at 208 and 222 nm, consistent with the spectrum expected for an α-helical protein. The ratio of the mean residue ellipticities of the peaks at 222 and 208 nm ([θ]222/[θ]208) at low temperatures is approximately 1.1 for [(AAAQ)5(AAAE)(AAAQ)5]2, which is consistent with values of 1.1–1.3 that have been previously reported for highly helical, hydrated, unaggregated alanine-rich peptides. 45–47 The mean residue ellipticity (MRE) of [(AAAQ)5-(AAAE)(AAAQ)5]2 at 222 nm ([θ]222) at 4 °C is approximately −53 900 deg cm2 dmol−1, which is significantly larger than [θ]222 values of approximately −44 000 deg cm2 dmol−1 previously reported for alanine-rich peptides 45 amino acids in length47 and for other long helical polypeptides.48
Figure 2.
Circular dichroic spectra of [(AAAQ)5(AAAE)(AAAQ)5]2 (25 µM) in 10 mM phosphate buffer, pH 2.3. CD spectra were collected in increments of 4 °C, with a range of 4–80 °C.
Helicity in alanine-rich proteins is length-dependent, and current theoretical models predict a maximum value of [θ∞]222 of approximately −61 000 deg cm2 dmol−1 for infinitely long alanine-rich helices.47 The increased helicity of the artificial protein (88 amino acids in the alanine-rich sequence) relative to that of alanine-rich peptides is consistent with these predictions. The fractional helicity for the artificial protein can be calculated according to a commonly employed function relating fractional helicity to experimental [θ]222:
| (2) |
where [θn]222 represents the MRE of an idealized 100% helical peptide of length n, [θ∞]222 is the limit of [θn]222 at very large n, and x is a length correction value that is set to 2.5.49,50 If values of −61 000 deg cm2 dmol−1 for [θ∞]222 and 88 for n are employed, [θn]222 is calculated as approximately −59 300 deg cm2 dmol−1 for an alanine-rich sequence comprising 88 amino acid residues. On the basis of this calculated limiting value of [θn]222, the fractional helicity for [(AAAQ)5(AAAE)(AAAQ)5]2 is approximately 91% under these experimental conditions. This experimentally determined fractional helicity appears to be consistent with theoretically predicted values that are determined via the Lifson-Roig formalism and that are based on commonly reported values for the helical propensity of alanine.47 The consistencies between this experimental data and theoretical prediction, combined with the form of the CD spectrum, suggest that the protein has similar properties as highly helical alanine-rich peptides at these concentrations, although contributions from any tertiary or quaternary structures adopted by these longer proteins cannot be excluded and may complicate this comparison.
Wavelength scans of the protein solutions, shown in Figure 2, were recorded from 260 to 190 nm at varying temperatures between 4 and 80 °C (at increments of 4 °C) to observe the temperature dependence of the conformational behavior of the protein. As the temperature is increased from 4 to 44 °C, the absolute values of the molar ellipticity for each observed peak decrease, but the spectra maintain the characteristic double minima of the α-helical conformation. At temperatures above 44 °C, there is a significant change in the form of the CD spectrum, with a loss of intensity at 222 nm and a slight blue shift in the minimum at 208 nm, indicating an increase in the random coil character of the protein. Indeed, the presence of an isoellipsoidal region at approximately 203 nm, for the spectra at lower temperatures (4–64 °C), suggests that in this temperature range the CD spectra are approximated by linear combinations of unordered and helical spectra.51 As the temperature is raised above 68 °C, however, the isoellipsoidal region is lost, which is consistent with a change in protein conformation that is corroborated by the marked change in the CD spectrum. The shift in the maximum from 195 to 198 nm and the development of a single minimum at 218 nm are consistent with spectral features expected for β-sheet structure. The β-sheet formation is irreversible, as the CD spectrum does not change upon reduction of the temperature to 4 °C (dotted line in Figure 2); the increased breadth of the minimum at 218 nm may suggest a small degree of reversibility under these conditions. The conversion to β-sheet structure is not unexpected for these proteins, given that the adoption of β-sheet conformation at elevated temperatures and concentrations by alanine-rich peptides52 and alanine-rich regions in silk proteins5,53–55 is well-known. The increased tendency of the proteins to adopt β-sheet structures with increasing concentration was confirmed via infrared spectroscopy (see Supporting Information). The conversion of [(AAAQ)5-(AAAE)(AAAQ)5]2 from an α -helical conformation to a β-sheet conformation may offer the opportunity to manipulate the conformation of the protein and alter its functional behavior.
