Abstract
Magnetic resonance imaging (MRI) is a technique used in both clinical and experimental settings to produce high resolution images of opaque organisms without ionizing radiation. Currently, MR imaging is augmented by contrast agents and the vast majority these small molecule Gd(III) chelates are confined to the extracellular regions. As a result, contrast agents are confined to vascular regions reducing their ability to provide information about cell physiology or molecular pathology. We have shown that polypeptides of arginine have the capacity to transport Gd(III) contrast agents across cell membranes. However, this transport is not unidirectional and once inside the cell the arginine-modified contrast agents efflux rapidly, decreasing the intracellular Gd(III) concentration and corresponding MR image intensity. By exploiting the inherent disulfide reducing environment of cells, thiol compounds, Gd(III)-DOTA-SS-Arg8 and Gd(III)-DTPA-SS-Arg8, are cleaved from their cell penetrating peptide transduction domains upon cell internalization. This reaction prolongs the cell-associated lifetime of the chelated Gd(III) by cleaving it from the cell transduction domain.
INTRODUCTION
The advent of wide-bore, high field instruments with improved coil designs and pulse sequences has made magnetic resonance imaging (MRI) a powerful tool for observing developmental and physiological events (1–4). This versatile modality is non-invasive and allows 3D-imaging of whole animals (5). This imaging technique is supplemented with an array of exogenous Gd(III) contrast agents conjugated to scaffolds (nanoparticles, antibodies, and proteins), that can detect physiological events (metal ion concentration, enzyme activation, and pH), and can be cell membrane permeable (via cell penetrating peptides or receptor uptake) (6–17). The focus of this work is to develop cell-permeable contrast agents that can be trapped inside cells via a cleavable disulfide linkage between the agent and the transduction moiety.
The biological role of thiol-containing molecules has been actively explored and a great deal is known about the active species and relative concentrations (18). The pseudo-tripeptide of glutathione (GSH) is recognized as the most abundant and subsequently most active thiol species in biological environments (19, 20). Previous work has shown that the intracellular GSH concentration of living cells (~ 1–10 mM) is significantly higher than that of the surrounding extracellular plasma (~ 2 µM). Therefore, thiols can be exploited as a selective chemical release switch for intracellular delivery of cargo (21–23). We have applied this strategy to our reported intracellular MR contrast agents in order to reduce cellular leaching (24). As a result, we have prepared a class of MR probes that are cell permeable while displaying increased intracellular lifetimes. These probes provide a method to passively label and non-invasively track the fate of transplanted tissues and cells.
It has been shown that an octamer of arginine residues allows cell membrane transduction of various cargo molecules, including fluorophores and contrast agents (25–27). However, the ability of these probes to cross cell membranes is not unidirectional. Not surprisingly, arginine-modified agents are able to leach out of the cells displaying a concentration decay curve of intracellular contrast agent and consequently decreasing the observed MR signal over time (28, 29).
Therefore, contrast agents containing a disulfide bond between the Gd(III) chelate and the transduction moiety (e.g., an octamer of arginine residues) were synthesized to enhance cell-associated retention (Figure 1). Exploiting the inherent disulfide reducing environment of cells, these new agents are cleaved from the cellular transduction domains upon entrance into the cytoplasm. This cleavage of the arginine transport domain shows prolonged cellular lifetime of the MR probe by dissociation of the transport peptide.
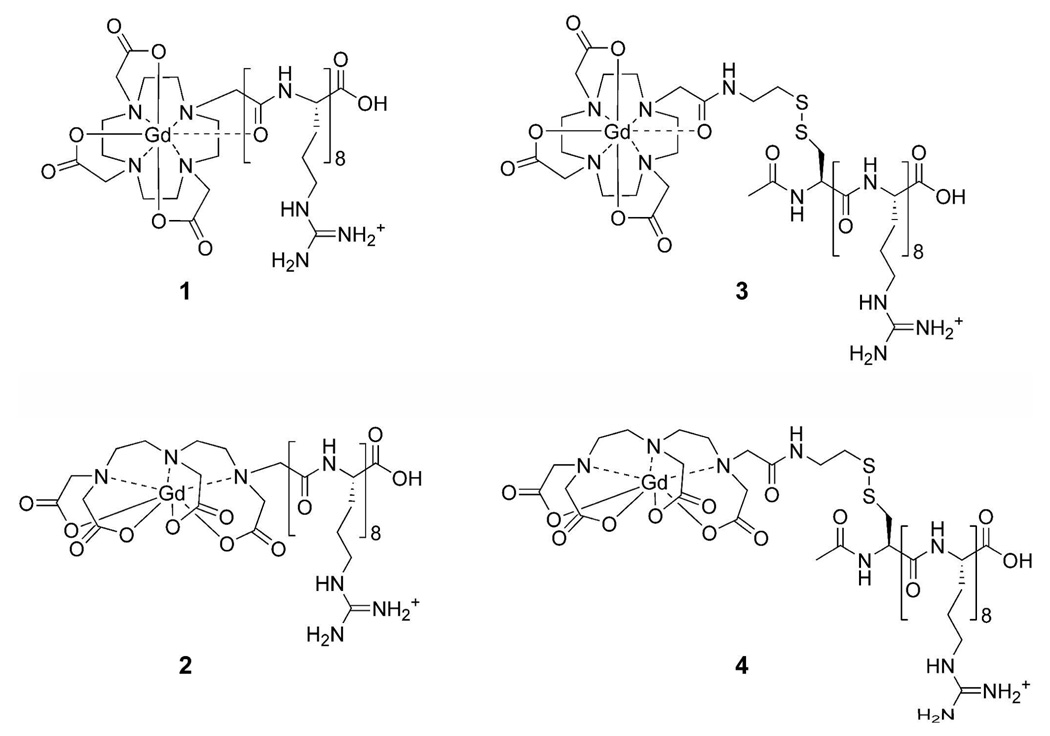
Figure 1.
Structures of the arginine-modified (cell-permeable contrast agents) and disulfide bridged (cell-permeable contrast agents) respectively. (1) Gd(III)-DOTA-Arg8, (2) Gd(III)-DTPA-Arg8, (3) Gd(III)-DOTA-SS-Arg8, and (4) Gd(III)-DTPA-SS-Arg8.
EXPERIMENTAL PROCEDURES
All reagents and solvents were of the highest purity available from Sigma-Aldrich (Milwaukee, WI) and Fluka (Sigma-Aldrich Chemie; GmbH, Switzerland) unless otherwise noted. Modified Wang resin and amino acids were purchased from Novabiochem (San Diego, CA). DMEM (with 4 mM L-glutamine modified to contain 4.5 g/L glucose and 1.5 g/L sodium carbonate), EMEM (with Earle’s BSS and 2 mM L-glutamine modified to contain 1.0 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 1.5 g/L sodium bicarbonate), NIH/3T3 cells, FBS, CBS, and 0.25% trypsin/EDTA solutions were purchased from the American Type Culture Collection (ATCC; Manassas, VA). Vent-cap flasks, multiwell plates, cell scrapers, and DPBS w/o calcium and magnesium were purchased from Fisher Scientific (Pittsburgh, PA).
1H and 13C NMR spectra were obtained on a Varian (Walnut Creek, CA) Inova spectrometer at 500 and 125 MHz, respectively. Compounds were dissolved in D2O (4.80 ppm used as internal references for NMR spectra). Due to the minimal solubility associated with the peptide-chelate conjugates, 13C NMR spectra were collected in D2O, and the first carbon resonance was assigned a value of 24.42 ppm. Mass spectrometry samples were analyzed using ESI, single quadrupole mass spectrometry on a Varian 1200L spectrometer or MALDI-TOF mass spectrometry on an Applied Biosystems (Foster City, CA) Voyager-DE Pro. Results reported for m/z are for [M+H+]+ or [M-H+]− unless stated otherwise. Elemental Analyses were performed at Desert Analytics Laboratory (Tuscon, AZ).
ICP-MS was performed on a computer-controlled Thermo Elemental (Waltham, MA) PQ ExCell Inductively Coupled Plasma Mass Spectrometer. All standards and samples contain 5 ng/mL of a multi-element internal standard (Spex CertiPrep; Metuchen, NJ) consisting of Bi, Ho, In, Li, Sc, Tb, Y and 3% nitric acid (v/v). Gadolinium standards were prepared in concentrations of 0.05, 0.10, 0.25, 0.50, 1.0, 5.0, 10, 25, and 50 ng/mL. Analysis was accomplished with 1 survey and 3 main scans (peak jumping, 100 sweeps per run) using 156Gd and 157Gd isotopes and interpolating through 209Bi and 115In internal standards.
HPLC-MS
Analytical reverse phase HPLC-MS was performed on a computer controlled Varian Prostar system consisting of a 410 autosampler equipped with a 100 µL sample loop, two 210 pumps with 5 mL/min heads, a 363 fluorescence detector, a 330 photodiode array (PDA) detector, and a 1200L single quadrupole ESI-MS. All separations were executed with a 1.0 mL/min flow rate using a Waters 4.6 × 250 mm 5 µm Atlantis C18 column, with a 3.1:1 split directing one part to the MS and 3.1 parts to the series-connected light and fluorescence detectors. Mobile phases consisted of Millipore Synthesis grade water (solvent A) and HPLC-grade MeCN (solvent B). Preparative HPLC was accomplished using a Varian Prostar system. Two Prostar 210 pumps with 25 mL/min heads supplied a 5 mL manual inject sample loop. Detection was performed after a 20:1 split by a two-channel Prostar 325 UV-visible detector and, on the low-flow side, a HP 1046A fluorescence detector. The mobile phases were Millipore Synthesis grade water and HPLC-grade MeCN. Preparative runs were on a Waters 19 × 250 mm 10 µm Atlantis C18 column.
Relaxivity (r1)
A 2 mM stock solution of 1–4, Gd(III)-DTPA, or Gd(III)-DOTA in 100 mM PBS buffer at pH 7.41 were serially diluted to give 500 µL of each of the five approximate concentrations for each compound: 0.15, 0.3, 0.5, 1.0, and 2.0 mM. The T1 of each sample was determined at 60 MHz (1.5 T) and 37 °C using an inversion recovery pulse sequence on a Bruker mq60 Minispec (Bruker Canada; Milton, Ontario, Canada) and at 600 MHz (14.1 T) and ambient temperature using a saturation recovery pulse sequence on a Bruker Omega 600WB spectrometer (Bruker BioSpin; Billerica, MA). Reproducibility of the T1 data was ± 0.3%. Ten microliters of each sample was analyzed for exact gadolinium concentration using ICP-MS. The inverse of the longitudinal relaxation time (T1) was plotted against the concentration obtained from ICP-MS and fit to a straight line. Lines were fit with R2 > 0.998 and each relaxivity was assessed in duplicate.