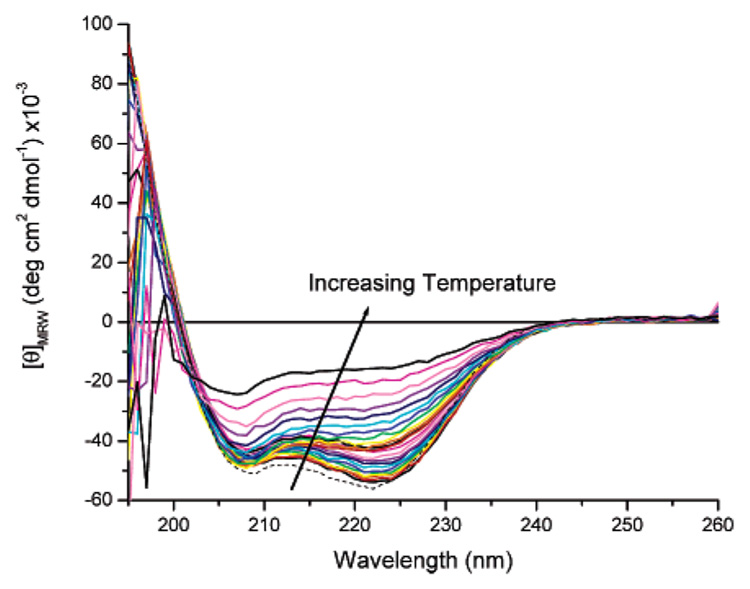
The conformation of the proteins was also monitored under isotonic salt concentrations (10 mM phosphate and 150 mM NaCl) at a pH of 2.3 to determine the impact of increasing salt concentration on the conformation of the proteins and to assess the feasibility of employing modified helical proteins under the ionic strength conditions of biological environments. The representative results of this characterization are shown for [(AAAQ)5(AAAE)(AAAQ)5]2 in Figure 3 and show that the protein remains highly helical under these conditions; spectra are recorded from 260 to 195 nm in these experiments owing to noise in the spectra at lower wavelengths in the presence of salt. The mean residue ellipticity values for the protein in 10 mM phosphate and 150 mM NaCl buffer at pH 2.3 are similar to those measured for the protein in 10 mM phosphate buffer at pH 2.3, with [θ]222 values of approximately −58 000 deg cm2 dmol−1 at 4 °C. Furthermore, the temperature-dependent CD spectra for the protein from 4 to 60 °C are essentially unchanged from those shown in Figure 2. This lack of salt concentration dependence of the CD spectra of the protein suggests that there are not significant charge effects on the helix and that the decahistidine tag (which is positively charged at pH 2.3) has no significant impact on the helical structure of the protein. In contrast to the data in Figure 2, at temperatures above 68 °C, the CD spectrum continues to show the double minimum characteristic of the α-helical conformation, indicating that helix melting is incomplete up to 92 °C under these buffer conditions. Accordingly, upon reduction of temperature from 92 to 4 °C, the CD spectrum (dotted line) is identical to the original wavelength scan at 4 °C, indicating the reversible recovery of the α-helical conformation. Although the marked impact of salt on the conformational behavior of the protein is somewhat surprising given the low concentrations of salt employed in these investigations, it has been previously reported that high salt concentrations can stabilize short helices in water.56,57
Figure 3.
Circular dichroic spectra of [(AAAQ)5(AAAE)(AAAQ)5]2 (10 µM) in 10 mM phosphate and 150 mM NaCl buffer, pH 2.3. CD spectra were collected in increments of 4 °C, with a range of 4–92 °C.