Determination of q by Luminescence Lifetime Measurements
Complexes (10–13) were dissolved in H2O and D2O. Emission was monitored at 544 nm with excitation at 229 nm on a Hitachi F4500 Fluorescence Spectrophotometer operating in phosphorescence lifetime mode. Twenty-five scans were averaged and fit to a monoexponential decay (R2 > 0.98) to give the phosphorescent lifetimes which were entered into this equation (corrected for one amide oscillator, where k is given in ms−1): q = 5.0 (kH2O -kD2O - 0.06) (30, 31).
Determination of τm by 17O Transverse Relaxation Rate Measurements
Samples of 1–4 were prepared at 15–20 mM concentrations in 1% 17O enriched water (Medical Isotopes, Inc.; Pelham, NH) adjusted to pH 7.40. Lock was achieved by means of an external D2O standard. 17O spectra were obtained at 54 MHz (number of averaged transients was 160 – 320 and relaxation delay was 400 ms) at temperatures ranging from 1 °C to 86 °C in 5 °C increments. The 17O transverse relaxation rate was determined by obtaining the line width (in Hz) at half of the peak height, Δν1/2, of the 17O water signal and later fitting the data (32–34). Using known sample concentrations and q values, the relaxation data were fit to these four parameters at 25 °C: τm (water exchange rate), ΔH‡ (activation enthalpy), T1e (electronic relaxation rate), and ΔET1e (activation energy of T1e) (35).
Cell Culture Conditions
All cell lines, media, sera, buffers, and dissociation reagents were purchased from ATCC. NIH/3T3 cells (CRL-1658) were grown in DMEM (with 4 mM L-glutamine, 4,5 g/L glucose, 1.0 mM sodium pyruvate, 1.5 g/L sodium carbonate, and phenol red supplemented with 10% CBS). Cells were grown in a humidified incubator at 37 °C and 5% CO2. Cells were washed with DPBS (without calcium and magnesium) and dissociated with a 0.25% trypsin-EDTA solution. All flasks and multi-well plates were Corning brand, tissue culture treated, and sterile. The studies for each contrast agent and time point were performed in triplicate
Cells were counted and viability assessed using a Guava EasyCite Mini Personal Cell Analyzer (Guava Technologies; Hayward, CA). Cell samples were diluted 10 fold with Guava ViaCount reagent (total volume of 200 µ L), allowed to stain at room temperature for at least 5 minutes (but no longer than 20 minutes), vortexed, and measured. Each sample was measured using the Guava ViaCount software module using 1000 trials per run doing 2 separate runs per sample. Instrument reproducibility was assessed daily following the manufacturer’s suggested protocol using GuavaCheck reagent and the corresponding software module.
ICP-MS assessment of the cell associated gadolinium was determined by collecting and processing all cell and media samples via this procedure: addition of 70% (v/v) nitric acid in water, vortexing (~ 5 min), and digestion at 65 °C for at least four hours. Samples were added to a 15 mL conical tube along with internal standard (5 ng/mL of indium and holmium) and water to give a final nitric acid concentration of 3% (v/v).
Synchrotron Radiation X-Ray Fluorescence (SR-XRF) Analysis
NIH/3T3 cells were grown to 65% confluency on silicon nitride windows (membrane thickness of 500 nm and frame size of 5.0 mm × 5.0 mm, Silson Ltd.; Northampton, England) in Corning brand 24-well cell culture treated plates. Each well had 2 silicon nitride windows (one incubation sample and one leach sample). Cells were incubated with 3.0 mM of compounds 1–4 for 4 hours. Samples were rinsed three times with DPBS and one window was removed for processing (time=0, named No leach sample) while one window was allowed to incubate with fresh media for 4 more hours followed by processing (named Leached sample). Windows were processed by cell fixation in 3.7% formalin for 5 minutes at room temperature, rinsing with DPBS (1×), rinsing with Millipore water (1×), rinsing with 100% ethanol (1×), and drying overnight (18 h, room temp.). The silicon nitride windows were mounted onto a kinematic specimen holder for both visible light and X-ray fluorescence microscopy. The samples were examined on a light microscope (Leica DMXRE), and the cells to be scanned with SR-XRF were located on the window relative to a reference point (one of the four window corners) using a high spatial resolution motorized x/y stage (Ludl Bioprecision).
Scanning SR-XRF microscopy was carried out at the 2-ID-E beamline of the Advanced Photon Source at Argonne National Laboratory (IL, USA). Hard X-rays from an undulator source were monochromatized using a single bounce Si <111> monochromator. The energy was selected to allow for efficient excitation of the Gd L-lines, and also to enable the detection of the P, S, Fe, and Zn K-lines. A Fresnel zone plate (320 µm diameter, focal length f = 250 mm, X-radia; Concord, CA) was used to focus the monochromatic X-ray beam to a spot size of ~ 0.4 × 0.3 µm2 on the specimen. The sample was raster scanned through the beam at room temperature under a helium atmosphere. At each scan position, a full fluorescence spectrum was acquired using an energy dispersive germanium detector (Ultra-LEGe detector, Canberra; Meriden, CT). Elemental content was determined by comparison of fitted sample spectra with NBS thin film standards 1832 and 1833 (NIST; Gaithersburg, MD) using MAPS software supplemented with fitting of fluorescence spectra at every pixel (36).
MR Imaging and T1 Analysis
MR measurements were performed on a General Electric/Bruker Omega 600WB 14.1 T imaging spectrometer fitted with Accustar shielded gradient coils at ambient temperature (~ 25 °C). Spin lattice relaxation times (T1) were measured using a saturation recovery pulse sequence with static TE (10.18 ms) and variable TR (127.5, 165, 200, 300, 500, 1000, 2000, 5000, 7500, 10000 ms) values. Two independent trials were run per sample and at least 5 slices were quantified per trial. Student’s t-tests were calculated in Origin 7 SR2 (Origin Lab; Northampton, MA) at a 95% confidence level with 8 degrees of freedom and a null hypothesis value (minimum acceptable time difference) of 100 ms. Images were acquired using a T1-weighted, spin-echo pulse sequence (TE = 10.18, TR = 750 ms) on freshly harvested cells that were spun down (at 250g) within glass capillary tubes for 5 min prior to image acquisition. Total time from harvest to image acquisition was 1–2 h.
Synthesis
Complex Gd(III)-DOTA-Arg8 (1), Gd(III)-DTPA-Arg8 (2), and (S-(2-Aminoethylthio)-2-thiopyridine hydrochloride) (AETP) were synthesized following previously published procedures (14, 24, 37) and (1,4,7-tris(acetic acid-tert-butyl ester)-10-acetic acid-1,4,7,10-tetraazacyclododecane) (DOTA-tris-TB) was purchased from Macrocyclics (Dallas, TX).
Synthesis of end-capped RRRRRRRRC (5)
Polystyrene-based Wang resin containing an Fmoc protected arginine residue (2.00 g, 0.580 mmol/g) was swelled in CH2Cl2 for 10 min (× 3) and washed with peptide synthesis grade DMF (4 × 10 min). The resin was treated three times with a solution of 20% piperdine in DMF (10 min) and the deprotected resin was washed with DMF (4 × 10 min). In a separate vial, Fmoc protected Pbf-arginine (1.51 g 2.32 mmol), HATU (0.881 g, 2.32 mmol), and DIPEA (0.748 g, 5.80 mmol) were dissolved in approximately 3 mL of DMF. The resulting solution was added to the deprotected resin and nitrogen was bubbled through the mixture for 6–8 h. The peptide solution was removed from the resin which was subsequently rinsed with DMF (4 × 10 min). This procedure was repeated a total of seven times in order to achieve the synthesis of an 8 amino acid, polyarginine oligomer bound to the Wang resin (3.96 g).
The Arg8 functionalized resin (2.00 g, 0.290 mmol/g, 0.580 mmol) was deprotected with the piperdine solution and washed with DMF as described above. In a separate vial, Fmoc protected Trt-cysteine (0.725 g, 1.16 mmol) and 2, 4, 6-trimethylpyridine (0.154 g, 1.27 mmol) were dissolved in a minimal amount of DMF. The resulting solution was added to the deprotected resin and nitrogen was bubbled through the mixture for 6–8 h. The peptide solution was removed and the resin was washed with DMF (4 × 10 min each). The resin bound Arg8-Cys oligomer was deprotected with the piperdine solution, and washed with DMF. The resin was resuspended in a solution of acetic anhydride (0.118 g, 1.16 mmol), DIPEA (0.374 g, 2.90 mmol) and DMF (3 mL). Bubbling nitrogen through this mixture for 4 h effectively capped the peptide’s N-terminus. Upon removal of the capping solution, the resin was washed with DMF, CH2Cl2, and MeOH (4 × 10 min each). Following the methanol washes, the resin was dried under vacuum. A solution of 95% TFA, 2.5% EDT, 1.5% H2O, and 1.0% TIS (50 mL) was added to the resin and nitrogen was bubbled through the mixture for 1 h. The resin was filtered and to the filtrate was added MTBE (40 mL) to precipitate a white solid that was subsequently washed with MTBE (3 ×). The solid was dissolved in 30 mL of water and freeze-dried to yield white flaky crystals of the peptide, Arg8-Cys (0.692 g, 84%). 1H NMR-spectrum (D2O @ pH 1.0): δ = 1.22–1.40 (m, 16H), 1.50 (dd, J = 8.5, 5.5 Hz, 16H), 1.59–1.69 (m, 2H), 1.74 (d, J = 6.0 Hz, 3H), 2.89 (m, 16H), 3.95–4.10 (m, 8H); 13C NMR (D2O @ pH 1.0): δ = 24.42, 27.50, 28.03, 39.36, 40.52, 52.26, 53.25, 156.57, 172.61, 173.44, 174.84; with ESI-MS (m/z): 1413.58; calc. for C53H105N33O11S+H+: 1413.68.
Synthesis of 1,4,7-Tris(carboxymethyl)-10-(N-[2-(pyridin-2-yldithio)ethyl]acetamide)-1,4,7,10-tetraazacyclododecane (6)
DOTA-tris-TB (0.600 g, 1.05 mmol), HATU (1.20 g, 3.15 mmol), and anhydrous DIPEA (0.683 g, 5.25 mmol) were dissolved in 4.0 mL of anhydrous DMF and allowed to stir for 10 min under a nitrogen atmosphere while AETP (0.466 g, 2.10 mmol) was dissolved in 2.0 mL of anhydrous DMF. After a 10 min activation time, the AETP solution was added via syringe and the mixture was allowed to stir for 5 h while being monitored by TLC [KNO3–H2O–MeCN (1:9:90)]. Upon reaction completion the solution was evaporated in vacuo. The resulting yellow residue was stirred overnight in 95% TFA, 2.5% TIS, and 2.5% H2O (30 mL). The reaction mixture was concentrated to 10 mL by application of a steady stream of dry nitrogen across the top of the vessel. The crude product was precipitated by addition of MTBE (40 mL) and was subsequently washed with MTBE (3 ×). The yellow solid was dried in vacuo to yield 1.12 g of crude product.