Conformational behavior identical to that shown in Figure 2 and Figure 3 was observed via circular dichroic spectroscopy for the longer protein [(AAAQ)5(AAAE)(AAAQ)5]6 under both sets of buffer conditions (see Supporting Information). The forms of the CD spectra are identical, and on average, the absolute MRE values for [(AAAQ)5(AAAE)(AAAQ)5]6 are greater than those for [(AAAQ)5(AAAE)(AAAQ)5]2 (Supporting Information), which is consistent with the known length dependence of helicity. Quantitative comparisons of the MRE values of the two proteins, however, are complicated by the dependence on purification conditions of the conformational behavior of the longer protein. Specifically, variations in purification and handling conditions permitted observation of either α-helix conformations of differing helicity or β-sheet conformations for the longer protein at 4 °C. These observations are consistent with the ability of these proteins to form β-sheet structure under certain solvent conditions, as described above, and suggest that the longer protein has a greater tendency for β-sheet formation, as might be expected. Nevertheless, the data indicate that both proteins can adopt highly helical structures under certain solution conditions, as desired. Details of the more complicated conformational behavior and helix–sheet transition for [(AAAQ)5(AAAE)(AAAQ)5]6 are currently under investigation and will be reported at a later date.
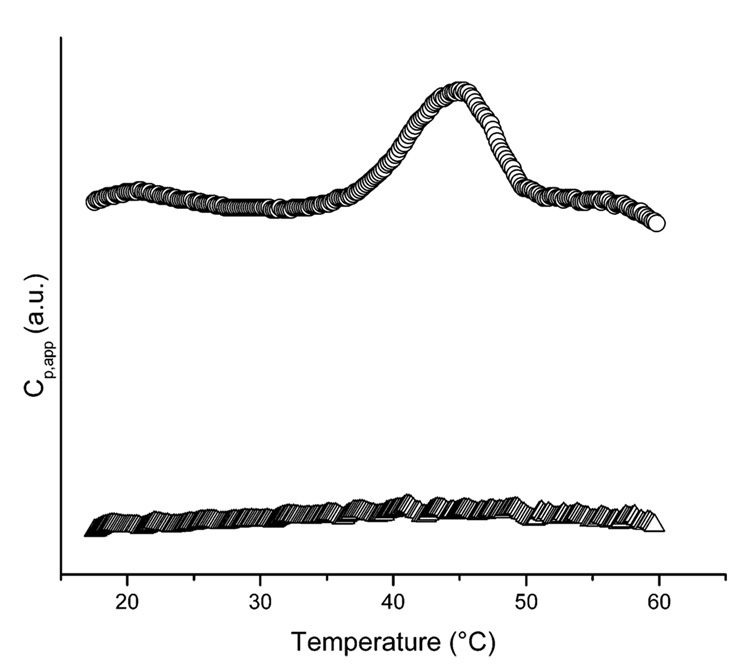
The thermal stability of [(AAAQ)5(AAAE)(AAAQ)5]2 was also monitored via DSC investigations to confirm the temperature of the initial transition observed in the CD spectra. Figure 4 presents the calorimetric data for the protein sample at a concentration of 100 µM in 10 mM phosphate buffer at a pH value of 2.3 (for comparison to the CD data in Figure 2) and in 10 mM phosphate and 150 mM NaCl buffer at a pH of 2.3 (for comparison to the CD data in Figure 3). In the former case, the DSC scan shows a positive peak at 45 °C, indicating an endothermic cooperative transition, which can be attributed to the helix to coil transition seen in CD at comparable temperatures and is consistent with previous reports of endothermic unfolding transitions in long alanine-rich peptides.46 The value of the average calorimetric enthalpy change (ΔHcal) calculated via integration of the endothermic peak is 2.75 kcal/mol, which is low in comparison to ΔHcal values reported for alanine-rich peptides stabilized by electrostatic interactions between charged glutamic acid and lysine residues.46 The small observed enthalpy change for the protein relative to the peptides is consistent with the lack of solvated charged groups of [(AAAQ)5(AAAE)(AAAQ)5]2 at low pH and the fact that the protein is not fully unfolded at temperatures below 50 °C. The energetic contributions of exposing ionic and hydrophobic groups are consequently reduced, thereby reducing the calorimetric enthalpy. As the temperature is raised above 55 °C, the excess heat capacity (i.e., the differential power normalized by the temperature scan rate and buffer background) shows a large exotherm, decreasing rapidly to levels well below the pretransition baseline and indicating conversion to a state of substantially lower enthalpy (see data in Figure 5). By analogy with other proteins that show such postpeak exotherms,58,59 this behavior was initially attributed to protein aggregation at the higher protein concentrations employed in the DSC investigations. Comparison of the DSC-monitored transitions and the CD-monitored transitions, however, suggests that this exotherm corresponds to the formation of β-sheet structure. Accordingly, when a similar DSC experiment was conducted on the protein in buffer containing 150 mM NaCl (Figure 4), no appreciable exothermic or endothermic transitions were observed, which suggests the lack of a cooperative transition and corroborates the lack of structure change to β-sheet observed via CD characterization of the protein in buffer containing isotonic saline.
Figure 4.
Differential scanning calorimetric data for [(AAAQ)5(AAAE)-(AAAQ)5]2 in 10 mM phosphate buffer, pH 2.3 (○), and in 10 mM phosphate and 150 mM NaCl buffer, pH 2.3 (Δ).
Figure 5.
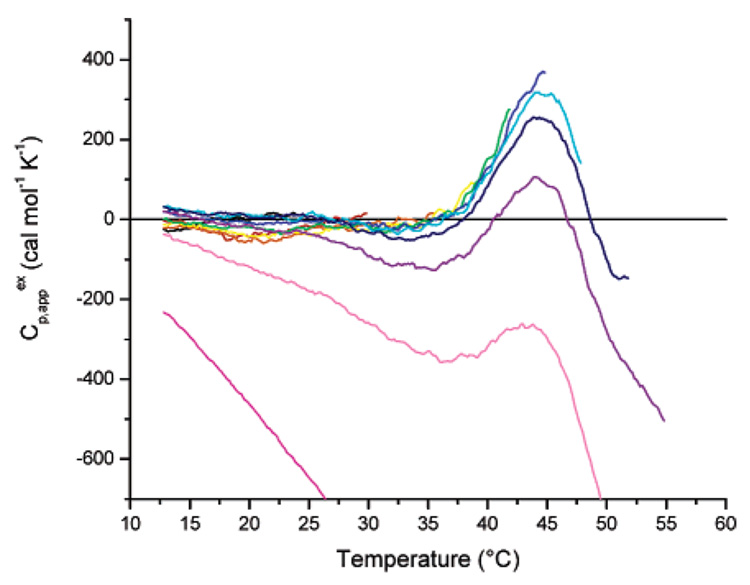
Differential scanning calorimetric characterization of the reversibility of conformational changes of [(AAAQ)5(AAAE)(AAAQ)5]2 in 10 mM phosphate buffer, pH 2.3.
After the transition to the β-sheet structure in the DSC sample was complete (confirmed via CD spectroscopy), the aggregation state of the protein was assessed via SEC. In these experiments, the protein sample was analyzed via the use of a Waters ProteinPak 125 column (7.8 × 300 mm) at a flow rate of 1 mL/min with isocratic elution in 0.5% (v/v) phosphoric acid, pH 2.5. These conditions afford resolution of molecular masses differing by a factor of 2 over a range of 2–80 kDa, which will permit the detection of aggregates of this protein. Only one peak was observed upon elution with an elution time and peak shape identical to that of the protein that had not been heat-treated. While this analysis may not definitively rule out the possibility of lower-order association (e.g., dimers and trimers) after heating, it does provide compelling evidence that the aggregation state of these proteins does not significantly change even after heat treatment and conversion to β-sheet conformation.
In an effort to determine specific conditions under which irreversible conformational changes may be minimized, the hysteresis behavior of the helix to coil transition for [(AAAQ)5(AAAE)(AAAQ)5]2 was studied at varying temperatures via both DSC and CD spectroscopy. Thermal cycling experiments were performed via DSC for protein samples with a concentration of 100 µM in 10 mM phosphate buffer, pH 2.3. In these experiments, the sample was initially heated from 12 to 24 °C, cooled to 12 °C, and then reheated over various temperature ranges, with the maximum temperature for each scan increased incrementally from 24 to 80 °C. Figure 5 displays the apparent heat capacity curves after buffer baseline subtraction for [(AAAQ)5(AAAE)-(AAAQ)5]2. The calorimetric curves overlap, as would be expected for a reversible system, when the sample is heated to various temperatures to a maximum temperature of 45 °C (the midpoint of the transition between helix and coil observed in CD experiments). When the sample is heated above this transition temperature, subsequent scans show negative deviations from the earlier curves, indicating appreciable (but incomplete) loss of reversibly refolded [(AAAQ)5(AAAE)(AAAQ)5]2 at the higher temperatures. For the lattermost scans, in which the sample had previously been exposed repeatedly to temperatures above ca. 50 °C, the entire curve follows a steep negative baseline, which is consistent with the formation of β-sheet structure at temperatures greater than 55 °C, suggested in the above discussion of Figure 4.