The crude mixture was submitted to preparatory HPLC using a Waters Atlantis column. The solid was brought up in H2O and purified using the following method: begin at 0% B, ramp to 3% B over 1 min, hold at 3% B for 15 min, ramp to 20% B over 7 min, and hold for 8 min, followed by a wash to 100% B before returning to 0% B. The desired fractions (retention time: 32.20 min by UV spectroscopy at 235, 280 nm) were collected and freeze-dried to yield the product as a white solid (0.410 g, 68%). 1H NMR spectrum (D2O ): δ = 2.96–3.10 (m, 11H), 3.41–3.51 (m, 13H), 3.79 (q, J = 16.5 Hz, 4H), 7.33 (t, J = 4.5 Hz, 1H), 7.88 (d, J = 3.5 Hz, 2H), 8.40 (d, J = 4.5 Hz, 1H); 13C NMR spectrum (D2O): δ = 37.05, 38.13, 48.22, 48.46, 50.94, 51.64, 53.73, 55.51, 56.59, 121.92, 122.26, 139.56, 148.50, 158.48, 170.02, 171.96, 175.07; ESI-MS (m/z): 573.26; calc. for C23H36N6O7S2 +H+: 573.70; with Anal. Calcd. for C23H36N6O7S2 • Na • 1.5 H2O: C, 44.36; H, 6.31; N, 13.50; S, 10.30. Found: C, 44.20; H, 6.17; N, 13.38; S, 10.98.
Synthesis of [(2-{carboxymethyl-[2-(carboxymethyl-{[2-(pyridin-2-yldisulfanyl)-ethylcarbamoyl]-methyl}-amino)-ethyl]-amino}-ethyl)-amino]-acetic acid (7)
DTPA dianhydride (2.00 g, 5.60 mmol) was dissolved in 100 mL of anhydrous DMSO and allowed to stir under a nitrogen atmosphere while AETP (0.672 g, 3.04 mmol) was dissolved in a stirring solution of anhydrous DMSO (25 mL) and anhydrous DIPEA (1.07 g, 8.21 mmol). The AETP solution was slowly added to the DTPA dianhydride solution via syringe pump over 5 h. Upon complete addition, the resulting solution was stirred for an additional hour before being quenched with H2O (100 mL) and concentrated in vacuo. The resulting residue was brought up in H2O and submitted to preparatory HPLC with the Waters Atlantis column using the following method: begin at 0% B, ramp to 100% B over 35 min followed by a wash at 100% B for 5 min before returning to 0% B. The desired fractions (retention time: 10.28 min by UV at 235, 280 nm) were collected and freeze-dried to yield the product as a white solid (1.07 g, 63%). 1H NMR (D2O ): δ = 3.04 (t, J = 5.5 Hz, 2H), 3.24–3.35 (m, 6H), 3.54 (t, J = 6.0 Hz, 2H), 3.59 (t, J = 6.0 Hz, 2H), 3.69 (s, 2H), 3.75–3.90 (m, 8H), 7.57 (t, J = 6.5 Hz, 1H), 8.05 (d, J = 8.5 Hz, 1H), 8.15 (t, J = 8.5 Hz, 1H), 8.53 (d, J = 5.5 Hz, 1H); 13C NMR (D2O): δ = 37.45, 37.94, 50.30, 51.24, 52.52, 52.09, 54.48, 56.52, 57.04, 57.21, 123.29, 123.95, 142.71, 145.62, 157.07, 168.74, 170.56, 172.17, 172.78; ESI-MS (m/z): 560.19; calc. for C21H31N5O9S2-H+: 560.63; with Anal. Calcd. for C21H31N5O9S2 • Na: C, 43.14; H, 5.34; N, 11.98; S, 10.97. Found: C, 42.70; H, 5.21; N, 12.02; S, 11.29.
Synthesis of DO3A-SS-Arg8 (8)
Compound 6 (0.160 g, 0.306 mmol) was added to end-modified Arg8-Cys (5) (0.432 g, 0.306 mmol) dissolved in 30 mL of degassed phosphate buffer (10 mM, pH 8.0). The solution was stirred under nitrogen atmosphere at room temperature. The reaction was monitored by analytical HPLC and found to be complete after 72 h. An aliquot (2.5 mL) of the solution was removed for characterization while the remaining material (27.5 mL) was carried onto the synthesis of Gd(III)-DOTA-SS-Arg8 (3). Dialysis (Mwt cut-off 1,000) against Millipore H2O (4 L × 4 days), yielded 3 mL of a translucent liquid that was 0.2 μm syringe filtered and freeze-dried to yield a white solid (0.389 g, 74%). 1H NMR spectrum (D2O) shows the disappearance of all aromatic protons. 13C NMR spectrum (D2O): δ = 24.42, 28.16, 28.88, 37.11, 38.17, 40.65, 40.79, 48.44, 50.71, 51.90, 53.59, 54.91, 55.52, 56.66, 156.86, 170.04, 172.19, 172.80, 173.75, 175.41; ESI-MS (m/z): 1875.31; calc. for C70H136N38O18S2+H+: 1875.21.
Synthesis of DTPA-SS-Arg8 (9)
Compound 7 (0.175 g, 0.312 mmol) was added to end-modified Arg8-Cys (3) (0.440 g, 0.312 mmol) dissolved in 30 mL of degassed phosphate buffer (10 mM, pH 8.0). The solution was stirred under a nitrogen atmosphere at room temperature. The reaction was monitored by analytical HPLC and found to be complete after 72 h. An aliquot (2.5 mL) of the solution was removed for characterization while the remaining material (27.5 mL) was carried onto the synthesis of Gd(III)-DTPA-SS-Arg8 (4). Dialysis (Mwt cut-off 1,000) against Millipore H2O (4 L × 4 days), yielded 4 mL of a translucent liquid that was 0.2 µm syringe filtered and freeze-dried to yield a white solid (0.351 g, 66%). 1H NMR (D2O) shows the disappearance of all aromatic protons. 13C NMR (D2O): δ = 24.42, 28.20, 38.79, 40.66, 50.53, 52.19, 53.44, 53.53, 54.94, 55.19, 57.88, 58.91, 156.84, 172.70, 172.97, 173.60, 174.32, 178.33; ESI-MS (m/z): 1864.21; calc. for C69H131N37O20S2+H+: 1864.14.
Synthesis of Gd(III)-DOTA-SS-Arg8 (3)
To an aliquot of DO3A-SS-Arg8 (8) (27.5 mL, 0.285 mmol), was added Gd(OH)3 (0.064 g, 0.314 mmol). All metalations were performed with a 1.1:1.0 molar excess of metal salt to unmetalated complex to minimize unmetalated final product. The reaction mixture was brought to pH 6.0 with 1.0 M NaOH and allowed to stir for 7 d at room temperature under a nitrogen atmosphere. Dialysis (Mwt cut-off 1,000) against Millipore H2O (4 L × 4 days), yielded 30 mL of a translucent liquid that was brought to pH 11.0 with concentrated NH4OH to precipitate excess free gadolinium. This suspension was 0.2 µm syringe filtered and freeze-dried to yield a white powder which was found to be (3) (0.453 g, 78%) by ESI-MS (m/z): 2029.44 (m/1), 1014.70 (m/2), 677.43 (m/3) 508.26 (m/4) with Gd isotope pattern; calc. for C71H133GdN38O18S2+H+: 2029.44.
Synthesis of Gd(III)-DTPA-SS-Arg8 (4)
To an aliquot of DTPA-SS-Arg8 (9) (27.5 mL, 0.286 mmol), was added Gd(OH)3 (0.064 g, 0.315 mmol). The reaction mixture was brought to pH 6.0 with 1.0 M NaOH and allowed to stir for 7 d at room temperature under a nitrogen atmosphere. Dialysis (Mwt cut-off 1,000) against Millipore H2O (4 L × 4 days), yielded 30 mL of a translucent liquid that was brought to pH 11.0 with concentrated NH4OH to precipitate excess free gadolinium. This suspension was 0.2 µm syringe filtered and freeze-dried to yield a white powder which was found to be (4) (0.484 g, 84%) by ESI-MS (m/z): 2017.30 (m/1), 1008.55 (m/2), 672.99 (m/3), 505.12 (m/4), 404.38 (m/5) with a Gd isotope pattern; calc. for C69H127GdN37O20S2+H+: 2017.36.
Synthesis of Tb(III)-DOTA-Arg8 (10)
The Tb(III) analogue of 1 was synthesized using a previously outlined metalation procedure and substituting Tb(OH)3 for Gd(OH)3 (14). The resulting white solid was found to be 10 (45.7 mg, 42%) by ESI-MS (m/z): 1810.85 (m/1), 906.07 (m/2), 604.24 (m/3), 453.47 (m/4); calc. for C64H121N36O16Tb+H+: 1810.80.
Synthesis of Tb(III)-DTPA-Arg8 (11)
The Tb(III) analogue of 2 was synthesized using a previously outlined metalation procedure and substituting Tb(OH)3 for Gd(OH)3 (24). The resulting white solid was found to be 11 (42.0 mg, 52%) by ESI-MS (m/z): 1798.63 (m/1), 899.91 (m/2), 600.26 (m/3), 450.47 (m/4); calc. for C62H115N35O18Tb+H+: 1798.73.
Synthesis of Tb(III)-DOTA-SS-Arg8 (12)
The Tb(III) analogue of 3 was synthesized using compound 8 (10.0 mg, 0.005 mmol), Tb(OH)3 (1.23 mg, 0.006 mmol), and the same metalation procedure previously described for 3 (6.0 mg, 60%). ESI-MS (m/z): 2031.01 (m/1), 1016.00 (m/2), 677.73 (m/3) 508.54 (m/4), 407.12 (m/5); calc. for C71H133N38O18S2Tb+H+: 2031.12.
Synthesis of Tb(III)-DTPA-SS-Arg8 (13)
The Tb(III) analogue of 4 was synthesized using compound 9 (10.0 mg, 0.005 mmol), Tb(OH)3 (1.23 mg, 0.006 mmol), and the same metalation procedure described for 4 (5.9 mg, 55%). ESI-MS (m/z): 2019.02 (m/1), 1010.01 (m/2), 673.64 (m/3) 505.43 (m/4), 404.65 (m/5); calc. for C69H127N37O20S2Tb+H+: 2019.04.