The complete reversibility of the structural changes at lower temperatures and subsequent conversion to β-sheet structure at elevated temperatures was also confirmed via an analogous CD experiment; data from these experiments are shown in the Supporting Information. Taken together with the DSC results in Figure 5, the CD-monitored reversibility of the structure of [(AAAQ)5(AAAE)(AAAQ)5]2 in the low-temperature range suggests that the transition to the β-sheet structure is a slow kinetic process, which does not occur on a relevant time scale for most assays of protein conformation and activity unless the sample is subjected to elevated temperatures.
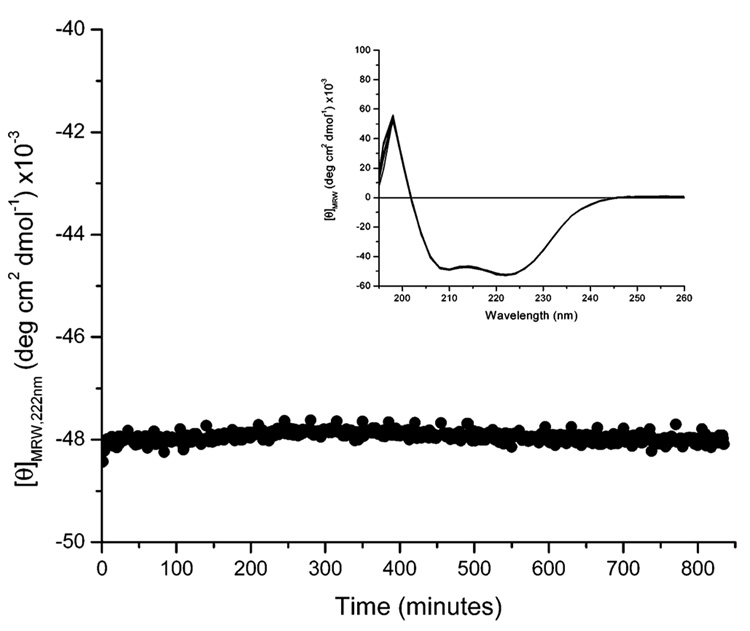
Although the CD and DSC experiments demonstrate that there are no significant irreversible conformation changes for the proteins near physiological temperatures, the DSC data indicate a kinetically slow conversion to β-sheet for [(AAAQ)5(AAAE)(AAAQ)5]2 at elevated temperatures under certain solvent conditions. To confirm the stability of the helical conformation of the proteins at physiological temperatures and salt concentrations over extended periods of time, the conformational behavior of [(AAAQ)5(AAAE)-(AAAQ)5]2 in 10 mM phosphate and 150 mM NaCl buffer, pH 2.3, was monitored via CD spectroscopy at 37 °C; results for [(AAAQ)5(AAAE)(AAAQ)5]2 are shown in Figure 6. The [θ]222 was monitored as a function of time, and full-wavelength scans were recorded every 30 min to observe any conformational changes that may not be detected at 222 nm. A compilation of these spectra is shown in the Figure 6 inset. As shown in Figure 6, the [θ]222 at 37 °C remains constant at approximately −48 000 deg cm2 dmol−1 for up to 14 h, and the full-wavelength scans taken every 30 min are identical. Identical stability was observed for the proteins of higher molecular weight (see Supporting Information).
Figure 6.
Circular dichroic characterization of [(AAAQ)5(AAAE)-(AAAQ)5]2 at 37 °C in 10 mM phosphate and 150 mM NaCl buffer, pH 2.3. The protein maintains helical structure for up to 14 h with no apparent loss in helicity.
These data definitively demonstrate that the proteins maintain a significantly helical structure (80% fractional helicity) at physiological temperatures and salt concentrations over extended periods of time. Taken together, the CD and DSC characterization of the [(AAAQ)5(AAAE)(AAAQ)5]x proteins at physiologically relevant temperatures and saline concentrations indicate that proteins of this family may find use for studying structure–function relationships in biological environments.
An overall goal of these investigations was the production of artificial protein polymers with controlled functionalization and conformational properties. The sequences reported here offer a chemically neutral polymer backbone on which chemically reactive functional groups have been regularly placed, offering opportunities for the elucidation of specific structure–function relationships in polymeric materials. The reported biomacromolecules adopt highly helical conformations that are sensitive to variations in temperature and solution conditions, which may offer additional advantages for the purposeful variation of structure. Additionally, the glutamic acid groups in these polymers are competent for chemical modification via straightforward strategies, which will be the focus of future reports.
Conclusions
The alanine-rich amino acid sequence [(AAAQ)5(AAAE)-(AAAQ)5]x, with architectural features modeled after naturally occurring alanine-rich proteins and synthetic alanine-rich peptides, was produced via protein engineering strategies. Solution characterization of the proteins via CD spectroscopy confirms that these proteins are highly helical in dilute aqueous solution, with MRE values among the highest previously reported for helical proteins and peptides. The proteins undergo an irreversible, thermally induced transition to β-sheet structure at temperatures greater than approximately 55 °C when dissolved in 10 mM phosphate buffer, pH 2.3, but do not exhibit such a transition when dissolved in 10 mM phosphate and 150 mM NaCl buffer, pH 2.3. Interestingly, the proteins show little tendency toward high-order aggregation at low concentrations in either the α-helical or β-sheet form. Furthermore, the proteins maintain a high degree of helicity (80%) at physiological temperatures and salt concentrations without any detectable conformational change for extended periods of time. The stability of the helical structures under these conditions suggests the potential for these molecules in studying and manipulating specific biological interactions at low concentrations. Furthermore, the ability to control the conformation of the protein with variations in temperature and solution conditions also suggests that macromolecules of this class may be useful in studies relating architectural variables to function.
Acknowledgment
This work was supported in part by grants from the Army Research Office (DAAD19-02-1-0173), the University of Delaware Research Foundation, and the National Institutes of Health (1-P20-RR17716-01). We are grateful to Nichole Goeden-Wood and Jay Keasling for donation of the modified expression plasmids employed in these experiments. We thank Brian Morrison at the Molecular Analysis Facility at the Univerisity of Iowa for performing amino acid analysis. Vincent Conticello and Kent Kirshenbaum are thanked for helpful discussions, and Christopher Roberts is thanked for assistance with DSC experiments.
Footnotes
Supporting Information Available.
Energy minimization calculations to estimate distance between functional groups; representative purification results obtained via SDS–PAGE analysis; amino acid analysis; HPLC analysis, and MALDI-TOF analysis, IR spectra showing the tendency of proteins to adopt β-sheet structures; CD spectra for [(AAAQ)5-(AAAE)(AAAQ)5]6, showing conformational behavior and stability; and CD experiments showing reversibility of structural changes at lower temperatures. This information is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Krejchi MT, Atkins EDT, Waddon AJ, Fournier MJ, Mason TL, Tirrell DA. Science. 1994;265:1427–1432. doi: 10.1126/science.8073284. [DOI] [PubMed] [Google Scholar]