RESULTS
Synthesis
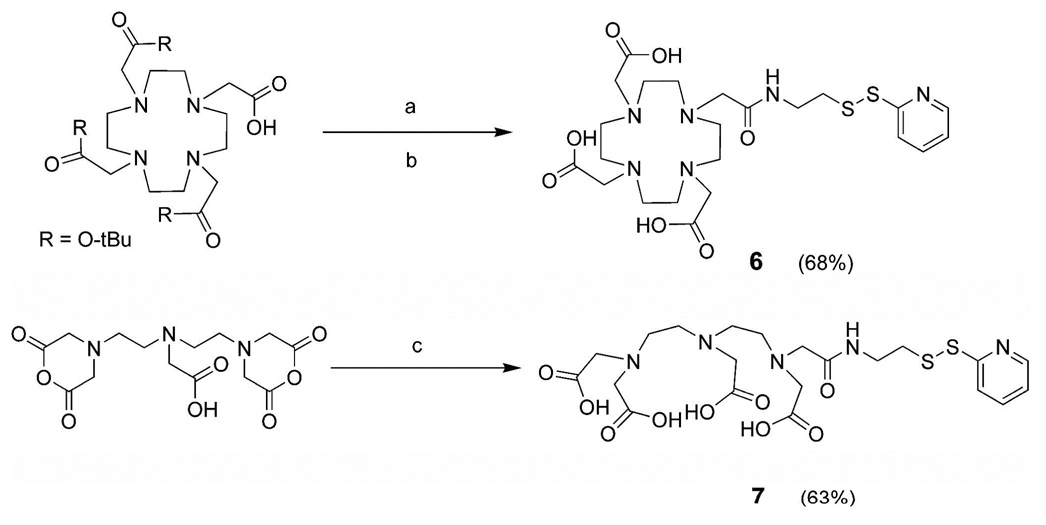
The synthesis of the pyridyl-activated Gd(III) chelators was accomplished by coupling an amine-terminated AETP to an activated form of either the DTPA or DOTA chelate (Scheme 1). Commercially available DTPA-dianhydride is amidated forming amines, while the DOTA-tris-TB was activated in situ by the commonly used uranium salt, HATU. Both reactions required an excess of DIPEA in order to deprotonate the hydrochloride salt starting material (AETP) and to aid in amide bond formation (24). Upon purification via preparative RP-HPLC, compounds 4 and 5 were afforded with yields of 68 and 63%, respectively. The peptide RRRRRRRRC was synthesized without automation using standard Fmoc solid-phase synthesis methodology on pre-loaded Wang resin (24, 25). After the addition of the cysteine residue, the resin was treated with acetic anhydride to end-cap the peptide’s terminal amine. To prepare peptide sequences with minimal structural differences as compared to the previously published Arg8-MR contrast agents, (e.g. charge and no free terminal amine) this end-capping step was performed prior to cleavage and deprotection from the resin (24, 25).
Scheme 1.
Synthesis of the pyridyl-activated Gd(III) Chelators a
aConditions: (a) AETP, HATU, DIPEA, DMF ; (b) TFA/H2O/TIS, followed by MTBE precipitation. (c) AETP, DMSO, and H2O.
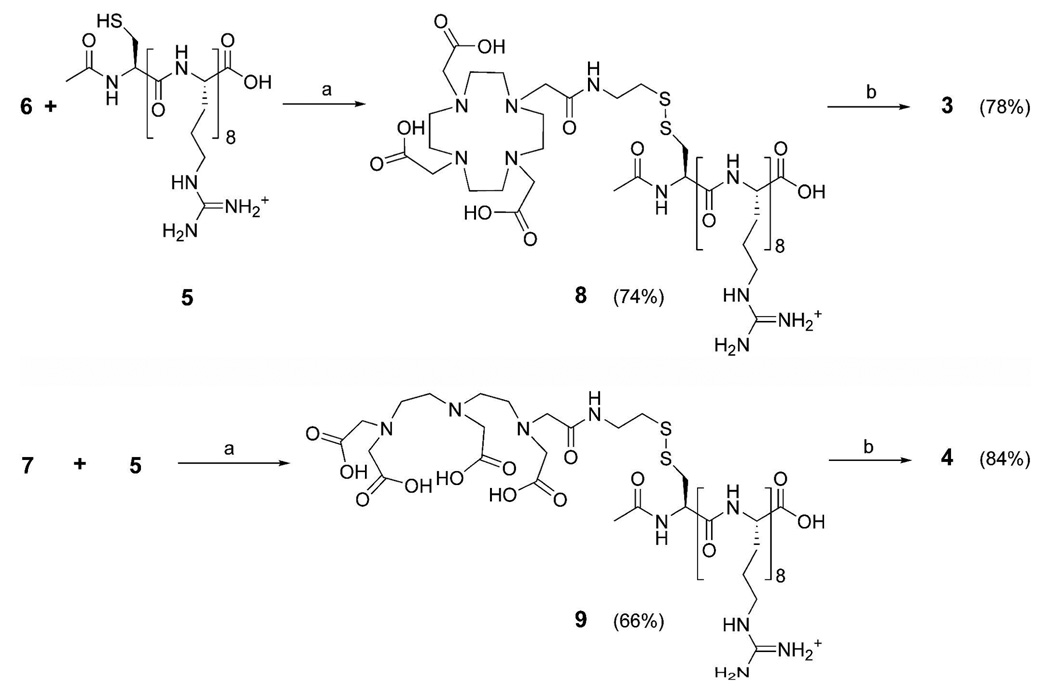
In order to optimize the thiol exchange reaction required to produce 8 and 9, the pyridyl-activated chelates (6 or 7) were quickly added to stirring solutions of the end-capped Arg8-Cys (5) oligomer under a nitrogen atmosphere in degassed phosphate buffer at pH 8.0 (Scheme 2). It was found that when 6 and 7 were added in equimolar quantities to 5, synthetic yields were maximized. We speculate that adding higher amounts of the free thiol (5) lowered the overall yield by increasing oxidation and disulfide exchange of the desired product. This effect has been observed in previous disulfide exchange reactions (38). Metalation of the cell-permeable contrast agents was achieved by adding Gd(OH)3 to the phosphate buffered solutions of 8 and 9. To ensure the complete insertion of Gd(III) the buffered solutions were lowered to pH 6.0 and allowed to stir for 7 days under a nitrogen atmosphere. The proposed structures of the peptide conjugates 5, 8, and 9 were confirmed by NMR spectroscopy, ESI-MS, and HPLC, while the unconjugated, small molecule chelates were characterized by NMR spectroscopy, ESI-MS, HPLC and EA.
Scheme 2.
Synthesis of disulfide bridged, intracellular contrast agents (3) Gd(III)-DOTA-SS-Arg8 and (4) Gd(III)-DTPA-SS-Arg8. a
aConditions: (a) stirring for 3 days under N2 atmosphere in 10 mM phosphate buffer at pH 8.0; (b) Gd(OH)3 , 5 days, pH 6.0.
Relaxivity, q, and τM
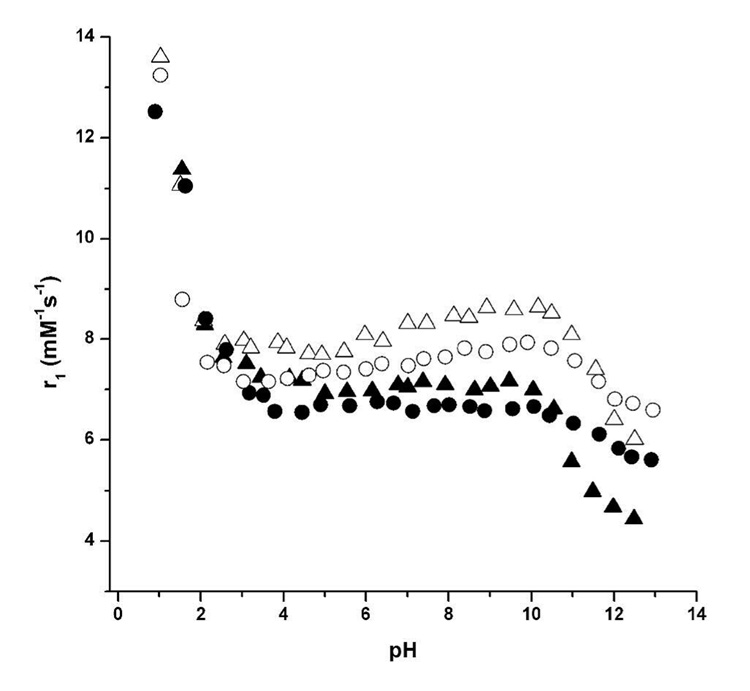
The relaxivity measurements (r1, mM−1s−1) provided in Table 1 show the relative enhancement of water proton relaxation rates per Gd(III) ion for complexes 1–4. These rates were measured at 1.5 and 14.1 T in order to provide data at both clinical and research field strengths. As expected, the r1 rates are strongly field dependant; however, 1–4 maintain higher relaxation rates than currently available contrast agents, Gd(III)-DOTA and Gd(III)-DTPA, at both 1.5 and 14.1 T. The relaxation rates of 1–4 were tested over a wide pH range (1.0–12.5) in order to determine the efficacy of these agents to relax water molecules at biologically relevant pH levels (Figure 3). At low pH (~ 1–4), the relaxivity for 1–4 is consistent with Gd(III) diassociation from the chelating cage. Conversely, at high pH (~ 9–13), the relaxivity shows a decrease that is most likely attributable to a structure that allows carbonate ion binding or association of the metal center to the slowly exchanging hydroxyl ions (23, 39, 40). However, in the physiologically relevant range of pH 4–8, all four of the cell-permeable agents display constant relaxivity values.
Table 1.
Relaxometric Properties of Complexes 1–4 at pH 7.4 and 37 °C
Figure 3.
The millimolar relaxivities (r1, mM−1s−1) of complexes 1–4 plotted against pH. Values were measured at 60 MHz and 37 °C. The contrast agents are represented as: 1 (closed triangles), 2 (open triangles), 3 (closed circles), and 4 (open circles). Comparison of the closed data points allows correlation of the two DOTA-based ligands. Comparison of the open data points allows correlation of the two DTPA-based ligands.
The Tb(III) analogs of complexes 1–4 (complexes 10–13) were subjected to fluorescence measurements so that the number of complex-associated water molecules could be determined. These measurements provided the following data (ms−1): 10: kH2O = 0.566, kD2O = 0.315; 11: kH2O = 0.635, kD2O = 0.331; 12: kH2O = 0.644, kD2O = 0.397; 13: kH2O = 0.671, kD2O = 0.390. These values were inserted into the equation listed in the Experimental Procedures section, resulting in the q values shown in Table 1. Typical of most small molecule DOTA and DTPA-based contrast agents where one carboxylic acid arm has been replaced with an amide, these values reflect one bound water molecule within acceptable experimental error of ±0.5 (41, 42).
To evaluate the mean residence lifetime (τm) of the water molecule coordinated to the metal center of complexes 1–4, temperature dependent 17O-NMR measurements were performed (32, 34). Plotting the paramagnetic contribution of the observed 17O-NMR transverse relaxation rate of water gave the expected bell-shaped curves for each complex that were subjected to the fitting parameters outlined in the Experimental Procedures section (Supporting Figure S1) (32–34). The fitted 17O vs. temperature (T) profiles revealed the τm values displayed in Table 1.
The τm values for 1–4 are similar, and decidedly higher than the previously reported optimal value of 10 ns for Gd(III) MR contrast agents (41). These bound-water lifetimes are substantially slower than those reported for DOTA and DTPA; however, this observation can be explained. Substitution of an acetate arm to an amide linkage (as in the case of 1–4) decreases the steric crowding of the lanthanide and lowers the overall molecular charge of the metalated complexes (1, 43). Both factors affect τm, and such values are similar to those obtained for other amidated DOTA and DTPA structures (44–46). Typically, a structural modification that lengthens τm should be avoided, however, the relaxivity of these small molecule chelates at high fields (> 20 MHz) is dominated by another parameter, the rotational correlation motion (τR) (47). Although the contrast agent is attached to the transport peptide via a flexible linker, τR for 1–4 must be substantially higher than Gd(III)-DOTA and Gd(III)-DTPA. 1–4 have the same q, T1e, and less than optimal τm values compared to Gd(III)-DOTA and Gd(III)-DTPA yet exhibit a 2.0 – 2.5 fold increase in overall relaxivity. Therefore, the increase in is likely due to an increase in τR (through an increase in molecular weight). In summary, this system is highly dependent on τR, allowing relaxivity values to remain acceptable in spite of varying, less-than-optimal τm values (1).
In Vitro Cleavage
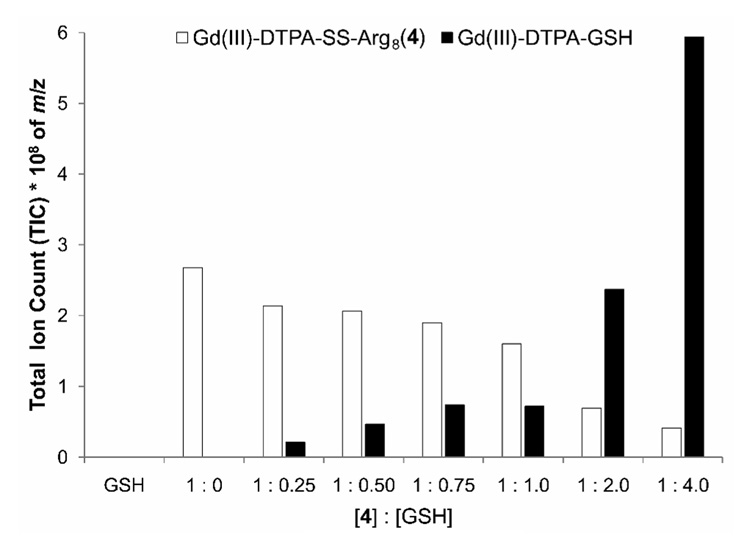
To assess thiol-mediated release of the chelated Gd(III) from the disulfide-linked arginine oligomer in solution, varying equivalents of GSH were added to a solution of 4 and the cleavage product was quantified via HPLC-MS. The samples were prepared by adding increasing amounts of GSH, from 0.25:1 to 4:1 (GSH:4), to a stock solution of 4 (1.0 mM in DPBS). These samples were combined with incrementally delayed starting times so that each could be incubated in a 37 °C waterbath for 4 h prior to LC-MS analysis (this is consistent with the cell culture conditions). Separation was accomplished by a gradient ramp starting with 0% MeCN which reached 60% MeCN at 40 min before returning to 0% MeCN. The proposed cleavage reaction was followed by plotting the total ion count (TIC) of the desired product (Gd(III)-DTPA-GSH, m/z = 912+ amu) and the starting material (4, m/z = 3, 673+ amu) as their concentrations changed in response to GSH addition (Supplemetnal Figure S2). The molecular ions chromatograms (912+ and 673+) were integrated so that the total area under the curve could be calculated (Figure 2).
Figure 2.
Quantification of in vitro cleavage of Gd(III)-DTPA-SS-Arg8 (4) via disulfide exchange with GSH. 4 was incubated with increasing amounts of GSH for 4 hours in 1.0 mM DPBS. The exchange was monitored by plotting the total ion count (TIC) of the desired product (Gd(III)-DTPA-GSH, m/z = 912+ amu, black bars) and the starting material (4, m/z = 3, 673+ amu, white bars). The data represents the calculated total area under the curve of the integrated molecular ion chromatograms (912+ and 673+). 4 decreases with increasing amounts of GSH which corresponds to an increase in Gd(III)-DTPA-GSH representing in vitro disulfide exchange. GSH had no discernable peak to integrate.
Cellular Retention
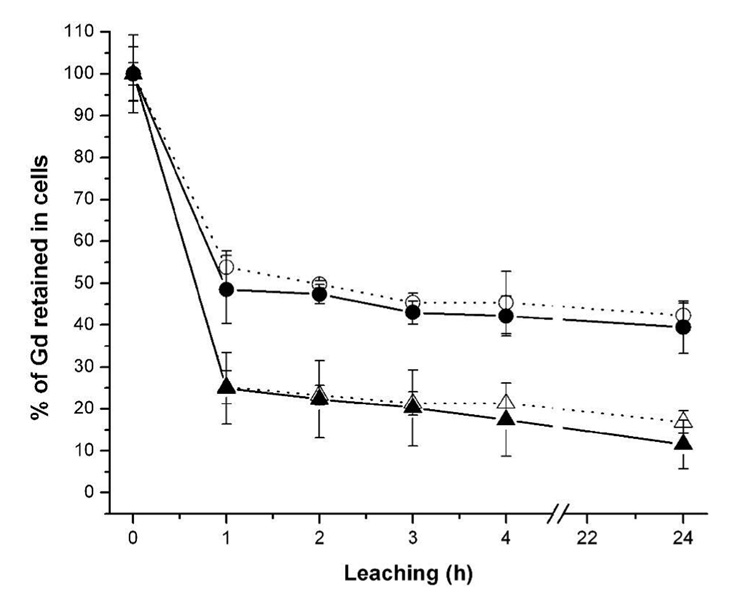
To elucidate cellular uptake and retention of complexes 1–4, 80% confluent cultures of NIH/3T3 cells were incubated with 0.10 mM of 1–4 for 4 h. After a 4 h incubation, the cells were washed three times with DPBS (time = 0, Initial Amt.), placed in fresh media, and allowed to equilibrate for 1 h. Three cell samples and aliquots of the corresponding cell media were removed for analysis. This procedure was repeated 4 times for a total of 5 washes where the last wash was allowed to leach for 24 h.
Each cell culture well consists of two Gd(III)-containing phases: (i.) the cells and (ii.) the external media. ICP-MS provided absolute quantities of accumulated, retained, and leached Gd(III) for the cells at five time points. This data shows that prior to leaching of the agent, non-disulfide complexes 1 and 2 show increased uptake efficacy over 3 and 4 (Supporting Table S1). However, Figure 4 shows that after washes, cells treated with 1 and 2 lose a more significant amount of the agent than those treated with 3 and 4. The absolute quantities (Supporting Table S1) were divided by their respective initial concentrations (those collected at time = 0) in order to elucidate the relative amount of each agent retained within the cells. Incorporation of a disulfide linkage between the transduction moiety and Gd(III) chelate of these cell-permeable contrast agents drastically improves cellular retention (Figure 4; e.g. from 15 → 40% in the case of 1 vs. 3 in the NIH/3T3 cell line).
Figure 4.
The percentage of Gd(III) associated with the NIH/3T3 cells as a function of the initial uptake concentration (data taken directly from Supporting Table S1). Cells were washed with DPBS at each time point displayed. The contrast agents are represented as: 1 (closed triangles), 2 (open triangles), 3 (closed circles), and 4 (open circles). Comparison of the closed data points allows correlation of the two DOTA-based ligands. Comparison of the open data points allows correlation of the two DTPA-based ligands.
SR-XRF Analysis
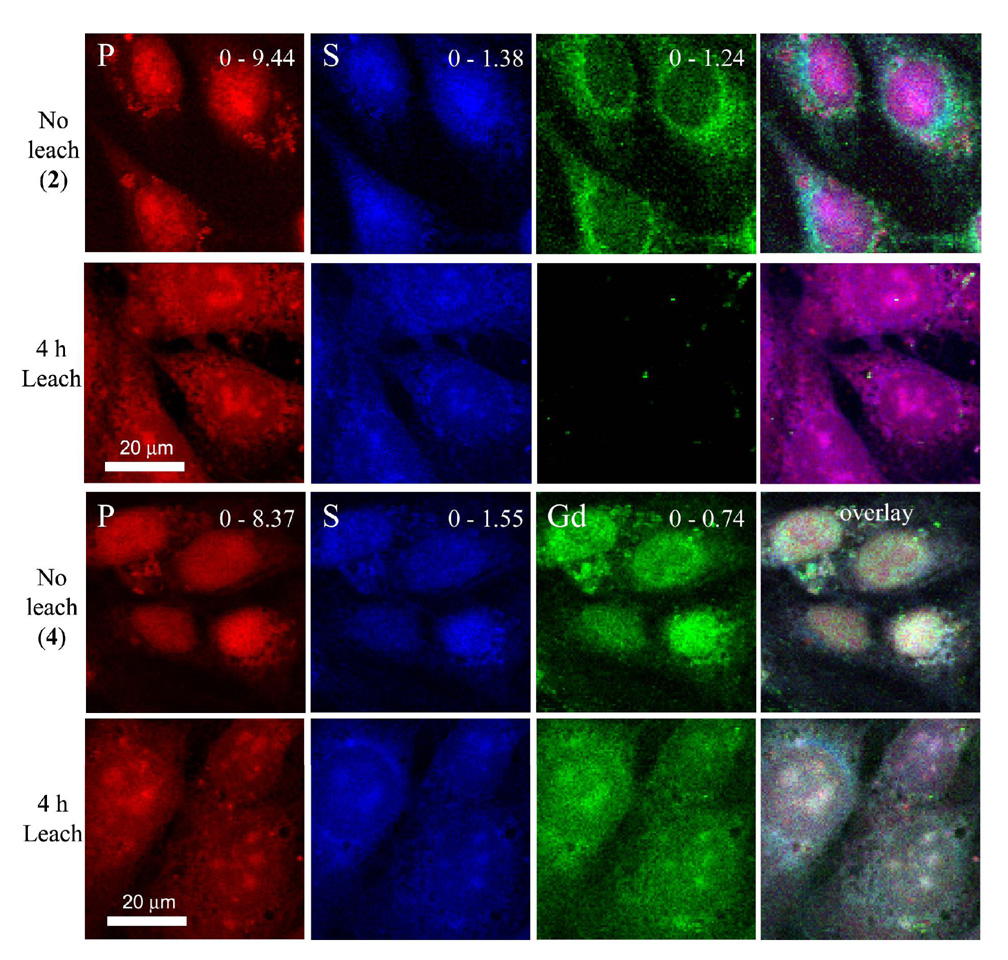
Synchrotron based XRF analysis was performed to determine the cellular association of complexes 1–4. For each sample, three to five randomly chosen areas were raster-scanned to acquire a multi-cell containing image with high resolution (60 × 60 µm with 0.25 µm2 pixel size). These high-resolution elemental maps provide pixel by pixel data sets that globally confirm, map, and quantify the Gd distribution within each sampled area (24). An example of this data is displayed in Figure 5 where the leaching and non-leaching data of 2 and 4 in the NIH/3T3 cell line is compared. Fluorescence maps of inherent biological elements (e.g., P and S) accompany each gadolinium image to prove cellular existence and provide boundaries. The overlay column combines the data from the three elemental maps (P, S, and Gd columns) to provide information on the areas of multi-element overlap. Owing to the proximity of the Gd and Fe fluorescence energy levels, it is important to deconvolute potential peak overlap between Fe and Gd fluorescence. This was accomplished using modified gaussian curves that were fitted at each scan position to the acquired X-ray fluorescence spectra (36). Supporting Figure S3 contains images of complexes 1 and 3. The top two rows of images displayed in Figure 5 show the extent to which leaching has depleted the NIH/3T3 cells of associated gadolinium when treated with the non-disulfide contrast agent (2). Conversely, when the cells are treated with a disulfide linked contrast agent (4), they retain a large amount of gadolinium post-leach (bottom 2 rows, Figure 5).