- 2.Ferrari FA, Cappello J. In: Protein-Based Materials. McGrath K, Kaplan D, editors. Boston: Birkhauser; 1997. pp. 37–60. [Google Scholar]
- 3.Yu SM, Conticello V, Zhang G, Kayser C, Fournier MJ, Mason TL, Tirrell DA. Nature. 1997;389:187–190. doi: 10.1038/38254. [DOI] [PubMed] [Google Scholar]
- 4.Petka WA, Hardin JL, McGrath KP, Wirtz D, Tirrell DA. Science. 1998;281:389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 5.Winkler S, Szela S, Avtges P, Valluzzi R, Kirschner DA, Kaplan D. Int. J. Biol. Macromol. 1999;24:265–270. doi: 10.1016/s0141-8130(98)00088-9. [DOI] [PubMed] [Google Scholar]
- 6.Szela S, Avtges P, Valluzzi R, Winkler S, Wilson D, Kirschner D, Kaplan DL. Biomacromolecules. 2000;1:534–542. doi: 10.1021/bm0055697. [DOI] [PubMed] [Google Scholar]
- 7.McMillan RA, Conticello VP. Macromolecules. 2000;33:4809–4821. [Google Scholar]
- 8.Zhou YT, Wu SX, Conticello VP. Biomacromolecules. 2001;2:111–125. doi: 10.1021/bm005598h. [DOI] [PubMed] [Google Scholar]
- 9.Chilkoti A, Dreher MR, Meyer DE. Adv. Drug Delivery Rev. 2002;54:1093–1111. doi: 10.1016/s0169-409x(02)00060-1. [DOI] [PubMed] [Google Scholar]
- 10.Meyer DE, Chilkoti A. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 11.Wright ER, McMillan RA, Cooper A, Apkarian RP, Conticello VP. Adv. Funct. Mater. 2002;12:149–154. [Google Scholar]
- 12.Megeed Z, Cappello J, Ghandehari H. Adv. Drug Delivery Rev. 2002;54:1075–1091. doi: 10.1016/s0169-409x(02)00063-7. [DOI] [PubMed] [Google Scholar]
- 13.Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. Biomacromolecules. 2003;4:602–607. doi: 10.1021/bm0201082. [DOI] [PubMed] [Google Scholar]
- 14.Panitch A, Yamaoka T, Fournier MJ, Mason TL, Tirrell DA. Macromolecules. 1999;32:1701–1703. [Google Scholar]
- 15.Lee TAT, Cooper A, Apkarian RP, Conticello VP. Adv. Mater. 2000;12:1105–1110. [Google Scholar]
- 16.Fahnestock SR, Irwin SL. Appl. Microbiol. Biotechnol. 1997;47:23–32. doi: 10.1007/s002530050883. [DOI] [PubMed] [Google Scholar]
- 17.Qu Y, Payne SC, Apkarian RP, Conticello VP. J. Am. Chem. Soc. 2000;122:5014–5015. [Google Scholar]
- 18.Welsh ER, Tirrell DA. Biomacromolecules. 2000;1:23–30. doi: 10.1021/bm0002914. [DOI] [PubMed] [Google Scholar]
- 19.Di Zio K, Tirrell DA. Macromolecules. 2003;36:1553–1558. [Google Scholar]
- 20.Tang A, Wang C, Stewart RJ, Kopecek J. J. Controlled Release. 2001;72:57–70. doi: 10.1016/s0168-3659(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 21.Lazo ND, Downing DT. J. Biol. Chem. 1999;274:37340–37344. doi: 10.1074/jbc.274.52.37340. [DOI] [PubMed] [Google Scholar]
- 22.Davies PL, Baardsnes J, Kuiper MJ, Walker VK. Philos. Trans. R. Soc. London, Ser. B. 2002;357:297–935. doi: 10.1098/rstb.2002.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding MM, Ward LG, Haymet ADJ. Eur. J. Biochem. 1999;264:653–665. doi: 10.1046/j.1432-1327.1999.00617.x. [DOI] [PubMed] [Google Scholar]
- 24.Chakrabartty A, Baldwin RL. Adv. Protein Chem. 1995;46:141–176. [PubMed] [Google Scholar]
- 25.Scholtz JM, Baldwin RL. Annu. Rev. Biophys. Biomol. Struct. 1992;21:95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- 26.Goeden-Wood NL, Conticello VP, Muller SL, Keasling JD. Biomacromolecules. 2002;3:874–879. doi: 10.1021/bm0255342. [DOI] [PubMed] [Google Scholar]
- 27.Short Protocols in Molecular Biology. 4th. New York: John Wiley & Sons; 1999. [Google Scholar]
- 28.Marqusee S, Baldwin RL. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shalongo W, Dugad L, Stellwagen E. J. Am. Chem. Soc. 1994;116:8288–8293. [Google Scholar]
- 30.Cochran DAE, Penel S, Doig AJ. Protein Sci. 2001;10:463–470. doi: 10.1110/ps.31001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirshenbaum K, Tirrell DA. Unpublished results. [Google Scholar]
- 32.Richardson JS, Richardson DC. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 33.Scholtz JM, Qian H, Robbins VH, Baldwin RL. Biochemistry. 1993;32:9668–9676. doi: 10.1021/bi00088a019. [DOI] [PubMed] [Google Scholar]
- 34.Aurora R, Rose GD. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penel S, Morrison RG, Mortishire-Smith RJ, Doig AJ. J. Mol. Biol. 1999;293:1211–1219. doi: 10.1006/jmbi.1999.3206. [DOI] [PubMed] [Google Scholar]
- 36.Vila JA, Ripoll DR, Scheraga HA. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13075–13079. doi: 10.1073/pnas.240455797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen C, Parry AD. Proteins: Struct., Funct., Genet. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 38.Hodges RS. Biochem. Cell Biol. 1996;74:133–154. doi: 10.1139/o96-015. [DOI] [PubMed] [Google Scholar]
- 39.Schneider JP, Lombardi A, DeGrado WF. Folding Des. 1998;3:R29–R40. doi: 10.1016/S1359-0278(98)00011-X. [DOI] [PubMed] [Google Scholar]
- 40.Micklatcher C, Chmielewski J. Curr. Opin. Chem. Biol. 1999;3:724–729. doi: 10.1016/s1367-5931(99)00031-9. [DOI] [PubMed] [Google Scholar]
- 41.Acharya A, Ruvinov SB, Gal J, Moll JR, Vinson C. Biochemistry. 2002;41:14122–14131. doi: 10.1021/bi020486r. [DOI] [PubMed] [Google Scholar]
- 42.Schnarr NA, Kennan AJ. J. Am. Chem. Soc. 2003;125:667–671. doi: 10.1021/ja027489b. [DOI] [PubMed] [Google Scholar]
- 43.Huang CY, Getahun Z, Wang T, DeGrado WF, Gai F. J. Am. Chem. Soc. 2001;123:12111–12112. doi: 10.1021/ja016631q. [DOI] [PubMed] [Google Scholar]
- 44.McMillan RA, Lee TAT, Conticello VP. Macromolecules. 1999;32:3643–3648. [Google Scholar]
- 45.Merutka G, Shalongo W, Stellwagen E. Biochemistry. 1991;30:4245–4248. doi: 10.1021/bi00231a020. [DOI] [PubMed] [Google Scholar]
- 46.Scholtz JM, Marqusee S, Baldwin RL, York EJ, Stewart JM, Santoro M, Bolen DW. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2854–2858. doi: 10.1073/pnas.88.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JS, Kennedy RJ, Kemp DS. J. Am. Chem. Soc. 2002;124:945–962. doi: 10.1021/ja011726d. [DOI] [PubMed] [Google Scholar]
- 48.Yu M, Nowak AP, Deming TJ, Pochan DJ. J. Am. Chem. Soc. 1999;121:12210–12211. [Google Scholar]
- 49.Scholtz JM, Qian H, York EJ, Stewart JM, Baldwin RL. Biopolymers. 1991;31:1463–1470. doi: 10.1002/bip.360311304. [DOI] [PubMed] [Google Scholar]
- 50.Chakrabartty A, Kortemme T, Baldwin RL. Protein Sci. 1994;3:843–852. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holtzer ME, Holtzer A. Biopolymers. 1992;32:1675–1677. doi: 10.1002/bip.360321209. [DOI] [PubMed] [Google Scholar]
- 52.Blondelle SE, Forood B, Houghten RA, Perez-Paya E. Biochemistry. 1997;36:8393–8400. doi: 10.1021/bi963015b. [DOI] [PubMed] [Google Scholar]
- 53.Fahnestock S. In: Biopolymers, Vol 8: Polyamides and Complex Proteinaceous Materials II. Fahnestock S, Steinbuchel A, editors. Weinheim, Germany: Wiley-VCH; 2003. pp. 47–79. [Google Scholar]
- 54.van Beek JD, Hess S, Vollrath F, Meier BH. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. Biomaterials. 2003;24:401–416. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 56.Satoh M, Kato F, Komiyama J. Biopolymers. 1993;33:985–993. [Google Scholar]
- 57.Kemp DS, Allen TJ, Oslick SL, Boyd JG. J. Am. Chem. Soc. 1996;118:4240–4248. [Google Scholar]
- 58.La Rosa C, Milardi D, Grasso D, Guzzi R, Sportelli L. J. Phys. Chem. 1995;99:14864–14870. [Google Scholar]
- 59.Arriaga P, Menendez M, Villacorta JM, Laynez J. Biochemistry. 1992;31:6603–6607. doi: 10.1021/bi00143a034. [DOI] [PubMed] [Google Scholar]