Figure 5.
XRF images of NIH/3T3 cells treated with contrast agents 2 (top 8 images) and 4 (bottom 8 images) at time = 0 (No leach) and after 4 h (Leach). Phosphorus fluorescence is red, sulfur is blue, and gadolinium is green. Note that each column of images is scaled to its respective maximum value (displayed at the upper right corner and given in µg/cm2).
MR Imaging and T1 Analysis
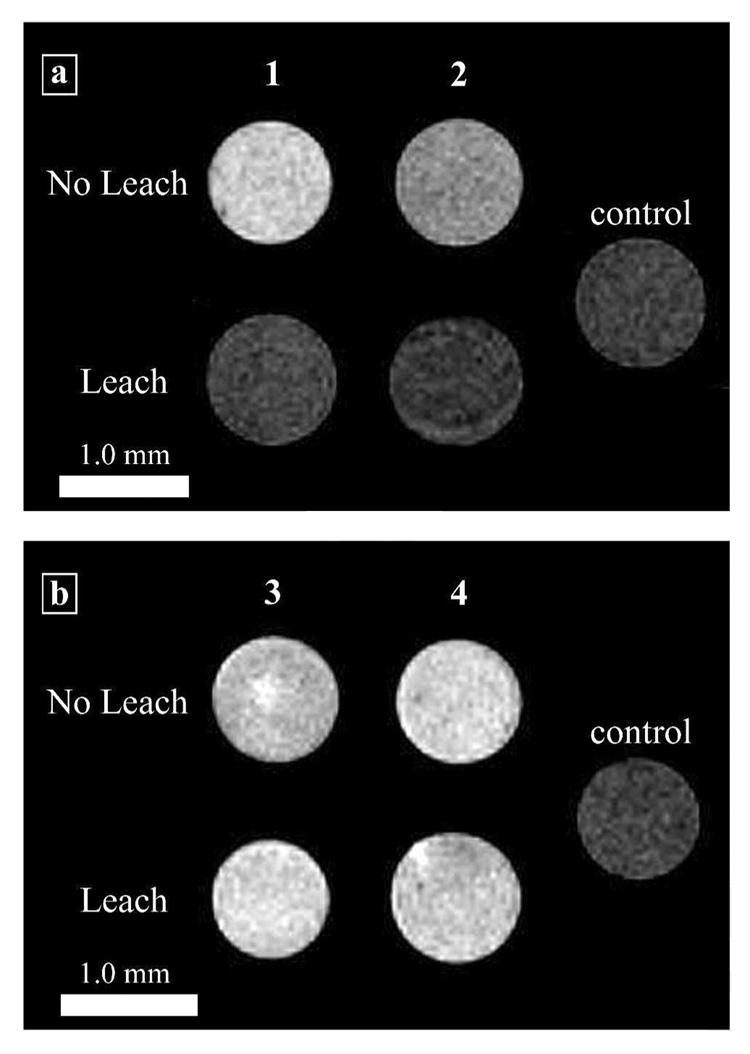
In order to assess the feasibility of the disulfide linkage to increase cell retention and maintain MR image contrast over an extended time period, NIH/3T3 cells were incubated with 1.0 mM of 1–4. After the cells were harvested and placed in glass capillary tubes, MR images were acquired (Figure 6A). Cells incubated with complexes 1 and 2 exhibit a significantly brighter signal than control cells. However, after the washing procedure outlined for the SR-XRF and ICP-MS experiments (4 h, with washes at 1 hour intervals) the cells incubated with 1 and 2 have a significantly lower intensity in the MR image. This loss of image contrast (due to leaching of the contrast agent) is much less in the case of the cells incubated with complexes 3 and 4 (Figure 6B). The signal intensities of the leached cells are similar to their respective non-leached samples, and show little loss of signal intensity (MR image intensity differences were mathematically confirmed via student t-tests of the corresponding T1 values, Supporting Table S2).
Figure 6.
T1-weighted MR images of NIH/3T3 cells incubated with complexes 1–4. The ‘No Leach’ rows are the cell populations that were not allowed to leach. The ‘Leach’ rows are the cell populations that were allowed to leach for 4 h with washes of DPBS at the hours. The control cells were not incubated with contrast agent but were harvested and packed following the same procedure. (a) Cells incubated with Gd(III)-DOTA-Arg8 or Gd(III)-DTPA-Arg8 (1 and 2). (b) Cells incubated with Gd(III)-DOTA-SS-Arg8 or Gd(III)-DTPA-SS-Arg8 (3 and 4). Images were acquired at 14.1 T with a FOV of 10 × 10 mm (matrix size 256 × 256) and a slice thickness of 1.0 mm. The scale bar represents 1.0 mm. T1 values and student t-tests confirming statistically significant differences can be found in Supporting Table S2.
DISCUSSION
Arginine-bound cell-permeable contrast agents 3 and 4 were prepared via thiol-activated pyridyl intermediates to create cell-permeable MR conjugates linked to cell transduction moieties via a disulfide bond. These agents (3 and 4), although structurally similar to 1 and 2, must be coupled to their respective Gd(III) chelating moieties off-resin and under strict pH and equivalence control. Once conjugated to the end-capped Arg8-Cys, compounds 8 and 9 are exposed to Gd(OH)3 to provide cell-permeable MR contrast agents with anti-leaching properties (Scheme 2).
The structural changes made to 1 and 2 in order to incorporate the disulfide bond were tested to determine their affect on proton relaxation. As seen in Table 1, the coordination environment of the Gd(III) ion (expressed by the q, τm) and relaxivity measurements, has not been significantly changed in spite of the structural differences between non-disulfide (1 and 2) and disulfide (3 and 4) agents. Even with varying pH, the observed relaxivities remain relatively constant (Figure 3). This structural stability and documented characterization make contrast agents 1–4 excellent choices for labeling cells cultured in a non-leaching environment (Supporting Table S1).
In order to quantify the efficiency of in vivo disulfide cleavage we used a model disulfide exchange reaction in vitro. Disulfide bridged complexes undergo exchange or reduction with endogenous thiols, especially GSH (18). As a result, the Gd(III) chelate will be cleaved from its cell-penetrating polyarginine oligomer and consequently prolonging the intracellular lifetime of the chelated Gd(III). The lifetime of the disulfide linkage was assessed by incubating agent 4 at 37 °C in the presence of increasing quantities of GSH (Figure 2). A qualitative depiction of these results is displayed in Supporting Figure S2. As the concentration of GSH is increased, the amount of desired product (Gd(III)DTPA-GSH) detected via quantitative HPLC-MS is increased. Conversely, as the concentration of GSH is increased, the amount of starting material (Gd(III)DTPA-SS-Arg8, 4) decreases. This trend continues through ratios of [GSH]:[4] that reach 4:1 and implies that while disulfide cleavage begins at very low GSH concentrations (0.25 equivalents); complete cleavage is not obtainable even at GSH concentrations that are 4 times that of the contrast agent.
When cells are incubated with the same initial concentration of agent, cell uptake of 1 and 2 is higher in NIH/3T3 cells than 3 and 4 (Initial Amount, Supporting Table S1). The initial uptake values of 1 and 2 show that NIH/3T3 cells prefer the negatively charged DTPA over the neutral DOTA chelate with the same transduction moiety. Conversely, with agents 3 and 4, the neutral DOTA chelate is preferred over the charged DTPA. We have previously shown that small changes in structure or charge have significant effects on the uptake efficiency multiple contrast agents (24). In contrast to initial uptake, the percentage of Gd(III) retained in cells post-leaching implies that incorporation of the disulfide bond into 3 and 4 has a positive effect on long term contrast agent retention. Addition of the reactive disulfide linkage (3 and 4) increases cell retention of the Gd(III) chelate 3.5 and 2.5 more than their non-disulfide counterparts (1 and 2), respectively (Figure 4).
To co-validate ICP-MS measurements at the single cell level, cells incubated with 1–4 were evaluated by SR-XRF. SR-XRF microprobes alleviate the difficulties associated with visualizing intracellular lanthanides and allow direct gadolinium imaging without covalent attachment of organic fluorophores that alter cell transport properties. Additionally, SR-XRF offers sub-micron resolution and quantifiable elemental data of sub-cellular regions of interest that are displayed in two-dimensional, intensity weighted images (48). As shown, cells labeled with 2 and 4 (fixed at time = 0) show a significant amount of associated gadolinium (No leach, rows 1 and 3, Figure 5). In contrast, rows 2 and 4 have significant differences in the amount of gadolinium (4 h Leach for 2 and 4, respectively). The 4 h leach images for complex 2 show only trace amounts of gadolinium while the corresponding images of cells treated with 4 display a relatively large amount of gadolinium comparable to non-leached cells. These results are in agreement with the ICP-MS measurements and can be attributed to the incorporation of the disulfide linkage between the Gd(III) chelate and the arginine transduction moiety.
The data from ICP-MS and SR-XRF images reveals that incorporation of the disulfide linkage increases the cell retention of contrast agents 3 and 4. However, since ICP-MS and SR-XRF are highly sensitive towards gadolinium concentration, the utility of 1–4 to increase MR image intensity was examined (before and after the leaching). This assessment was accomplished by incubating 1–4 (1.0 mM) on 2 populations of NIH/3T3 cells, one subjected to the leaching protocol outlined in the Experimental Procedures section and one that was not. Visual comparison of the images in Figure 6A to 6B clearly show that cells incubated with 3 and 4 retain their image contrast enhancement to a greater extent than those incubated with 1 and 2 (e.g. 88% signal retention for complex 3 over the leaching period). These images show that the SR-XRF and ICP-MS data have translated to MRI whereby increased cell retention of the Gd(III)-based contrast agents prolongs image contrast. We performed T1 analysis to validate that the image intensity changes are due to changes in T1. As shown in Supporting Table S2, the shortest T1s correspond to the highest image intensities.
Complexes 1–4 undergo similar cell uptake, the disulfide exchange reactions possible for 3 and 4 must significantly alter their intracellular fate from 1 and 2. If 1 and 2 are non-specifically bound to the internal membrane or sequestered in lysosomes, the image intensity and resultant T1 values will decrease due to reduced Gd(III) access to the bulk water. This explains the high T1 values and reduced MR image intensity in Figure 6A (Leach) despite the Gd(III) measurements displayed in Supporting Table S1. Such encapsulation allows the contrast agent access to only a small number of rapidly exchanging inner sphere water molecules. This phenomenon has been observed within artificial systems such as those involving LIPOCEST contrast agents, and more importantly, within certain sub-cellular compartments (49, 50).
The SR-XRF images display a similar loss of gadolinium signal after the leaching process. We propose that 1 and 2 have been processed differently than 3 and 4 leaving the Gd(III) chelate non-specifically bound to the recycling membrane (either endosomal or lysosomal). Thus, non-specific contrast agent binding was only removed when the cells were subjected to harsh conditions (i.e. the formaldehyde fixation and dehydration necessary for SR-XRF sample preparation). Therefore, the SR-XRF images supplement the MR images while supporting our current hypothesis of membrane internalization and non-specific binding of complexes 1 and 2.
In conclusion, we have synthesized Gd(III)-DOTA-SS-Arg8 (3) and Gd(III)-DTPA-SS-Arg8 (4), cell-permeable contrast agents that show increased cell retention via disulfide bond incorporation which may be a result of intracellular thiol concentrations. The agents’ relaxometric properties, pH stability, in vitro cleavage kinetics, and cell retention have been investigated. As expected, 3 and 4 show increased cell retention (ICP-MS), single cell association (SR-XRF), and viability as long term MRI contrast agents (MR images). These agents could be used to ectopically label cell populations and provide longterm contrast enhancement for cell tracking and lineage analysis using MR imaging.
Supplementary Material
The variable temperature 17O data accompanied by the τm fitting curves, HPLC-MS ion count of disulfide exchange, quantitation of cell uptake and leaching, additional SR-XRF images, and MR image T1 values and student t-test results for the MR images are available free of charge via the World Wide Web at http://pubs.acs.org.
ACKNOWLEDGMENT
We thank Prof. Matthew J. Allen for helpful discussions. This research was supported by the National Institutes of Health under Grant Number 1 R01 EB005866-01, the Nanomaterials for Cancer Diagnostics and Therapeutics under 5 U54 CA1193 41-02, and the Center for Excellence in Transitional Human Stem Cell Research under 1 P50 NS0542 78–01. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract Number W-31-109-ENG-38. MR images were acquired on a 14.1 T-WB imaging spectrometer operated by the CBMRR under NIH/NCRR Grant Number 1 S10 RR13880-01.
Abbreviations
- MRI
magnetic resonance imaging
- GSH
glutathione
- DOTA
1,4,7,10-tetraacetic acid-1,4,7,10-tetraazacyclododecane
- DTPA
Diethylenetriaminepentaacetic acid
- DMEM
Dulbecco’s modified Eagle’s medium
- EMEM
Eagle’s minimum essential medium
- DPBS
Dulbecco’s phosphate buffered saline
- FBS
fetal bovine serum
- CBS
calf bovine serum
- EDTA
ethylenediaminetetraacetic acid
- DOTA-tris-TB
1,4,7-tris(acetic acid-tert-butyl ester)-10-acetic acid-1,4,7,10-tetraazacyclododecane
- MALDI-TOF
matrix assisted laser desorption ionization time of flight
- ESI
electrospray ionization
- RP
reverse-phase
- MeCN
acetonitrile
- ICP-MS
Inductively Coupled Plasma Mass Spectrometry
- Fmoc
fluorenyl-methoxy-carbonyl
- DMF
N,N- dimethylformamide
- Pbf
pentamethyldihydrobenzofuran-5-sulfonyl
- HATU
N,N,N',N'-tetramethyl-O-(7-azabenzotriazol-1-yl)uraniumhexafluorophosphate
- DIPEA
N,N-diisopropylethylamine
- Trt
trityl
- TFA
trifluoroacetic acid
- EDT
1,2-ethanedithiol
- TIS
triisopropylsilane
- MTBE
methyl tert-butyl ether
- AETP
S-(2-Aminoethylthio)-2-thiopyridine hydrochloride
- DMSO
dimethylsulfoxide
- SR-XRF
synchrotron radiation X-ray fluorescence
References
- 1.Merbach AE, Toth E. The Chemistry of Contrast Agents in Medical Magnetic Resonance Imaging. 2001. and Editors. [Google Scholar]
- 2.Aime S, Cabella C, Colombatto S, Geninatti Crich S, Gianolio E, Maggioni F. Insights into the use of paramagnetic Gd(III) complexes in MR-molecular imaging investigations. J. Magn. Reson. Imaging. 2002;16:394–406. doi: 10.1002/jmri.10180. [DOI] [PubMed] [Google Scholar]
- 3.Hu X, Norris DG. Advances in high-field magnetic resonance imaging. Annu. Rev. Biomed. Eng. 2004;6:157–184. doi: 10.1146/annurev.bioeng.6.040803.140017. [DOI] [PubMed] [Google Scholar]
- 4.Collins Christopher M, Wang Z, Mao W, Fang J, Liu W, Smith Michael B. Array-optimized composite pulse for excellent whole-brain homogeneity in high-field MRI. Magn. Reson. Med. 2007;57:470–474. doi: 10.1002/mrm.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter PM, Caruthers SD, Wickline SA, Lanza GM. Molecular imaging by MRI. Curr. Cardiol. Rep. 2006;8:65–69. doi: 10.1007/s11886-006-0013-2. [DOI] [PubMed] [Google Scholar]
- 6.Endres PJ, Paunesku T, Vogt S, Meade TJ, Woloschak GE. DNA-TiO2 nanoconjugates labeled with magnetic resonance contrast agents. J. Am. Chem. Soc. 2007;129:15760–15761. doi: 10.1021/ja0772389. [DOI] [PubMed] [Google Scholar]
- 7.Towner RA, Smith N, Doblas S, Tesiram Y, Garteiser P, Saunders D, Cranford R, Silasi-Mansat R, Herlea O, Ivanciu L, Wu D, Lupu F. In vivo detection of c-Met expression in a rat C6 glioma model. J. Cell. Mol. Med. 2008;12:174–186. doi: 10.1111/j.1582-4934.2008.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caravan P, Cloutier NJ, Greenfield MT, McDermid SA, Dunham SU, Bulte JWM, Amedio JC, Jr, Looby RJ, Supkowski RM, Horrocks WD, Jr, McMurry TJ, Lauffer RB. The interaction of MS-325 with human serum albumin and its effect on proton relaxation rates. J. Am. Chem. Soc. 2002;124:3152–3162. doi: 10.1021/ja017168k. [DOI] [PubMed] [Google Scholar]
- 9.Major JL, Parigi G, Luchinat C, Meade TJ. The synthesis and in vitro testing of a zinc-activated MRI contrast agent. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13881–13886. doi: 10.1073/pnas.0706247104. S13881/1-S13881/9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louie AY, Huber MM, Ahrens ET, Rothbacher U, Moats R, Jacobs RE, Fraser SE, Meade TJ. In vivo visualization of gene expression using magnetic resonance imaging. Nat. Biotechnol. 2000;18:321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 11.Kalman FK, Woods M, Caravan P, Jurek P, Spiller M, Tircso G, Kiraly R, Bruecher E, Sherry AD. Potentiometric and relaxometric properties of a gadolinium-based MRI contrast agent for sensing tissue pH. Inorg. Chem. 2007;46:5260–5270. doi: 10.1021/ic0702926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saborowski O, Simon GH, Raatschen H-J, Wendland MF, Fu Y, Henning T, Baehner R, Corot C, Chen M-H, Daldrup-Link HE. MR imaging of antigen-induced arthritis with a new, folate receptor-targeted contrast agent. Contrast Med. Mol. Imag. 2007;2:72–81. doi: 10.1002/cmmi.128. [DOI] [PubMed] [Google Scholar]
- 13.Allen MJ, Meade TJ. Magnetic Resonance Contrast Agents for Medical and Molecular Imaging. In: Sigel A, Sigel H, editors. Met. Ions Biol. Syst. New York: Marcel Dekker Inc; 2004. pp. 1–28. [PubMed] [Google Scholar]
- 14.Allen MJ, Meade TJ. Synthesis and visualization of a membrane-permeable MRI contrast agent. JBIC, J. Bio. Inorg. Chem. 2003;8:746–750. doi: 10.1007/s00775-003-0475-2. [DOI] [PubMed] [Google Scholar]
- 15.Frullano L, Meade Thomas J. Multimodal MRI contrast agents. J. Biol. Inorg. Chem. 2007;12:939–949. doi: 10.1007/s00775-007-0265-3. [DOI] [PubMed] [Google Scholar]
- 16.Datta A, Hooker JM, Botta M, Francis MB, Aime S, Raymond KN. High Relaxivity Gadolinium Hydroxypyridonate-Viral Capsid Conjugates: Nanosized MRI Contrast Agents. J. Am. Chem. Soc. 2008;130:2546–2552. doi: 10.1021/ja0765363. [DOI] [PubMed] [Google Scholar]
- 17.Aime S, Castelli DD, Lawson D, Terreno E. Gd-Loaded Liposomes as T1,Susceptibility, and CEST Agents, All in One. J. Am. Chem. Soc. 2007;129:2430–2431. doi: 10.1021/ja0677867. [DOI] [PubMed] [Google Scholar]
- 18.Meister A, Anderson ME. Glutathione. Annu. Rev. Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 19.Franco R, Schoneveld OJ, Pappa A, Panayiotidis MI. The central role of glutathione in the pathophysiology of human diseases. Arch. Physiol. Biochem. 2007;113:234–258. doi: 10.1080/13813450701661198. [DOI] [PubMed] [Google Scholar]
- 20.Pastore A, Federici G, Bertini E, Piemonte F. Analysis of glutathione: Implication in redox and detoxification. Clin. Chim. Acta. 2003;333:19–39. doi: 10.1016/s0009-8981(03)00200-6. [DOI] [PubMed] [Google Scholar]
- 21.Hong R, Han G, Fernandez JM, Kim B-j, Forbes NS, Rotello VM. Glutathione-mediated delivery and release using monolayer protected nanoparticle carriers. J. Am. Chem. Soc. 2006;128:1078–1079. doi: 10.1021/ja056726i. [DOI] [PubMed] [Google Scholar]
- 22.Han G, Chari Nandini S, Verma A, Hong R, Martin Craig T, Rotello Vincent M. Controlled recovery of the transcription of nanoparticle-bound DNA by intracellular concentrations of glutathione. Bioconjug. Chem. 2005;16:1356–1359. doi: 10.1021/bc050173j. [DOI] [PubMed] [Google Scholar]
- 23.Carrera C, Digilio G, Baroni S, Burgio D, Consol S, Fedeli F, Longo D, Mortillaro A, Aime S. Synthesis and characterization of a Gd(III) based contrast agent responsive to thiol containing compounds. Dalton T. 2007:4980–4987. doi: 10.1039/b705088g. [DOI] [PubMed] [Google Scholar]
- 24.Endres PJ, MacRenaris KW, Vogt S, Allen MJ, Meade TJ. Quantitative imaging of cell-permeable magnetic resonance contrast agents using X-ray fluorescence. Mol. Img. 2006;5:485–497. [PubMed] [Google Scholar]
- 25.Allen MJ, MacRenaris KW, Venkatasubramanian PN, Meade TJ. Cellular delivery of MRI contrast agents. Chem. Biol. 2004;11:301–307. doi: 10.1016/j.chembiol.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Futaki S. Membrane-permeable arginine-rich peptides and the translocation mechanisms. Adv. Drug Deliver. Rev. 2005;57:547–558. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. Arginine-rich peptides: An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 28.Allen MJ. Department of Chemistry. Pasadena, CA: California Institute of Technology; 2004. p. 181. [Google Scholar]
- 29.Endres PJ. Department of Chemistry. Evanston, IL: Northwestern University; 2008. p. 204. [Google Scholar]
- 30.Beeby A, Clarkson IM, Dickins RS, Faulkner S, Parker D, Royle L, de Sousa AS, Williams JAG, Woods M. Non-radiative deactivation of the excited states of europium, terbium and ytterbium complexes by proximate energy-matched OH, NH and CH oscillators: An improved luminescence method for establishing solution hydration states. J. Chem. Soc., Perkin Trans. 1999;2:493–504. [Google Scholar]
- 31.Quici S, Cavazzini M, Marzanni G, Accorsi G, Armaroli N, Ventura B, Barigelletti F. Visible and near-infrared intense luminescence from water-soluble lanthanide [Tb(III),Eu(III), Sm(III), Dy(III), Pr(III), Ho(III), Yb(III), Nd(III), Er(III)] complexes. Inorg. Chem. 2005;44:529–537. doi: 10.1021/ic0486466. [DOI] [PubMed] [Google Scholar]
- 32.Terreno E, Botta M, Boniforte P, Bracco C, Milone L, Mondino B, Uggeri F, Aime S. A multinuclear NMR relaxometry study of ternary adducts formed between heptadentate Gd(III) chelates and L-lactate. Chem-Eur. J. 2005;11:5531–5537. doi: 10.1002/chem.200500129. [DOI] [PubMed] [Google Scholar]
- 33.Aime S, Botta M, Crich SG, Giovenzana G, Pagliarin R, Sisti M, Terreno E. NMR relaxometric studies of Gd(III) complexes with heptadentate macrocyclic ligands. Magn. Reson. Chem. 1998;36:S200–S208. [Google Scholar]
- 34.Swift TJ, Connick RE. NMR (nuclear magnetic resonance)-relaxation mechanisms of O17 in aqueous solutions of paramagnetic cations and the lifetime of water molecules in the first coordination sphere. J. Chem. Phys. 1962;37:307–220. [Google Scholar]
- 35.Caravan P, Parigi G, Chasse JM, Cloutier NJ, Ellison JJ, Lauffer RB, Luchinat C, McDermid SA, Spiller M, McMurry TJ. Albumin binding, relaxivity, and water exchange kinetics of the diastereoisomers of MS-325, a Gadolinium(III)-based magnetic resonance angiography contrast agent. Inorg. Chem. 2007;46:6632–6639. doi: 10.1021/ic700686k. [DOI] [PubMed] [Google Scholar]
- 36.Vogt S. MAPS: A set of software tools for analysis and visualization of 3D X-ray fluorescence data sets. J. Phys. IV. 2003;104:635–638. [Google Scholar]
- 37.Ebright YW, Chen Y, Kim Y, Ebright RH. S-[2-(4-azidosalicylamido)ethylthio]-2-thiopyridine: Radioiodinatable, cleavable, photoactivatable crosslinking agent. Bioconjug. Chem. 1996;7:380–384. doi: 10.1021/bc9600168. [DOI] [PubMed] [Google Scholar]
- 38.Glogard C, Stensrud G, Aime S. Novel radical-responsive MRI contrast agent based on paramagnetic liposomes. Magn. Reson. Chem. 2003;41:585–588. [Google Scholar]
- 39.Botta M, Aime S, Barge A, Bobba G, Dickins RS, Parker D, Terreno E. Ternary complexes between cationic Gd(III) chelates and anionic metabolites in aqueous solution: An NMR relaxometric study. Chem-Eur. J. 2003;9:2102–2109. doi: 10.1002/chem.200204475. [DOI] [PubMed] [Google Scholar]
- 40.Urbanczyk-Pearson LM, Femia FJ, Smith J, Parigi G, Duimstra JA, Eckermann AL, Luchinat C, Meade TJ. Mechanistic investigation of b-galactosidase-activated MR contrast agents. Inorg. Chem. 2008;47:56–68. doi: 10.1021/ic700888w. [DOI] [PubMed] [Google Scholar]
- 41.Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Gadolinium(III) chelates as MRI contrast agents: Structure, dynamics, and applications. Chem. Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 42.Chan KW-Y, Wong W-T. Small molecular gadolinium(III) complexes as MRI contrast agents for diagnostic imaging. Coord. Chem. Rev. 2007;251:2428–2451. [Google Scholar]
- 43.Jaszberenyi Z, Moriggi L, Schmidt P, Weidensteiner C, Kneuer R, Merbach AE, Helm L, Toth E. Physicochemical and MRI characterization of Gd3+-loaded polyamidoamine and hyperbranched dendrimers. JBIC, J. Bio. Inorg. Chem. 2007;12:406–420. doi: 10.1007/s00775-006-0197-3. [DOI] [PubMed] [Google Scholar]
- 44.Kubicek V, Rudovsky J, Kotek J, Hermann P, Vander Elst L, Muller RN, Kolar ZI, Wolterbeek HT, Peters JA, Lukes I. A bisphosphonate monoamide analogue of DOTA: A potential agent for bone targeting. J. Am. Chem. Soc. 2005;127:16477–16485. doi: 10.1021/ja054905u. [DOI] [PubMed] [Google Scholar]
- 45.Laurent S, Parac-Vogt TN, Kimpe K, Thirifays C, Binnemansq K, Muller RN, Vander Elst L. Bis(phenylethylamide) derivatives of Gd-DTPA as potential receptor-specific MRI contrast agents. Eur. J. Inorg. Chem. 2007:2061–2067. [Google Scholar]
- 46.Zhang S, Jiang X, Sherry AD. Modulation of the lifetime of water bound to lanthanide metal ions in complexes with ligands derived from 1,4,7,10-tetraazacyclododecane tetraacetate (DOTA) Helv. Chim. Acta. 2005;88:923–935. [Google Scholar]
- 47.Lauffer RB. Paramagnetic metal complexes as water proton relaxation agents for NMR imaging: Theory and design. Chem. Rev. 1987;87:901–927. [Google Scholar]
- 48.Twining BS, Baines SB, Fisher NS, Maser J, Vogt S, Jacobsen C, Tovar-Sanchez A, Sanudo-Wilhelmy SA. Quantifying trace elements in individual aquatic protist cells with a synchrotron X-ray fluorescence microprobe. Anal. Chem. 2003;75:3806–3816. doi: 10.1021/ac034227z. [DOI] [PubMed] [Google Scholar]
- 49.Aime S, Castelli DD, Terreno E. Highly sensitive MRI chemical exchange saturation transfer agents using liposomes. Angew. Chem. Int. Edit. 2005;44:5513–5515. doi: 10.1002/anie.200501473. [DOI] [PubMed] [Google Scholar]
- 50.Bulte JWM, Douglas T, Witwer B, Zhang S-C, Strable E, Lewis BK, Zywicke H, Miller B, van Gelderen P, Moskowitz BM, Duncan LD, Frank JA. Magnetodendrimers allow endosomal magnetic labeling and in vivo tracking of stem cells. Nat. Biotechnol. 2001;19:1141–1147. doi: 10.1038/nbt1201-1141. [DOI] [PubMed] [Google Scholar]
- 51.Bryden CC, Reilley CN. Europium luminescence lifetimes and spectra for evaluation of 11 europium complexes as aqueous shift reagents for nuclear magnetic resonance spectrometry. Anal. Chem. 1982;54:610–615. [Google Scholar]
- 52.Powell DH, Ni Dhubhghaill OM, Pubanz D, Helm L, Lebedev YS, Schlaepfer W, Merbach AE. High-pressure NMR kinetics. Part 74. Structural and dynamic parameters obtained from 17O NMR, EPR, and NMRD studies of monomeric and dimeric Gd3+ complexes of interest in magnetic resonance imaging: An integrated and theoretically self-consistent approach. J. Am. Chem. Soc. 1996;118:9333–9346. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The variable temperature 17O data accompanied by the τm fitting curves, HPLC-MS ion count of disulfide exchange, quantitation of cell uptake and leaching, additional SR-XRF images, and MR image T1 values and student t-test results for the MR images are available free of charge via the World Wide Web at http://pubs.acs.org.